Duplex Allele-Specific PCR Assay Simultaneously Detecting Genotype of Osteoarthritis Susceptibility Loci Within GDF5 and COL11A1 Genes
Article Information
Svetlana B. Panina1*, Igor V. Kornienko1,2, Igor V. Krolevets3, Mikhail A. Zabrodin3, Natalia P. Milyutina1
1Academy of Biology and Biotechnology, Southern Federal University, 194/1 Stachki St., Rostov-on-Don, Russia
2Southern Scientific Center of the Russian Academy of Sciences, 41 Chekhova St., Rostov-on-Don, Russia
3Department of Traumatology and Orthopaedics, Rehabilitation and Sport Medicine, Rostov State Medical University, 29 Nakhichevanskii St., Rostov-on-Don, Russia
*Corresponding Author: Svetlana B. Panina, Academy of Biology and Biotechnology, Southern Federal University,194/1 Stachki St., Rostov-on-Don, Russia.
Received: 18 July 2019; Accepted: 29 July 2019; Published: 01 August 2019
Citation: Svetlana B. Panina, Igor V. Kornienko, Igor V. Krolevets, Mikhail A. Zabrodin, Natalia P. Milyutina. Duplex Allele-Specific PCR Assay Simultaneously Detecting Genotype of Osteoarthritis Susceptibility Loci Within GDF5 and COL11A1 Genes. Archives of Clinical and Biomedical Research 3 (2019): 281-286.
View / Download Pdf Share at FacebookAbstract
Introduction: Genome-wide association studies and meta-analyses have identified rs143383 GDF5 (growth and differentiation factor 5) and rs2615977 COL11A1 (collagen type XI alpha 1) as polymorphic loci among the most robust associations with osteoarthritis (OA). However, currently, there is a lack of available rapid, technically feasible, and costeffective methods to detect OA susceptibility genotype, not linked with the specific descent of the patients.
Methods: Genomic DNA was extracted from the blood of OA patients and healthy individual using standard methods. Primers specific at the 3'-end for the polymorphic sites were designed. Direct sequencing was used to determine the genotype of the study subjects.
Results and conclusion: Here we describe one-step allele-specific PCR assay simultaneously detecting genotype of rs143383 GDF5 and rs2615977 COL11A1. Duplex PCR enabled to obtain specific bands corresponding to the amplified product of interest. The genotypes called based on PCR results were consistent with the results of direct sequencing.
Keywords
Single nucleotide polymorphism; Osteoarthritis; Genetic susceptibility; Multiplex polymerase chain reaction;
Growth and differentiation factor 5
Article Details
Introduction
Osteoarthritis (OA) has a clear and detrimental impact on wellbeing, with up to one-fifth of affected individuals giving up work or retiring early [1]. Treatment options for severe steoarthritis (OA) beyond pain relief or total knee replacement are very limited. Because of this, attention has recently shifted to identifying which factors increase the risk of OA [2]. Among these factors, certain genetic susceptibility loci have been identified by association studies and meta-analyses [3]. More specifically, T-allele of polymorphic locus rs143383 GDF5 (growth and differentiation factor 5) is one of the most robust OA predisposition loci across multiple populations [4]. Another strong association uncovered by arcOGEN genome-wide association study (GWAS) is rs2615977 (A-allele) within intron 31 of COL11A1 gene (p = 1.2 × 10-5) [5]. The Arthrotest, a prognostic tool to predict progression towards severe knee osteoarthritis, has been developed based on genetic information from Spanish knee OA patients and is used at clinic [6]. However, currently, there is lack of available rapid, technically feasible, and cost-effective methods to detect OA susceptibility genotype independently upon genetic ancestry. We were keen therefore to develop duplex PCR assay simultaneously detecting genotype of rs143383 GDF5 and rs2615977 COL11A1.
Materials and Methods
DNA samples
We collected samples of peripheral blood from patients diagnosed with severe knee OA (n=3) and healthy donor (n=1). The basic characteristics of the subjects are shown in Table 1. The study was approved by the local research ethics board (Rostov State Medical University Ethics Committee, Rostov-on-Don, Russia). Genomic DNA was isolated from peripheral blood leukocytes using a commercial kit (Litech, Moscow, Russia) according to manufacturer’s instructions.
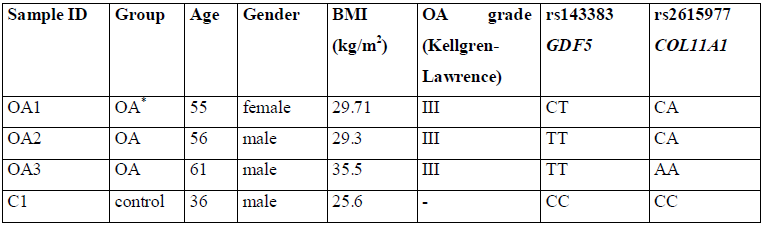
*OA - patients diagnosed with osteoarthritis; control - healthy individual.
Table 1: Basic characteristics of the subjects
Allele-specific PCR
The primers, designed to be specific for either 3'-end rs143383 GDF5 or rs2615977 COL11A1, are shown in Table 2 (synthesized by Eurogene, Moscow, Russia). In order to develop duplex PCR, the amplicon sizes were designed to be 307 bp for GDF5 and 99 bp for COL11A1.
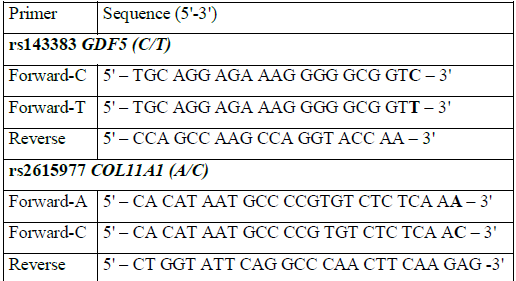
Table 2: Primers used in the study
First, we identified the most robust conditions to detect genotype of either rs143383 GDF5 or rs2615977 COL11A1. PCR was carried out using GeneAmp® PCR System 9700 thermal cycler (ThermoFisher, Waltham, MA, USA). The reaction mixture (total of 25 µl) contained 1x PCR buffer (2.5 µl), 1.5 mM Mg2+ (GDF5) / 2 mM Mg2+ (COL11A1), 200 µM (COL11A1) / 300 µM (GDF5) each of dNTPs, 1.5 U thermostable Taq polymerase (all reagents by Syntol, Moscow, Russia), 1 µM each forward and reverse primers, and 2 µl of DNA solution. PCR conditions were as follows: initial denaturation for 3 min at 95?C, then 30 cycles of 95?C for 10 s, 64 ?C (GDF5) / 66 ?C (COL11A1) for 10 s, and 72 ?C for 30 s. Amplicons were analyzed in 2% agarose gels using GelDoc System (Bio-Rad, Hercules, CA, USA).
Modified PCR conditions (duplex allele-specific reaction)
Next, we defined the most robust conditions for simultaneous genotyping of GDF5 and COL11A1. The reaction mixture (total of 25 µl) contained 1x PCR buffer (2.5 µl), 2 mm Mg2+, 200 µM each of dNTPs, 2 U thermostable Taq polymerase (Syntol), 1 µM each forward and reverse primers, and 2 µl DNA solution. PCR conditions were as follows: after the initial denaturation step for 3 min at 95 °C, we consecutively performed 6 cycles (10 s at 96 0C, 10 s at 70 0C, 30 s at 72 0C), 6 cycles (10 s at 95 0C, 10 s at 69 0C, 30 s at 72 0C), 6 cycles (10 s at 94 0C, 10 s at 68 0C, 30 s at 72 0C), followed by 17 cycles of 10 s at 94 0C, 10 s at 67 0C, and 30 s at 72 0C (35 cycles in total).
Direct sequencing
GDF5 and COL11A1 amplicons were purified using GeneJet PCR Purification Kit (ThermoFisher) and sequenced with the BigDyeTM Terminator V 1.1 Cycle Sequencing Kit (ThermoFisher) on an automatic ABI Prism 3130xl Genetic Analyzer (ThermoFisher) using Sequencing Analysis software version 5.4.
Results and Discussion
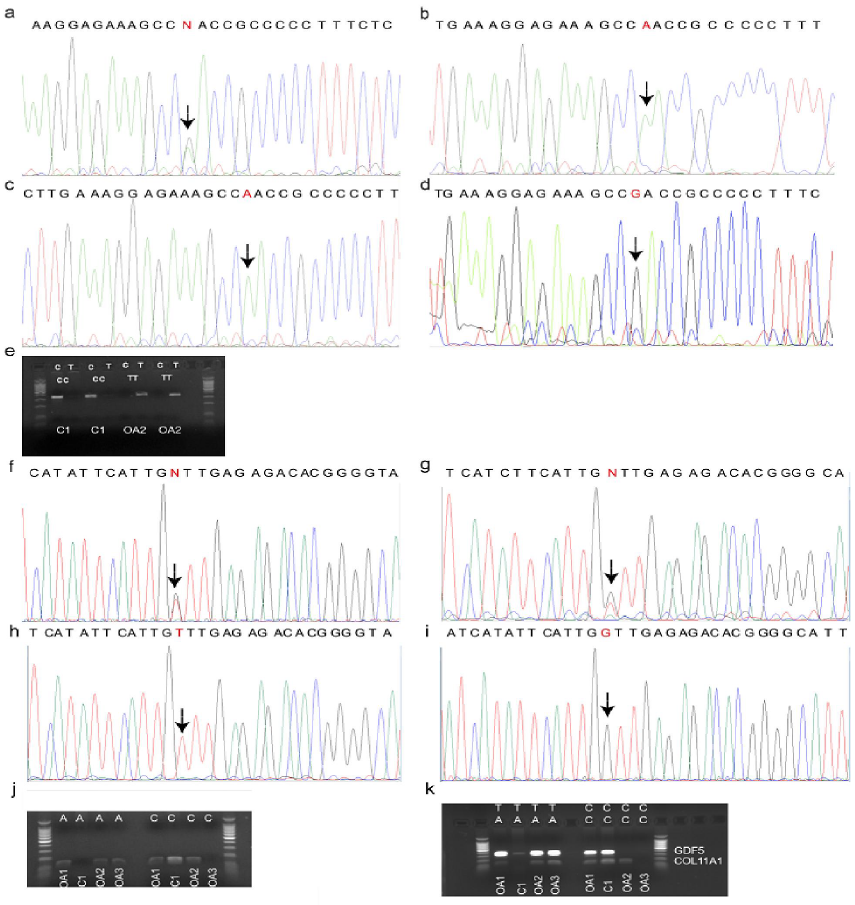
DNA sequences of target regions of interest for the samples OA1, OA2, OA3 (patients with OA), and C1 (healthy individual) are presented in Figure 1a-d (rs143383 GDF5) and Figure 1f-i (rs2615977 COL11A1), respectively. First, genotypes called based on uniplex PCR results for these two genes (Figure 1e, j) were compared with DNA sequencing
results, which confirmed the specificity of PCR (Table 1). After that, we proceeded with duplex PCR (Figure 1k), which could enable to obtain specific bands corresponding to both GDF5 (307 bp) and COL11A1 genes (99 bp). In case of GDF5, the weak band in the second lane can be referred to as non-specific amplification when compared to other much thicker bands corresponding to GDF5 alleles. In overall, this duplex PCR performed well to call the genotypes for analyzed samples.
Figure 1: Detection of OA susceptibility genotype. (a-e) C>T GDF5 (rs143383) genotypes: results of direct sequencing for OA patients (a) OA1, (b) OA2, (c) OA3 and healthy individual (d) C1. The positions of SNP are highlighted with arrows. (e) Uniplex PCR amplicons (rs143383) for the samples OA2 and C1 (in duplicates). (f-j) C>A COL11A1 (rs2615977) genotypes: results of direct sequencing for OA patients (f) OA1, (g) OA2, (h) OA3 and healthy individual (i) C1. (j) Uniplex PCR amplicons (rs2615977) for the samples OA1-3 and C1. (k) Duplex allele-specific PCR for all four samples OA1-3, C1. C and T – alternate alleles for rs143383 GDF5; C and A – alternate alleles for rs2615977 COL11A1.
Currently, multiple methods can be applied for genotyping known single nucleotide polymorphisms (SNPs): hydridization of allele-specific probes, primer extension, multiplex allele-specific and real-time PCR etc, each of them having its pros and cons [7]. Allele-specific PCR is a relatively simple technique where DNA polymerase amplifies target DNA only if allele-specific primers are perfectly complementary to it [7]. Despite allele-specific PCR being laborious and low throughput, it is a rapid, widely-used, and technically feasible genotyping method that is suitable for moderate-throughput laboratories [8]. Previously, multiplex allele-specific PCR assays have been used for detection of polymorphic loci associated with response to long-term anti-inflammatory treatment in patients with rheumatoid arthritis [9], with schizophrenia and Parkinson’s disease [10], Alzheimer’s disease [11]. Here, we developed and described duplex PCR assay detecting risk alleles of rs143383 GDF5 and rs2615977 COL11A1 genes, associated with OA. According to the recent reviews and meta-analyses, rs143383 GDF5 is significantly associated with radiographic defined knee OA [12], and probably, hip and hand OA [13], whereas rs2615977 COL11A1 has been mostly discussed as association with hip OA [5]. This duplex assay might be useful for developing more complex allele-specific genotyping methods.
Conflict of interest
The authors have no conflicts of interest to declare.
Acknowledgments
The study was supported by the Russian Fund for Assistance to Small Innovative Enterprises [N 6233GU/2015].
References
- Reynard LN, Loughlin J. The genetics and functional analysis of primary osteoarthritis susceptibility. Expert Rev Mol Med 15 (2013): e2.
- Gardiner BS, Woodhouse FG, Besier TF, Grodzinsky AJ, Lloyd DG, Zhang L, et al. Predicting Knee Osteoarthritis. Ann Biomed Eng 44 (2016): 222-233.
- Warner SC, Valdes AM. The Genetics of Osteoarthritis: A Review. Journal of Functional Morphology and Kinesiology 1 (2016): 140-153.
- Chapman K, Takahashi A, Meulenbelt I, Watson C, Rodriguez-Lopez J, Egli R, et al. A meta-analysis of European and Asian cohorts reveals a global role of a functional SNP in the 5' UTR of GDF5 with osteoarthritis susceptibility. Hum Mol Genet 17 (2008): 1497-1504.
- Panoutsopoulou K, Southam L, Elliott KS, Wrayner N, Zhai G, Beazley C, et al. Insights into the genetic architecture of osteoarthritis from stage 1 of the arcOGEN study. Ann Rheum Dis 70 (2011): 864-867.
- Blanco FJ, Möller I, Romera M, Rozadilla A, Sánchez-Lázaro JA, Rodríguez A, et al. Improved prediction of knee osteoarthritis progression by genetic polymorphisms: the Arthrotest Study. Rheumatology (Oxford) 54 (2015): 1236-1243.
- Kwok PY, Chen X. Detection of single nucleotide polymorphisms. Curr Issues Mol Biol 5 (2003): 43-60.
- Sistonen J, Fuselli S, Levo A, Sajantila A. CYP2D6 genotyping by a multiplex primer extension reaction. Clin Chem 51 (2005): 1291-1295.
- Schotte H, Schlüter B, Drynda S, Willeke P, Tidow N, Assmann G, et al. Interleukin 10 promoter microsatellite polymorphisms are associated with response to long term treatment with etanercept in patients with rheumatoid arthritis. Ann Rheum Dis 64 (2005): 575-581.
- Zahari Z, Salleh MR, Zahri Johari MK, Musa N, Ismail R. A Nested Allele-Specific Multiplex Polymerase Chain Reaction Method for the Detection of DRD2 Polymorphisms. Malays J Med Sci 18 (2011): 44-57.
- Darawi MN, Ai-Vyrn C, Ramasamy K, Hua PP, Pin TM, Kamaruzzaman SB, et al. Allele-specific polymerase chain reaction for the detection of Alzheimer's disease-related single nucleotide polymorphisms. BMC Med Genet 14 (2013): 27.
- Aghili K, Sobhan MR, Mehdinezhad-Yazdi M, Jafari M, Miresmaeili SM, Rastegar S, et al. Association of GDF-5 rs143383 polymorphism with radiographic defined knee osteoarthritis: A systematic review and meta-analysis. J Orthop 15 (2018): 945-951.
- Evangelou E, Chapman K, Meulenbelt I, Karassa FB, Loughlin J, Carr A, et al. Large-scale analysis of association between GDF5 and FRZB variants and osteoarthritis of the hip, knee, and hand. Arthritis Rheum 60 (2009): 1710-1721.


 Impact Factor: * 3.1
Impact Factor: * 3.1 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 