The Quantum microRNA Immunity in Human Virus-Associated Diseases: Virtual Reality of HBV, HCV and HIV-1 Infection, and Hepatocellular Carcinogenesis with AI Machine Learning
Article Information
Yoichi Robertus Fujii*
Kawada-Cho, 106-6, Atsuta-Ku, Nagoya, 456-0065, Japan
*Corresponding author: Yoichi Robertus Fujii, Kawada-Cho, 106-6, Atsuta-Ku, Nagoya, 456-0065, Japan
Received: 21 April 2020; Accepted: 28 April 2020; Published: 06 May 2020
Citation: Yoichi Robertus Fujii. The Quantum microRNA Immunity in Human Virus-Associated Diseases: Virtual Reality of HBV, HCV and HIV-1 Infection, and Hepatocellular Carcinogenesis with AI Machine Learning. Archives of Clinical and Biomedical Research 4 (2020): 089-129.
View / Download Pdf Share at FacebookAbstract
Objectives: Since virus-related hepatocellular carcinoma (HCC) is quite complexed, the etiology of virus-associated HCC is remained unclear. We have previously shown that the microRNA (miRNA) entangling sorter (METS) analysis with quantum miRNA/miRNA language is available for the etiology investigation in silico from miRNA biomarker panels of human cancers to predict carcinogenesis. To further investigate the etiology of human virus-associated diseases on the stage minus one (zero), host-virus miRNA interactions were investigated by computer simulation on METS analysis with artificial intelligence (AI) machine learning (MIRAI).
Materials and Methods: The information of the miRNA biomarker panels in hepatitis B virus (HBV), hepatitis C virus (HCV) and human immunodeficiency virus type 1 (HIV-1) infection, virus-related fibrosis (cirrhosis), and virus-associated HCCs was extracted from database. The miRNA hub in the panels was selected by both protein/protein interaction and carcinogenic protein function. The statistical analysis upon tumorigenesis was calculated by Prediction One.
Results: The etiology of infection, fibrosis or HCC was simulated by METS analysis with host miRNAs and viral miRNAs. Quite different strategy was shown as the host defense against HBV, HCV and HIV-1 infection under controlled by host miRNAs and viral mRNAs. HBV and HIV-1 was defensed by cell death through shutdown and cell cycle arrest, respectively. HCV was prevented by inhibition of virus production. Carcinogenesis by HBV was induced by repeated wound-healing in fibrosis (cirrhosis) programmed by host miRNA information, and host and viral miRNAs were implicated in tumorigenic activity upon HCV infection.
Conclusions: We found the third host defense neomechanism, named ‘the quantum miRNA immunity’ against human virus-related diseases.
Keywords
MicroRNA; Hepatitis B virus; Hepatitis C virus; Human immunodeficiency virus type 1; Hepatocellular carcinoma; Fibrosis; Quantum microRNA language; AI machine learning; VR; Covid-19
MicroRNA articles, Hepatitis B virus articles, Hepatitis C virus articles, Human immunodeficiency virus type 1 articles, Hepatocellular carcinoma articles, Fibrosis articles, Quantum microRNA language articles, AI machine learning articles, VR articles, Covid-19 articles
Article Details
1. Introduction
Hepatitis B virus (HBV) and C virus (HCV) are the critical carcinogens for hepatocellular carcinoma (HCC) [1-3]. Although 75-90% of liver cancers are HCC [4], liver cancer is the sixth common cancer and the fourth leading cause of cancer related death in GLOBOCAN 2018 (uicc.org/news/new-global-cancer-data-globocan-2018#). HBV and HCV are the most common risk factors of all HCC incidence (approximate 56% of HBV and 15% of HCV) [5]. As a viral factor, the HBV life cycle contributes for chronic inflammation as liver hepatitis by viral DNA integration into the human genome and for induction of host gene mutation [6]. About causes of HCC by HBV infection, the steps for integration of HBV DNA into the human genome is thought to be important for tumorigenesis [7], and after the integration, HBV x (HBx) transactivator protein expression from the HBV DNA in the human genome, not viral load, seems to have carcinogenetic effects to promote cell growth, anti-apoptotic effects and epigenetic modification [7]. However, despite the plethora of many experimental evidences published in in vivo mouse models, the precious etiology of HBx-associated carcinogenesis in human remains uncertain. On the contrary, HCV proliferation has also been speculated to be related with the risk of HCC. The risk of HCV-related HCC development was higher in patients with high titer of HCV RNA than with low titer [8] but the HCV RNA titer was low in patient with HCV-related liver cirrhosis, which is a risk factor of HCC [9]. HCV and HBV double infection was strongly correlated with HCC development in Egyptian [10]; however, HCV RNA levels was not correlated with HCC. Further, HCV RNA was detected in both HCC and surrounding non-tumor tissues [11]. Thus, it is uncertain whether HCV replication would be a cause of HCV-related HCCs or not; therefore, the carcinogenetic mechanisms by HCV replication itself are not completely elucidated.
Other risk factors of HCC are host liver cirrhosis and fibrosis, excessive alcohol consumption, incorporation of aflatoxin B1, nonalcoholic steatohepatitis, obesity etc. [12], and the most important causes for the development of liver fibrosis are chronic HBV or HCV infection, cirrhosis, chronic alcohol abuse and metabolic syndrome, which are similar with the risk factors of HCC. Therefore, both liver cirrhosis and fibrosis as host factors are important risk factors of HBV- and HCV-related HCC. Although HCC cells are derived from hepatocytes, hepatic fibrosis is induced by hepatic stellate cells as the major mesenchymal ones in the liver [13]. Upon activation of inflammation processes with mesenchymal cells after HBV or HCV infections, an excessive production of extracellular matrix proteins from liver stellate cells induces fibrosis as the results of a wound-healing response, and finally the loss of hepatocyte functions.
Liver cirrhosis is caused by repetitive and chronic liver damage, and is characterized by the development of regenerative nodules with fibrous connective tissues [14]. Therefore, cirrhosis is an advanced stage of liver fibrosis and fibrosis processes containe the protection of the host liver against infection and the liver regeneration from inflammation. It leads to liver matrix deposition, normal liver architecture destruction, parenchymal disruption, etc. Many cytokines and their related pathways are implicated in these processes. Although the liver fibrosis was examined using rodent models, it has been remained a deep gap between putative targets for human fibrosis therapy and murine pathways of complexed fibrosis in the liver [15, 16]. Liver fibrosis and cirrhosis are involved in chronic hepatitis by HBV and/or HCV in the bed side, therefore, the prevalence of histologic fibrosis and cirrhosis has deeply been associated with HBV- or HCV-associated HCC [17]. In Yan’s lab study, more than 80% of untreated HBV patients with HCC were HBV e antigen (HBe Ag)-negative. Although it is well known that the area of chronic HBV infection is geographically matched to that of HCC, Chen et al. [18] of the Taiwan group have found that inactive HBV carriers have 4.6 times high risk for HCC than individuals without HCV or HBV, and liver cirrhosis and loss of serum HBe Ag patients were correlated with the increasing risk of HCC development [19]. Contradictorily, the same Taiwan’s group had reported 8 years before that HBe Ag positive and no cirrhosis patients were associated with the risk of HCC [20]. Finally, above Taiwan’s group has recently shown that HBe Ag seroclearance by high levels of HBV core antibody are associated with the reduced risk of HCC [21]. Other papers showed that low HBV load patients compensated cirrhosis were not at low risk for HCC, and high HBV surface antigen (HBs Ag) levels and low viral load plus low HBe Ag were implicated in HCC, and high HBs Ag was related with prognosis after curative resection in the case of low viral load [22-25]. Therefore, the relation among liver fibrosis and cirrhosis, viral load and HBV antigen was far simplified in early HCC diagnosis and HCC development. About anti-HCV treatments, it has been reported that new anti-HCV strategies failed to progress carcinogenesis of the liver compared with the previous standard-of-care interferon and ribavirin [26] whereas an HCV cure and reduction of the risk in HCC have been developed by direct-acting antivirals (DAAs) treatment [27] and been achieved a sustained response of virus (SRV) [28]. Furthermore, although it is the same problem in HBV infection as in HCV one, data about the relation between HCV coding proteins and HCC development is also conflicting. It is quite difficult to elucidate complex causes of HBV- and HCV-related HCCs because the onset of HCC at stage minus one or zero was not be preciously identified by the readily prepared biomarker, finally, the etiology of carcinogenesis by HBV or HCV infection remains to be cleared [29].
The host microRNA (miRNA) has an important role for initiation, induction, development and metastasis of HCC [26, 30]. miRNAs have been reported as a biomarker for diagnosis, prognosis of HCC [31, 32] and hepatic fibrosis [33, 34]. We have studied in silico with the quantum miRNA language for the etiology of solid tumors, breast, lung, colorectal, pancreatic, esophageal and gastric cancers by using circulating diagnostic miRNA panels [35-37]. We found the hub miRNA in the miRNA biomarker panel and we showed that the miRNA/miRNA quantum language in the hub miRNA modulates carcinogenesis. To further understand function of miRNA biomarker panels in human infectious diseases and cancers, the etiologies of HBV or HCV infection including concomitant viral miRNA or its candidates, HBV-miRNA-2 or HCV-miRNA candidate1 and 2, chronic HBV- or HCV-infection associated liver fibrosis and HBV- or HCV-related HCC were computationally simulated to elucidate the mechanisms upon carcinogenesis of hepatocytes using miRNA memory package (MMP) in the biomarker miRNA panel, and quantum miRNA network simulation was performed by miRNA entangling target sorting (METS) with artificial intelligence (AI) machine learning (MIRAI). Further, a human immunodeficiency virus type 1 (HIV-1) infection case was also investigated as a non-solid tumorigenic virus in the control simulation with previous reported viral HIV-miR-N367. Subsequently, we found the quantum miRNA immunity against human hepatitis and immunodeficiency viruses.
2. Materials and Methods
2.1 Database usage
Google scholar (https://scholar.google.co.jp) was used for extraction of miRNA panel data. Total information content was 42,382 in HBV infection, 58,282 in HCV infection, 4,118 in HBV-associated fibrosis, 8,605 in HCV-associated fibrosis, 9,056 in HBV-induced HCC, 8,168 in HCV-induced HCC and 367 in HIV-1 infection. The gene function of protein was searched by GeneCards (www.genecards.org). Protein ontology was investigated by GO enrichment analysis in Geneontology (geneontology.org). Data mining about miRNA panels was performed by; 1) data from serum or plasma, 2) cleared in expression levels of up- and down-regulation.
2.2 METS in silico analysis
MMP calculation and METS analysis were performed by the computer processing as described previously [35, 38-42]. In short, MMP from miRNA biomarker panels was calculated by double miRNAs’ quantum energy levels of double nexus score (DNS) entangling single miRNA quantum energy levels of single nexus score (SNS) (Table 1). Data of multi-targets to a miRNA was extracted from TargetScan Human 7.2 (targetscan.org) and miRTarBase Ver. 8.0 (mirtarbase.cuhk.edu.cn). Target protein/protein interaction and cluster analysis were searched by STRING Ver. 11.0 (string-db.org).
2.3 Viral miRNA and functionally analogy analysis
Data of viral miRNA and viral genome was obtained from Viral Genomes (ncbi.nim.nih.gov) and miRBase Ver. 22.1 (miRbase.org). HCV subtype 1a sequence (NC_004102.1) was used for prediction of viral miRNA in the 5’UTR. The functional analogy between viral miRNA and host miRNA was performed as previously described [37, 43]. MiRCompare (160.80.35.140) was used for homology sequence search. The RNA secondary structure of RNA was computed by RNA Folding Form (unafold.rna.albany.edu).
2.4 AI machine learning
Prediction One Ver. 04.08.20 (Sony Network communications Inc. Tokyo, Japan) was used for AI machine learning. The area under the curve (AUC) in receiver operating characteristic (ROC), accuracy, precision and F values were calculated by Prediction One.
2.5 MIRAI
Previous METS analysis data in pancreatic, lung, colorectal, gastric and esophageal cancers [36, 37] was combined with the present data to produce the AUC data through AI machine learning.
3. Results and Discussion
3.1 Quantum energy in liver cancers
Quantum energy levels were calculated by MMP simulation and DNS frequency in HBV and HCV infection, HBV- and HCV-associated fibrosis (cirrhosis) or HCC compared with HIV-1 infection as a control of viral miRNA effects.
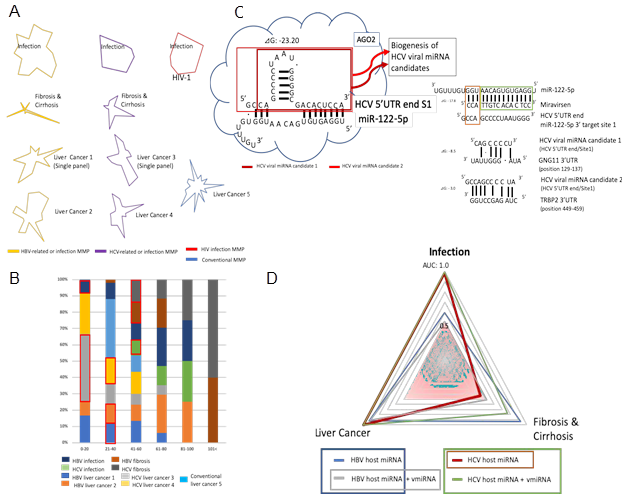
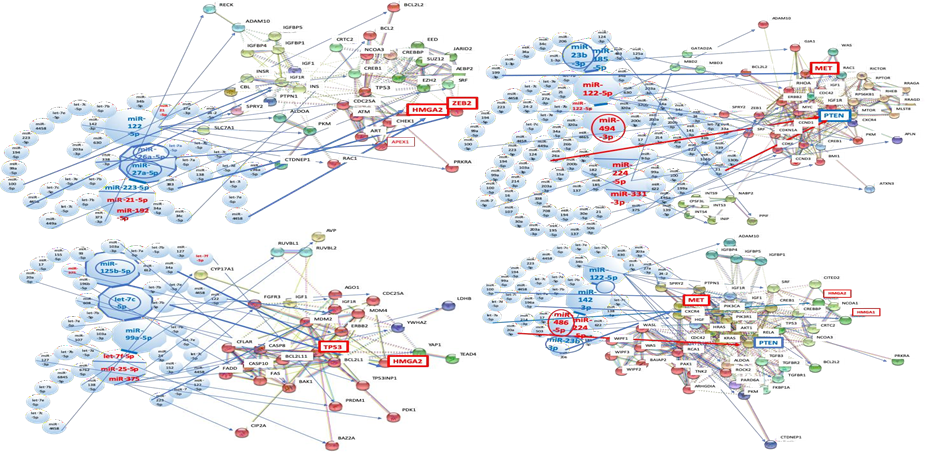
3.1.1 MMP maps in virus-associated liver diseases: Four to six miRNAs were collected for MMP calculation as the quantum energy level from a biomarker panel of HBV- and HCV-associated liver diseases by data mining (Table1 and Figure 1A). The quantum energy levels of the liver diseases clearly showed unique radar charts of MMP in HBV- and HCV-related human liver diseases, and HIV-1 infection (Figure 1A). Interestingly, MMPs from single- and multi-biomarker panels were also different in HCV- and HBV-related cancers.
3.1.2 DNS frequency of virus-associated liver diseases: According to the MMP characterization, the mining MMP data in all liver disease cases was used for a calculation of DNS frequency and each layer of the quantum core region (QCR) was determined (Figure 1B). Quantum energy frequency of virus-related liver disease was widely distributed among the layer of quantum level 0 to the layer of quantum level 144 (over 101). Proportion of DNS frequency percentage was also irregularly distributed among each disease. The hub miRNA layer about liver diseases was selected from the total QCR layers through the AI machine learning as described above (Table 1 and Figure 2B red square). The hub miRNA layer was restricted within low QCR layer limits, level 0-60.
3.2 METS analysis against liver carcinogenicity by AI
According to QCR layers as described above, METS analysis was performed by AI in HBV and HCV infection, and HBV- and HCV-related fibrosis and cirrhosis, and cancers. Since MMPs from single- and multi-panel data of biomarkers were different, METS data from single and multi-biomarker panels were cohered for statistical analysis by AI to preciously supply much data of carcinogenic effects. Quite recently, it has been shown that human AGO2 protein binds to the HCV site 1 with miR-122-5p [44]. Since AGO2 protein induces to catalyze endonucleolytic cleavage on target mRNA within partially paired miRNA/mRNA sequences [45], we predicted HCV-miRNA candidates from HCV 5’UTR site 1 (Figure 1C).
3.2.1 Prediction of human miRNA paralogues in viral miRNAs: To cohere HBV viral miRNA and HCV miRNA candidate effects, the seed paralogue of viral xentropic miRNAs (xenomiRNAs) and candidates were searched in that of human miRNAs. Since HBV viral xenomiRNAs have already been shown [46], the seeds of HBV-miR-1, HBV-miR-2, HBV-miR-3 and HBV-miR-4, and HCV-miR-candidate 0, HCV-miR-candidate-1 and HCV-miR-candiadte-2 were analyzed, and HBV-miR-2 were 75 and 62% homologous to those of hsa-miR-4436a and has-miR-5000-3p, respectively (Table 2). The seeds of HBV-miR-4, HCV-miR-candidate 0, 1, and 2 were 62, 62, 62, and 75% homologous to those of hsa-miR-3910, hsa-miR-5684, hsa-miR-652-5p and hsa-miR-3155b, respectively (Table 2). The human miRNA paralogues of HBV-miR-1 and -3 were not found by sequence homology analysis. In METS analysis of HBV-miRNAs, the cluster of protein/protein interaction was only observed in miR-4436a (the seed paralogue of HBV-miR-2). In METS analysis of HCV-miRNAs, the cluster of protein/protein interaction was found in miR-652-5p (the seed paralogue of HCV-miR-candidate 1) and miR-3155b (that of HCV-miR-candidate 2). The protein/protein interaction of HBV-miR-4, and HCV-miR-candidate 0 were not determined because too less data number (data not shown).
3.2.2 MIRAI for virus-associated liver diseases: The carcinogenic relation among infection, fibrosis and cirrhosis, and cancer cases was computed in METS analysis of virus-related liver diseases by AI (MIRAI) (Table 3). The AUC, accuracy, precision and F values were calculated in the presence or absence of viral miRNAs and viral miRNA candidates. As shown in Figure 1D, the significant AUC value of carcinogenicity in host miRNA plus viral miRNA (gray line) was a proper subset of the host miRNA AUC (blue line) in HBV-related liver diseases. On the contrary, in HCV-related liver diseases, the AUC value of host miRNA (brown line) was a proper subset of the host miRNA plus viral miRNA candidates AUC (green line) (Figure 1D). Thus, it is suggested by AI analysis that in HBV-related carcinogenesis, host miRNA alteration in fibrosis and cirrhosis has an important role for progression to liver carcinogenesis, and in HCV-related tumorigenesis, the first impact of HCV infection leads to carcinogenesis in the liver through fibrosis and cirrhosis. To further elucidate etiology of MIRAI analysis, quantum miRNA network analysis was performed preciously.
*miRNA in the bold grid: Hub miRNA
Table 1: miRNA biomarkers in virus-related liver diseases and HIV-1 infection.
Table 2: Viral miRNA and candidate seed paralogue in human miRNAs (*8 seed).
*Data of viral miRNA or candidates in Table 2 was integrated into the quantum miRNA network analysis; **Data was caluculated by Prediction One
Table 3: Statistical AI analysis in hepatitis virus-associated carcinogenesis.
3.3 HBV infection
HBV is a DNA virus. HBV has a partially double-stranded and circular genomic DNA (3020-3320 nts) and it contains 4 open reading frames (orfs) overlapped partially. These orfs encoded the reverse-transcriptase (RT)/polymerase (Pol), the capsid protein including core antigen (HBc) and pre-C plus core protein (HBe), three envelope proteins (L-HBs, M-HBs and S-HBs) and the trans-activator x (HBx) protein.
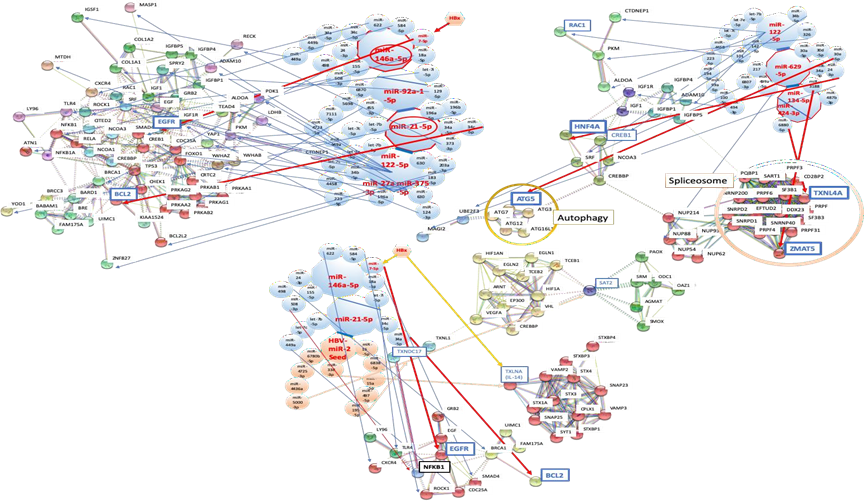
3.3.1 Network analysis of HBV infection with host miRNAs: After data mining, six miRNAs were selected as an MMP of HBV infection from biomarker panels of it [47, 48] (Figure 2A and 2B). miR-146a-5p, miR-92a-1-5p, miR-21-5p, miR-122-5p, miR-27a-3p and miR-146a-5p were upregulated (Table 1). These miRNAs shut down the expression of 34 proteins at once and these proteins are implicated in several cell functions, such as the cell proliferation (EGFR, IGF1R), cell cycle (CDC25A), transcription factors or repressor (FOXO1, SRF, CREB1, ATN1), apoptosis (BCL2, BCL2L2), inflammatory response (NFKB1, MTDH), innate immunity (TLR4), signal transduction system (ROCK1, YWHAZ, SMAD4), ubiquitin system (YOD1, BRCA1), cell adhesion (COL1A1) and metabolic pathways (PKM, LDHB, PDK1, ALDOA, CTDNEP1, PRKAB2). It may simultaneously induce acute liver injury with the hepatic acute-phase reaction (APR).
Although anti-apoptotic BCL2 was inhibited by miR-21-5p upregulation with miR-34a-5p and miR-34c-5p (Figure 2), upregulation of miR-21-5p, miR-34a-5p and miR-122-5p was observed in sera of non-alcoholic fatty liver [49]. Therefore, miR-21-5p and miR-34a-5p upregulation would be a key of liver injury. After HBV infection, a lethality of unfunctional liver cells could be modulated by themselves under the upregulation of these miRNAs and infected cells would be locked down. Subsequently, it is suggested that BCL2 suppression would induce apoptosis of hepatocytes and cell death of liver cells to protect HBV proliferation, and then would induce inflammation following liver injury [50]. The role of HBx protein remained controversial about apoptotic stimulation [51, 52]. Recently, Li et al. [53] have reported that HBx protein inhibits apoptosis; however, Kong et al. [54] have documented that increasing apoptosis in hepatoma cells were associated with increasing expression of tumor necrosis factor (TNF) receptor superfamily member 10b (TNFRSF10B, DR5) that mediated by HBx protein through NF-kB pathway. Since both experiments have been performed by cancer cells and on other cases, the results were obtained from experiments using mice, the effects of HBx protein to human liver cells have still not elucidated. Thus, escape from species bias and cultured tumor cell bias, quantum network simulation from the diagnostic miRNA panel is very useful to understand bona fide human hepatocyte and stromal cell conditions under HBV infection as described above. BCL2 inhibition was anti-virus and anti-carcinogenesis.
The epidermal growth factor (EGF) is known to be necessary in DNA synthesis of rat primary hepatocytes [55]. The continuous activation of EGF- epidermal growth factor receptor (EGFR) signaling is considered as a key factor of inflammation and development for cancers [56]. And expression of EGFR was enhanced by inhibition of SOCS5-mediated ubiquitination in the M1 polarized macrophages [57]. HBx protein inhibited the expression of EGFR via upregulation of miR-7-5p in HCC cells and HBx protein decreases cell proliferation of HCC cells [58]. In our simulation of HBV infection, miR-146a-5p upregulation inhibited EGFR with miR-7-5p (Figure 2A); therefore, inhibition of EGFR expression would also decrease proliferation of HBV-infected liver cells. In rat model, EFGR inhibitor erlotinib prevented hepatocyte proliferation, cirrhosis and hepatocellular carcinogenesis [59]. Further, EGFR was a host-entry cofactor when HBV binds to the cell surface [60], it is suggesting that during HBV infection, EGFR has an advantage in infection rather than in cell growth by EGF/EGFR, the receptor-ligand signaling. The internalization and downregulation of EGFR would rationally protect superinfection of HBV. Since EGFR polymorphisms was not associated with the risk of HBV-related hepatocellular carcinoma in China [61], EGFR downregulation would be implicated in virally invading steps on acute phase HBV infection. EGFR suppression has anti-virus and anti-tumor effects.
3.3.2 The quantum immunity and HBV viral miRNA: HBV-miR-2 inhibited spermidine N1-acetyltransferase family member 2 (SAT2) (Figure 2B). SAT is involved into hypoxia-related proteins and hypoxia-related enzymes, such as hypoxia inducible factor 1 subunit alpha (HIF1A) and ornithine decarboxylase (ODC) [62, 63]. Since SAT binds to HIF1A and RACK1 resulting in oxygen-independent HIF1A ubiquitination and degradation, inhibition of HIF1A levels via SAT contributed to the anti-tumor activity [64]. Since HBV-miR-2 inhibited SAT2 (Figure 2B), HBV-miR-2 would be decreasing hypoxic level. Therefore, HBV-miR-2 would be anti-tumor. Further, hypoxia induces hepatitis in general [65], while reducing hypoxic state by HBV-miR-2 would result anti-inflammation and anti-carcinogenesis. HBV-miR-2 inhibited thioredoxin domain containing 17 (TXNDC17, TRP14) and taxilin α (TXLNA) expression in our quantum simulation by using the seed paralogue search for host/viral miRNAs (data not shown). NFKB1 expression was inhibited by upregulation of miR-146a-5p with mir-155-5p and miR-508-3p (Figure 2B). TXNDC17 inhibited the TNF-α inducing NF-kB activation [66]. Although NF-kB was inhibited by upregulation of miR-146a-5p with miR-155-5p and miR-508-3p, downregulation of TXNDC17 by HBV-miR-2 may indirectly sustain the ability of NF-kB inducing inflammation and carcinogenesis. TXLNA, α-taxilin bound to syntaxin (STX) family and syntaxin binding proteins (STXBPs) [67] (Figure 2B). Their association is related with intracellular traffic machinery [68] and with HBV particle releasing from infected cells [69]. Therefore, downregulation of TXLNA would induce suppression of lethal viral releasing and would maintain persistent infection of HBV. On the contrary, TXLNA is also a cytokine, inteleukin-14 (IL-14) or B-cell growth factor (BCGF), which are mainly produced by T cells [70] and inhibition of IL-14 would suppresses inflammation. Further, HBx inhibits TXLNA gene transcription in T cell lines [71]. Thus, HBV-miR-2 and HBx would promote anti-inflammatory state and represses tumorigenesis in chronic infection state. In the case of HBV-miR-3 expressing infected cells, HBV-miR-3 represses HBV protein, such as HBx, and suppresses viral replication [46]. At that case under early phase, HBV-miR-2 with HBV-miR-3 would induce anti-inflammation and anti-carcinogenesis conditions. It is suggested that HBV-encoded miRNAs would affect to be distinctive between acute and chronic phases of HBV infection. Taken together, this is a first report of ‘quantum miRNA immunity’ that HBV infection would be preventive with programmed host defense mechanisms, such as shutdown and lockdown plus apoptosis, and it would be anti-carcinogenesis. The etiology of HBV infection was strongly supported by MIRAI in the quantum miRNA network analysis (Figure 1D).
3.4 HCV infection
HCV is a positive strand RNA virus. HCV genome (9600 nts) is not integrated into the human genome. The HCV orf is coding a 3000 amino acids polyprotein and it is processed into 3 structural proteins (core, E1 and E2) and 7 non-structural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B). Since HCV proteins are associated with host proteins, such as tumor suppressors, TP53, TP73, it is a possible idea that HCV infection would cause HCC.
3.4.1 HCV infection and liver specific miR-122: Liver specific miR-122-5p upregulation is well known as enhancement of propagation of HCV [72, 73]; however, let-7b-5p strongly inhibited HCV replication [74]. In our computer simulation, miR-122 upregulation was cooperated with let-7b-5p (Figure 2C). let-7a-5p in the let-7 family was downregulated in the plasma of chronic HCV infected patients [75]. Further, low expression of miR-122 did not allow HCV propagation to human primary synovial fibroblast [76]. On the other hand, HCV mutants were propagated in the miR-122-deficient PBMCs [77]. Although miR-122 seed region has an affinity to HCV internal ribosomal entry site (IRES) and site 1 plus 2 RNA in the 5’UTR [78, 79], quantum energy levels between miR-122 and the site 2 of 5’UTR HCV were very different among genotypes of HCV [80]. It is suggested that the enhancing effects of miR-122 on HCV propagation may deeply be dependent on viral subtypes. Thus, HCV propagation would be dependent on; 1) not only miR-122 levels but also other miRNA levels, such as let-7 family levels, 2) HCV RNA genomic mutation including HCV subtypes, 3) infectious host states including fibrosis and cirrhosis. Therefore, miravirsen, anti-miR-122, single therapeutic regimen may not be enough to treatment of HCV infection. The etiology of human diseases may have been usually searched about common miRNAs, such as miR-122 in HCV infection but the functions of biomarker miR-122-5p would be conflicting in previously reported papers.
3.4.2 Network analysis of HCV infection with host miRNAs: Four miRNAs were selected as an MMP of HCV infection from biomarker panels of it [81] (Figure 2C). miR-122-5p, miR-134-5p, miR-629-5p, and miR-424-3p were upregulated by HCV infection (subtype 1b/2a/3a/others; 76.9/10.2/5.1/7.7%) (Table 1). It is well known that after asymptomatic acute infection of HCV, HCV dominantly leads to persistent infection on a high proportion of infected individuals [82]. Therefore, the network scheme of HCV infection was very different from that of HBV infection. As miRNA-depend host defense machinery, it has been documented that host miRNAs directly target HBV and HCV genomic RNA [83, 84]; however, in HCV, miR-122-5p has only been showed to bind HCV 5’UTR site1 and site 2 [78, 79]. In our simulation, miR-122-5p with miR-142-3p and miR-101-3p inhibited Rac family small GTPase 1 (RAC1) expression (Figure 2C). RAC1 is a factor of HCV entry [85]. Furthermore, cAMP responsive element binding protein 1 (CREB1) was suppressed by upregulation of miR-122-5p with miR-33b-5p (Figure 2C). CREB1 was activated by HCV infection [86] and the CREB1 phosphorylation induced liver specific peroxisome proliferator-activated receptor gamma coactivator 1 alpha (L-PGC-1α) activation. The upregulation of L-PGC-1α enhanced replication of HCV [87]. Therefore, inhibition of CREB1 would indirectly block HCV replication. Thus, HCV entry and replication would be repressed by RAC1 and CREB1 downregulation as the quantum miRNA immunity.
3.4.3 Strong defense by the quantum miRNA immunity against HCV: HCV infection effectively reduced expression of hepatocyte nuclear factor 4 alpha (HNF4A) with a pinpoint. The hepatic transcriptional factor, HNF4A was downregulated by upregulation miR-629-5p with miR-34a-5p and miR-24-3p (Figure 2C), and autophagy related 5 (ATG5) was also inhibited by upregulation of miR-629-5p with miR-30a-5p, miR-30d-5p and miR-30e-5p (Figure 2C). HNF4A controls several hepatic genes and HNF4A-dependent gene expression downregulation is associated with alcoholic hepatitis in patients [88]. Upregulation of HNF4A is essential for viral late stage processing, such as assembly and secretion in liver carcinoma cells [89]. Upregulation of ATG5 participates viral replication [90]. These results suggested that host cells would defensed from HCV proliferation by quantum miRNA immunity. It would not be distinct from bona fide liver cell failure because prevention of acute injury of hepatocytes by HCV particle burst from the cell surface would be implicated in hepatitis with persistent HCV infection. HNF4A treatment inhibited proliferation of liver cancer stem cells [91] and the stress-induced HNF4A downregulation resulted in a long-term suppression of miR-122-5p, which increases the HCC risk [92]. Therefore, inhibition of HNF4A expression by the host defense against HCV infection stress would be highly carcinogenic. Further, human HCC is autophagy defective with HCV infection [93]. ATG5 is downregulated by increasing of miR-30e in Huh7.5 hepatoma cell line [94]. So, suppression of ATG5 may be tumorigenic.
Zinc finger Matrin-type 5 (ZMAT5) and thioredoxin like 4A (TXNL4A) expression were inhibited by miR-134-5p with human specific miRNA-3188 and miR-424-3p with human specific miR-6880-5p, respectively (Figure 2C). Both proteins are related with the pre-mRNA splicing machinery [95, 96] and HCV replication needs to hijack host splicing pathway [97, 98]. Therefore, suppression of function in the spliceosome would protect from HCV proliferation. These data showed a new idea on demand of HCV infection; namely, the first defense response of the host as quantum miRNA immunity could be elucidated beyond previous old researches, which have been explained in the replication steps of the HCV life cycle soon after absorption of viruses by the in vitro HCV experiments. This is a first report of quite early host defense response machinery against HCV infection. Although TXNL4A is a member of the U5 small ribonucleoprotein particle (snRNP), anti-U5 snRNP autoantibody was found in a patient of systemic sclerosis and polymyositis accompanied by large-cell lung carcinoma [99]. Therefore, spliceosome TXNL4A may be related with carcinogenesis. Given the early and programmed human defense reactions against HCV as the quantum miRNA immunity, it is suggested that the HCV preventive protein expression could developed to cancerous states in the host liver. The etiology of HCV infection was supported by MIRAI in the quantum miRNA network analysis (Figure 1D).
3.5 HIV-1 infection
HIV-1 is a twin positive strand RNA virus and infects to helper CD4+ T cells and macrophages. The HIV-1 genomic DNA is integrated into the host genome. Since HCV and HBV were mainly susceptible for the hepatocyte, HIV-1 infection in T cells and macrophages was monitored as a control for quantum miRNA network analysis as described previously [41], and it was followed by the updated database information.
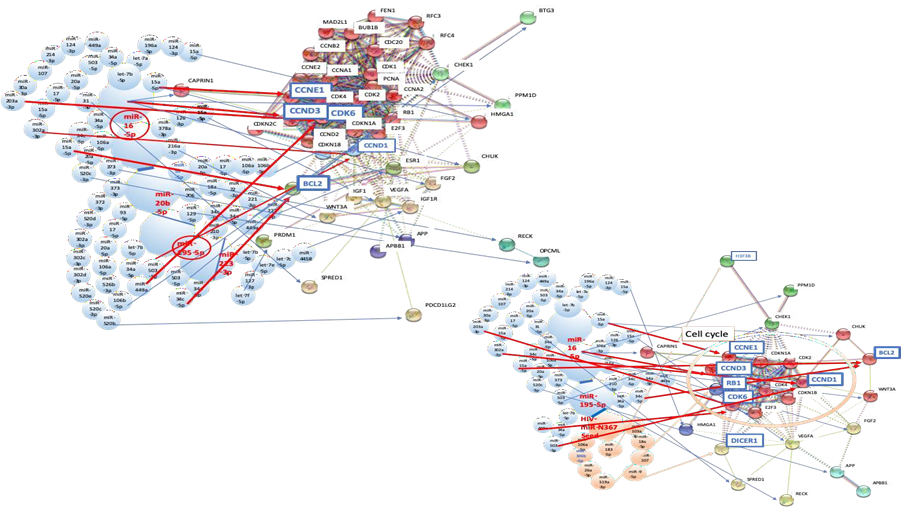
3.5.1 Network analysis of HIV-1 infection with host miRNAs: Four miRNAs were selected as an MMP of HIV-1 infection from biomarker panels of it [100-102] (Figure 3A and 3B). miR-16-5p, miR-20b-5p, miR-195-5p, and miR-213-3p were upregulated by HIV-1 infection (Table 1). miR-16-5p upregulation suppressed CDK6 with let-7b-5p, miR-34a-5p, miR-449a, miR-124-3p, miR-214-3p, miR-107, miR-30a-3p and miR-203a-3p (Figure 3A and B). Upregulation of miR-195-5p simultaneously inhibited CDK6 with let-7-5p, miR-34a-5p and miR-449a (Figure 3A and B). CCND3 expression was reduced by miR-16-5p upregulation and CCNE1 expression was blocked by increasing expression of miR-16-5p with 15a-5p (Fig. 3A and 3B). Cyclin dependent kinase 6 (CDK6), cyclin D3 (CCND3), cyclin D1 (CCND1) and cyclin E1 (CCNE1) are cell cycle-related proteins. CCND3 and CCND1 form a complex with CDK6, whose function is required for G1/S transition and is linked to HIV-1 susceptibility [103]. CCNE1 also makes a complex with CDK2 and its complex needs for G1/S transition. Therefore, HIV-1 infection would induce G1/S arrest of infected cells. Since infection of quiescent lymphocytes with HIV-1 does not produce progeny virus [104], the quantum simulation showed that infectious host CD4 T cells would be defended against HIV-1 infection by cell cycle inhibition. Further, infection of DNA viruses, RNA viruses or retroviruses has been implicated in G2/M arrest [105], therefore, host defense machinery of G1/S arrest against HIV-1 would be specific response to HIV-1. In macrophages, knockdown of CCND3 complexed with CDK6 inactivated SAM domain and HD domain-containing protein 1 (SAMHD1) and led to decreased dNTP levels, and inhibited the HIV-1 reverse transcription [106]. Further, HIV-1 Tat protein was phosphorylated by CDK2/CCNE1 and inhibition of its phosphorylation inhibited HIV-1 transcription via Tat in T lymphocytes [107] and non-proliferating macrophages [108]. In fact, pharmaceutical cyclin inhibitor r-roscovitin inhibited HIV-1 transcription by blocking of Tat function via inhibition of CDK2/CCNE1 activities in peripheral blood mononuclear cells (PBMCs) [109].
Aillet et al. [110] has reported that HIV-1 infection to T and monocyte cell lines decreased BCL2 expression level. As shown in Figure 3A, BCL2 gene expression was inhibited by upregulation of miR-16-5p with miR-34a-5p, miR-34c-5p plus miR-15a-5p and upregulation of miR-192-5p with miR-34a-5p and miR-34c-5p. Thus, cell cycle arrest and apoptosis would be a defense machinery to HIV-1 infection in PBMC and would be a cause of decrease of CD4+ T lymphocyte number in early HIV-1 infection period.
3.5.2 The quantum miRNA immunity and HIV-miR-N367: On the other hand, as described previously [41], HIV-miR-N367 inhibited RB transcriptional repressor 1 (RB1) with miR-106a-5p, miR-106b-5p, miR-26a-5p and miR-519a-3p (Figure 3B). HIV-miR-N367 has been found to inhibit transcription in trans and translation of HIV-1 proteins [111, 112]; however, downregulation of RB1 would progress T cell G1/S cell cycle, which would induce HIV-1 infection [103, 104]; therefore, HIV-1 nef/3’LTR function was both positive and negative viral factor in T lymphocytes [113]. In the case of macrophages, inhibition of RB1 in HIV-1 R5 infected human monocytes led to increasing apoptosis in vitro, therefore, RB1 upregulation would mediate apoptosis resistance feature in asymptomatic viremic HIV+ donors [114]. Thus, it is suggested that elevation of apoptosis by HIV-1-N367 may decrease macrophage number during the early infection [115]. Although Dicer expression inhibited HIV-1 replication in T cells and macrophages [116, 117], as shown Figure 3B, HIV-miR-N367 inhibited DICER1 expression with miR-103a-3p, miR-18a-5p, miR-107 and miR-9-5p. Therefore, downregulation of DICER1 by HIV-miR-N367 would induce inhibition of host miRNA biogenesis and high susceptibility of HIV-1 infection as a positive factor. Taken together, although the effects of host miRNAs could not make the latency, apoptotic effects by host miRNAs and HIV-miRNA would enhance susceptibility of HIV-1 infection and induce decreasing of T and macrophages in the early stage of infection. Subsequently, it causes immunodeficiency state of host, and then at the latency, HIV-1 replication would start being inhibited by HIV-miRNA in trans as a negative factor. Since HIV-miR-N-367 plus other host miRNAs were simulated to activate viral production, such as reactivation after the latency, HIV-1 nef/3’LTR region functions reciprocal without encoded Nef protein as described previously [41]. This updated data is also suggested that HIV-miR-N367 would be a curator of HIV-1 infection, which will not be complete without it for an acquired immunodeficiency with the latency. It was not statistically related to T cell and macrophage tumorigenesis (data not shown) while HIV-miR-N367 has previously been predicted as a tumor suppressor [41]. From the results of the quantum network analysis in HBV and HCV infection, it is strongly supported by MIRAI that anti-HIV-miR-N367 may cure HIV-1 infection with host quantum miRNA immunity as an HIV-1 miRNA vaccine in plant [37, 118].
3.6 Fibrosis and cirrhosis after hepatitis virus infection
Cirrhosis is an advanced stage of fibrosis. Cirrhosis is frequently indolent, asymptomatic and unsuspected until complication of liver disease present [14]. The diagnosis of asymptomatic cirrhosis is usually liver transaminases, radiologic tomography and liver biopsy when incidental screening tests. However, computerized tomography (CT) and magnetic resonance imaging (MRI) are not sensitive to diagnose cirrhosis. Therefore, more advanced diagnostic tools are necessary. The inflammation processes after HBV or HCV infections are deeply implicated in expression of miRNAs [119, 120]. Although clinical non-symptom of fibrosis and cirrhosis is similar among liver diseases [14], circulating miRNA profiles are very different from HBV-associated fibrosis to HCV-associated one. miRNA biomarker panel would be a good tool for early diagnosis of HCV-related fibrosis and cirrhosis. However, the etiologic schema of miRNA biomarker panels in virus-associated fibrosis remains to be proven. The liver fibrosis has mainly been examined in vivo using rodent models; therefore, there is a deep gap between putative targets for human fibrosis and murine fibrosis models [15, 16]. In their analytic data by single cell RNA sequencing, hepatic fibrosis was involved in the hepatic mesenchymal cells, therefore, therapeutic target for fibrosis has been determined in the functional zonation. It would not be suitable for precision medicine. For the establishment of miRNA biomarker in fibrosis and cirrhosis, quantum miRNA network analysis was performed to understand human hepatocyte information in liver fibrosis on demand.
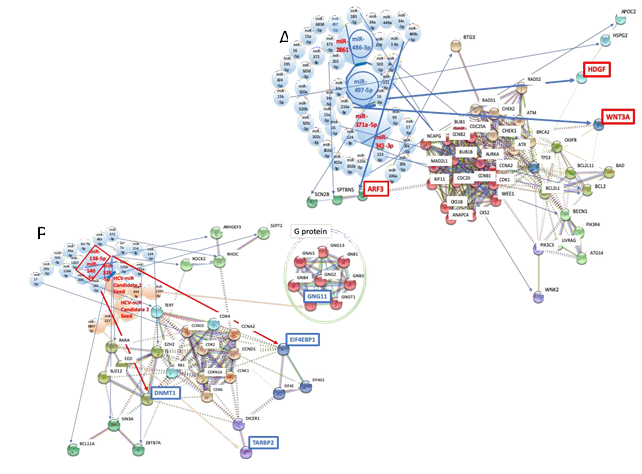
3.6.1 Network analysis of HBV-related fibrosis and cirrhosis: In human miRNA biomarker by liquid biopsy, five miRNAs were selected as an MMP of fibrosis after HBV and HCV infections from biomarker panels of them [13, 33] (Table 1). miR-2861, miR-371a-5p, and miR-345-3p were upregulated, and miR-486-3p and miR-497-5p were downregulated by fibrosis under HBV infection (Table 1). miR-138-5p, miR-143-5p, miR-140-5p, miR-325 and miR-328-5p were upregulated by fibrosis plus HCV infection (Table 1).
High level expression of hepatoma-derived growth factor (HDGF) is involved in liver fibrosis and carcinogenesis [121]. HDGF bound to nucleolin on the cell surface of liver cells and HDGF/nucleolin signal induced PI3K/Akt pathways in hepatoma cells. Upregulation of HDGF was induced by downregulation of miR-497-5 with miR-16-5p in fibrosis plus HBV infection (Figure 4A). Further, WNT3A was increased by upregulation of miR-497-5p with miR-216a-3p (Figure 4A). Reactivation of WNT/β-catenin pathway in the niche of liver cells is linked to the pathogenic disorders in liver fibrosis [122]. Blocking of WNT/β-catenin pathway by siRNA inhibited hepatic stellate cell activation in human hepatic tissues [123]. Simultaneously, activation of WNT/β-catenin pathway is related with liver cell carcinogenesis [124]. In 1996, Ihara et al. firstly demonstrated that WNT/β-catenin pathway overexpression was observed in hepatocellular carcinoma [125]. Thus, these data strongly suggested that fibrosis and carcinogenesis would be progressed in the liver at the same time about HBV-related fibrosis. The etiology of HBV-associated fibrosis was strongly supported by MIRAI in the quantum miRNA network analysis (Figure 1D). Since HBV-miR-2 would induce anti-inflammation and anti-carcinogenesis conditions (Figure 2B), HBV-miR-2 mimic may be able to use in treatment of HBV-induced fibrosis and cirrhosis.
3.6.2 Network analysis of HCV-related fibrosis and cirrhosis: In contrast to HBV-derived fibrosis, HCV-related moderate fibrosis in the early stage was in inactive or dormant state because of asymptomatic and chronic HCV infection [126]. It is common that some individuals have fibrosis even if HCV was asymptomatically infected because fibrosis and cirrhosis are asymptomatic in HCV infection. Expression of RHOC or ROCK2 was inhibited by upregulation of miR-138-5p, and expression of enhancer of zeste homolog 2 (EZH2) was also blocked by miR-138-5p upregulation with let-7a-5p, miR-214-3p and miR-124-3p (Figure 4B). High level expression of EZH2 was implicated in liver failure [127] and inhibition of EZH2 reduced fibrosis [128]. Activation of the RHO-associated serine/threonine kinase (ROCK) is required for multiple profibrotic responses [129]; therefore, RHOC and ROCK2 suppression would be anti-fibrosis. Since ROCK2 is known as the promoter of tumorigenesis [130], downregulation of ROCK2 by host miRNA would be anti-cancer. Although DNMT1 was downregulated by upregulation of miR-140-5p with miR-185-5p (Figure 4B), promoter hypomethylated genes in the genome-wide profiling were the gene cluster of the immune-related and defense response pathways but not cancer ones [131]. It is suggested that host defense and recovery systems would be dominant in fibrosis under HCV infection.
On the contrary, HCV NS5A protein bound to EIF4F and upregulated translation initiation [132]. It is well known that upregulation of translation initiation is generally implicated in tumorigenesis. The upregulation of host translation initiation has been involved in 4EBP1 (EIF4EBP1) inactivation. Therefore, inactivation of 4EBP1 in HCV infection may play a critical role in tumorigenesis, but the advantage of translation initiation would contribute to viral protein translation. Although EIF4EBP1 was reduced by upregulation of host miR-138-5p (Figure 4B), it is not certain whether downregulation of 4EBP1 by HCV NS5A or host miR-138-5p could be related to carcinogenesis or not.
It was analyzed by MIRAI that host miRNAs would not contribute for carcinogenesis in HCV-related fibrosis (Figure 1D). Therefore, the seed of HCV-miRNA candidate 1 and 2 were predicted and were analyzed in the quantum miRNA network by using the seed paralogue of host/viral miRNAs as previously described [37] (Table 2). HCV-miRNA candidate 1 targeted G protein subunit gamma 11 (GNG11) to downregulate with miR-493-5p and miR-1298-3p, and HCV-miRNA candidate 2 targeted TAR (HIV-1) RNA binding protein 2 (TARBP2) to downregulate with miR-217 and miR-6807-3p (Figure 1C and Figure 4B). Efficient HCV RNA replication requires TARBP and Dicer to produce mature miR-122-5p [133]. Therefore, suppression of TARBP would suppress viral replication. HCV-miRNA candidates would be implicated in suppression of HCV proliferation and subsequently in establishment of persistent and slow infection. On the other hand, downregulation of TARBP2 increased cancer stem cells (CSCs) and contributed for CSC clonogenicity, proliferation in Ewing sarcoma family tumors [134]. Furthermore, downregulation of TARBP2 exhibited properties of miRNA-independent regulation in cancer, such as sorafenib resistance in HCC [135]. Therefore, suppression of TARBP2 by HCV-miRNA candidate 2 would only be tumorigenic in HCV-induced fibrosis and cirrhosis. However, G protein upregulation was mediated renal fibrosis in heart failure [136] and aberrant activation of G protein-coupled receptor is implicated in prostate cancer progression [137]. Therefore, downregulation of GNG11 would be anti-inflammation and anti-cancer. Taken together, the aberrant balance of etiology was shown by host miRNAs in HCV-associated fibrosis and cirrhosis among anti-inflammation, anti-oncogenesis and viral producible. Only when the HCV-miRNA candidate 2 was presented, HCV-induced fibrosis and cirrhosis was occasionally carcinogenesis. The indefinite etiology of HCV-associated fibrosis and cirrhosis was supported by MIRAI in the quantum miRNA network analysis (Figure 1D, Table 3).
3.6.3 Miravirsen and HCV-miR-candidate: Anti-miRNA agents, miravirsen targets miR-122-5p (Figure 1C) and the liver specific miR-122-5p is essential for HCV propagation in general [72, 73]. Therefore, miravirsen inhibits HCV proliferation in the bedside [138] and miR-122-5p biogenesis was simultaneously blocked by miravirsen in vitro [139]. MIRAI data strongly suggests that eradication of HCV by miravirsen or RG-101 [140] could be effective for prevention of HCV infection and its progression of carcinogenesis. However, in our simulation, it is also suggested that anti-HCV-miR-candidate 2 according to HCV subtypes would be essential for enough treatment of HCV-related high carcinogenic viral infection.
3.7 HBV-related HCC
Since it has been reported in the meta-analysis of HCC miRNA biomarker panels from complexed HCC clinical conditions of patients as described above [48], simulations of HBV- and HCV-associated HCC were performed in both single panel data and multiple panel data included into the meta-analysis.
3.7.1 Network analysis of HBV-associated HCC: As the case one (stage I-II, 61%; III-IV, 25%), single panel of miRNA biomarker (liver specific miR-122-5p downregulated data) was applied for the quantum miRNA network analysis. Six miRNAs were selected as an MMP of HBV-related HCC from biomarker panel of it [141] (Fig. 5A and 5B). miR-21-5p and miR-182-5p were upregulated, and miR-122-5p, miR-26a-5p, miR-27a-5p and miR-223-5p were downregulated in HBV-associated HCC (Table 1). As the case two (stage I, 68.7%; II, 12.5%; III-IV, 18.8%), multiple panels of miRNA biomarker (no miR-122-5p in the panel) were used for the analysis [47, 48, 142, 143]. miR-125b-5p, let-7c-5p and miR-99a-5p were downregulated, and let-7f-5p, miR-25-5p and miR-375 were upregulated.
3.7.2 A common therapeutic target of HBV-associated HCC: High mobility group AT-hook 2 (HMGA2) expression was increased by down regulation of miR-26a-5p with let-7a-5p, let-7b-5p, let-7c-5p, let-7d-5p, let-7i-5p, let-7e-5p and miR-4458, and miR-26a-5p with let-7a-5p, let-7b-5p, let-7c-5p, let-7d-5p and miR-196a-5p in the case 01 (Figure 5A). HMGA2 was also augmented by downregulation of let-7c with let-7b in the case 02 (Figure 5B). It suggests that the relation between HMGA2 and let-7 family has an important role for oncogenesis in the hepatocytes. Previous our studies have demonstrated with quantum miRNA network that HMGA2 is implicated in carcinogenesis upon lung cancer from smoking, and gastric cancer stage I-IV at the etiological level [36, 37]. In HCC, HMGA2 overexpression induced invasion and metastasis [144]. Further, HMGA2 expression was significantly enhanced by tumorigenic HBx and HMGA2 upregulation augmented proliferation of HCC cells, metastasis and invasion [145]. Interestingly, although WNT3A was upregulated in HBV-associated fibrosis (Figure 4A), long noncoding RNA (lcnRNA), LSINCT5 induced HMGA2 expression and the oncogenic activities was related with interaction among LSINCT5 in HCC cell lines [146]. HMGA2 and WNT/β-catenin pathways were contributed to the oncogenic properties of endometrial carcinoma cell lines [147]. Proliferation of HCC cell line was blocked by propofol and propofol inhibited HMGA2 expression and WNT/β-catenin pathway [148]. Thus, it is newly suggested by our simulation that HBV-associated fibrosis would be related with HBV-associated tumorigenic progression upon the connection between both WNT3A and HMGA2 upregulation.
3.7.3 Case specific therapeutic targets on HBV-associated HCC: In the case 1, zinc finger E-box binding homeobox 2 (ZEB2) was upregulated by suppression of miR-27a-5p with miR-383-5p (Figure 5A), and in the case 2, tumor protein 53 (TP53) was increased by downregulation of miR-125b-5p with miR-612 and miR-34a-5p, respectively (Figure 5B). As the host defense mechanisms against viral infection and tumorigenesis, viral proliferation would be inhibited by expression of ZEB2 [149] and tumor proliferation would be inhibited by upregulation of tumor suppressor TP53, respectively. Although ZEB2 upregulation was observed in HCC tissues and cell lines [150], ZEB2 has been deeply related with oncogenesis of esophageal squamous cell carcinoma (ESCC) stage 0-I (AUC, 0.999) [37]. Further, overexpression of TP53 upregulated HBx expression levels [151]. HBx upregulation would lead to HMGA2 increasing and further proliferation of HCC cells as described above. In turn, tumor suppressor TP53 would be a specific driving force of oncogenesis in HBV-associated HCC. Therefore, host defense mechanisms would grow to induce cancerous and lethal state of the liver.
3.8 HCV-related HCC
As the case three (no cancer stage information), single panel of miRNA biomarker (liver specific miR-122-5p upregulated data) was used for the quantum miRNA network analysis.
3.8.1 Network analysis of HCV-associated HCC: Six miRNAs were selected as an MMP of HCV-related HCC from biomarker panel of it [152] (Figure 5C and 5D). miR-122-5p, miR-494-3p, miR-224-5p and miR-331-3p were upregulated, and miR-23b-3p and miR-185-5p were downregulated in HCV-associated HCC (Table 1). As the case four (cancer stage I), multiple panels of miRNA biomarker (miR-122-5p downregulated data) were integrated into the analysis [153-155]. miR-122-5p, miR-142-3p and miR-23b-3p were downregulated, and miR-486-5p and miR-224-5p were upregulated (Table 1).
3.8.2 Common therapeutic targets of HCV-associated HCC: Tumor suppressor, phosphatase and tensin homolog (PTEN) was inhibited by upregulation of miR-494-3p with miR-4465, miR-20b-5p and miR-20a-5p, and with miR-17-5p, miR-214-3p, miR-20a-5p, miR-222-3p, miR-106b-5p and miR-21-5p in the case 3 (Figure 5C). In the case of 4, PTEN was suppressed by upregulation of miR-486-5p with miR-17-5p, miR-214-3p and miR-20a-5p (Figure 5D). PTEN was downregulated in HCV-associated HCC cirrhotic tissues (63.1% of low level) compared with normal liver ones (91.3% of high level) [155]. As a cause of PTEN reduction in HCV-associated HCC, it has been documented that HCV core protein downregulated PTEN expression by activating NF-kB [157] and PTEN negatively modulated HCV replication [158]. Taken together, it is suggested that both host miRNAs and viral factors suppressed PTEN tumor suppressor, and would accelerate oncogenic progression of hepatocytes. HCV RNA information may make hepatocytes to be free from host defense machinery by controlling of miRNA quantum language.
Hepatocyte growth factor (HGF) receptor (MET) was increased by downregulation of miR-23b-3p with miR-206, miR-34c-5p, miR-34a-5p, miR-1-3p and miR-199-3p in the case 3, and downregulation of miR-23b-3p with miR-206 in the case 4 (Figure 5D). c-MET expression was augmented in 25-100% of HCC cells compared with normal liver ones [159]. Knockdown of c-MET decreased HCC cell proliferation in vitro and in vivo [160, 161]. Therefore, MET is a therapeutic target of HCC and the MET inhibitor capmatinib showed antitumor activity in phase I and II trials (HBV positive: 87%, HCV positive: 16%) [162, 163]. However, c-MET has not been specifically related with HCV replication. In the case 4, tumorigenic HMGA1 and HMGA2 were also upregulated by suppression of miR-142-3p with let-7b-5p, let-7a-5p and miR-196a-5p, and with let-7b-5p, let-7e-5p, let-7c-5p, miR-4458, let-7i-5p, let-7d-5p and let-7a-5p, respectively (Figure 5D).
3.9 Conventional HCCs
To further understand the difference between virus specific HCC and conventional HCC, the etiology analysis by conventional HCC miRNA biomarkers was performed by the quantum miRNA network analysis in the case five (stage I-II, 51.8%; III-IV, 28.0%). Seven miRNAs were selected from the miRNA biomarker panels in patient sera, and five of seven miRNAs were applied for computing simulation (Table 1) [165-166]. miR-92a-3p, miR-106b-5p, miR-107 and miR-21-5p were upregulated and miR-224-5p was downregulated. Although the five miRNAs were not contained in meta-analysis data [167], the panels using these five miRNAs AUC was over 0.950 and in the meta-analysis by using 34 studies the summarized AUC was 0.92. Therefore, the five miRNAs would be a candidate as a potential and diagnostically conventional biomarker for HCC. Since miR-3126-5p (downregulation) and miR-519d-5p (upregulation) were less reference and target score data in HCC, the network analysis has not been performed (data not shown).
Tumor suppressors, PTEN, runt-related transcription factor 3 (RUNX3) and retinoblastoma 1 (RB1) were suppressed by upregulation of miR-106b-5p with miR-20a-5p, miR-17-5p and miR-214-3p, with miR-532-5p, miR-106a-5p and miR-20a-5p, and with miR-106a-5p, respectively (data not shown). Further, cell cycle inhibitor, cyclin dependent kinase inhibitor 1A (CDKN1A) was repressed by upregulation of miR-106b-5p with miR-93-5p, miR-17-5p, miR-20a-5p, miR-20b-5p, miR-106a-5p, miR-526b-3p (data not shown). Tumor suppressor control protein E2F1 was reduced by upregulation of miR-106b-5p with miR-149-5p, miR-34a-5p, miR-20a-5p, miR-106a-5p, miR-17-5p (data not shown). PTEN downregulation was associated with HCC as described above. RUNX3 expression was decreased in HCC tissues and HCC cell lines and overexpression of RUNX3 reduced tumorigenesis of the cancer stem cell lines [168].
Although miR-106b/25 and miR-17/92 clusters were implicated in downregulation of tumor suppressor protein expression in the quantum miRNA network, serum miR-106b-5p upregulation has been identified as early diagnostic biomarker of HCC [165, 169]. miR-106b-5p overexpression promotes oncogenic progression and metastasis of HCC in vitro and in vivo [170, 171]. Finally, Gu et al. [172] showed that miR-106b-5p expression level was high in HCC tissues and was related with poor prognosis of patients. They found the target RUNX3 tumor suppressor. From these simulation data, the etiology of virus specific HCC could be distinguished from that of conventional HCC. It is suggested that the HCC therapeutic target should be separated between virus-related and conventional HCC in the precision medicine. Thus, the quantum miRNA network analysis by MIRAI would be useful for investigation of etiology in human HCC using miRNA biomarker panels.
(A) MMPs in HBV, HCV and HIV-1 infection, virus-associated fibrosis & cirrhosis, and HCCs; (B) Distribution of quantum energy in human hepatitis virus-related diseases. Frequency of DNS was calculated and the quantum core region (QCR) with the hub miRNA was identified by AI machine learning (red square); (C) HCV-miRNA candidates were presented. HCV-miRNAs were predicted (left upper panel) and their targets were extracted by the miRNA functionally analogy analysis (right lower panel); (D) METS analysis against liver carcinogenesis was performed by AI machine learning. AUC was calculated in infection, fibrosis & cirrhosis and cancer with host miRNAs or with host miRNAs plus virus-miRNAs, and the results were depicted with the radar.
(A) Network with protein clusters by METS simulation was presented in HBV infection; (B) Coherence of the hub miRNAs in the host and HBV-miR-2 was performed in the METS analysis; (C) Network with protein clusters by METS simulation was shown in HCV infection. miRNAs: red, upregulation. Proteins: blue, downregulation.
(A) Network with protein clusters by METS simulation was depicted in HIV-1 infection; (B) Coherence of the hub miRNAs in the host and HIV-1-miR-N367 was performed in the METS analysis. miRNAs: red, upregulation. Proteins: blue, downregulation.
(A) Network with protein clusters by METS simulation was presented in HBV-related fibrosis and cirrhosis; (B) Coherence of the host miRNAs and HCV-miRNA candidate 1 and 2 was performed in the METS analysis of fibrosis and cirrhosis, and the network was presented. miRNAs: blue, downregulation; red, upregulation. Proteins: red, upregulation; blue, downregulation.
(A) METS simulation of HBV-related HCC was performed in single panel data; (B) METS simulation of HBV-related HCC from multi-panel data; (C) METS simulation of HCV-related HCC from single panel data; (D) METS simulation of HCV-related HCC from multi-panel data. miRNAs: blue, downregulation; red, upregulation. Proteins: red, upregulation; blue, downregulation.
4. Conclusions
In general, HCC is the poorest prognostic tumor and the low prognosis is due to lack of effective diagnostic tools for early HCC. Recently, the combination of α-fetoprotein (AFP) and ultrasounds surveillance has been used for screening and detection of early HCC [173]. However, ultrasonography is low sensitivity of detection of HCC and often results in cirrhosis as misdiagnosis of HCC. AFP is low sensitivity and high false-positive rate even with tumor markers [174]. Therefore, conventional HCC diagnostic tool would be required for early diagnosis of HCC onset.
4.1 The quantum miRNA immunity
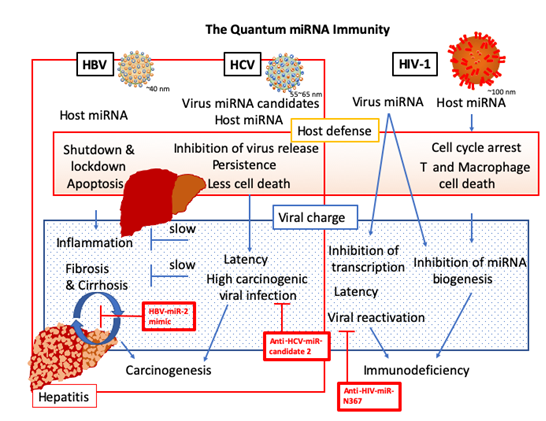
We have shown that miRNA biomarker panels would be etiologically useful for early diagnosis of several cancers. For liver viruses and HIV-1 infection, virus-related fibrosis or cirrhosis and virus-related human HCC, miRNA biomarker panels were also available. Especially, AI machine learning was harnessed from miRNA biomarker panels to serve the elucidation of causes in the complexed virus-related diseases. It is believed that host cells are hijacked by virus and it has been documented that in a non-immunologic manner, cellular miRNA aberration rationally induces HCC and immunodeficiency by viral infection [175]. However, our AI simulation data suggested that the virus infection was clearly controlled by a new host miRNA defense machinery as programmed ‘the quantum miRNA immunity’, and the quantum miRNA immunity was distinct from innate and adaptive immunity [176, 177], such as natural immunity, cellular immunity and humoral immunity (Figure 6). It was confirmed as the third immunity programmed by quantum miRNA language and the neo-mechanism was controlled by miRNAs with virus-specific. Although extracellular miRNAs have been reviewed to contribute for pathogenesis of viral infection [178], host defense mechanisms against viruses have been explained by RNA interference (RNAi)-related machinery in one to one of the two-dimensions [179]. It is a first report that the MMP program of host and virus miRNAs have an important role for early host defense machinery as the quantum miRNA immunity to inhibit and block virus infection and invasion in the integrated multi-dimensions. In the quantum miRNA immunity (Figure 6), HBV infected host participated in cellular function shutdown and cellular lockdown plus apoptosis through miRNA hub upregulation of miR-146-5p and miR-21-5p, on the other hand, HCV infected host was inhibited by suppression of virus production under less cell death and keep persistence in salt down through miRNA hub upregulation of miR-629-5p, miR-134-5p and miR-424-3p. HIV-1 infected host induced cell cycle arrest and then cell death of T plus macrophage through miRNA hub upregulation of miR-16-5p and miR-195-5p. Thus, the host defense mechanisms on the quantum miRNA immunity showed quite unique characters among three different viral infection. It was suggested that these systems are not aberration of miRNA expression and the miRNA expression is completely programmed according to the quantum language in host against exogenous quantum energy information of virus.
4.2 Interaction between host miRNA and viral miRNA
Given the quantum miRNA immunity in the host, the information in HBV and HIV-1 was charged by viral integration into host genome DNA. HBV-miR-2 from the own genomic DNA inhibited inflammation, fibrosis and cirrhosis through suppression of IL-14 and SAT2. As mentioned in Figure 4A, host miR-497-5p downregulation induced inflammation through upregulation of WNT3A and HDGF; therefore, wound and healing were repeated for the long term, which would induce fibrosis and cirrhosis. As WNT3A and HDGF upregulation would be carcinogenic, finally host miR-497-5p downregulation causes carcinogenesis in HBV-related fibrosis and cirrhosis. HIV-1 miRNA N367 from integrated provirus DNA blocked miRNA biogenesis through suppression of DICER1 and makes latency, and then prepared to reactivate viral replication (Figure 3). Since host miRNA decreased T and macrophage via cell cyclin suppression and apoptosis, such as CCND3 and CCNE1 by upregulation of miR-16-5p, during the latency, immunodeficiency was continued for the long term but not carcinogenic. On the contrary, HCV infection itself was quite carcinogenic while suppression of HNF4A by upregulation of host miR-629-5p is strongly carcinogenic. Therefore, HCV infection stress might be aimed to make tumorigenic and a part of HCV RNA, HCV-miRNA candidate 2 is also carcinogenic. It is suggested that HBV fibrosis (cirrhosis) and HCV infection would be the minus one stage of virus-associated HCC and carcinogenesis could be predicted in the minus on stage of cancer with miRNA biomarker panel [36]. Further, the risk of carcinogenesis would be remained in infectious host after hepatitis virus was eradicated. Thus, according to the host-virus interaction of miRNAs from miRNA biomarker panels, HBV-miR-2 mimic, anti-HCV-miRNA candidate 2, and anti-HIV-miR-N367 may be available for therapeutic agent development to prevent fibrosis and cirrhosis, carcinogenesis, and immunodeficiency, respectively (Figure 6).
HBV-, HCV-associated human diseases were shown as the new programmed host defense system ‘quantum miRNA immunity’ against virus infection. Anti-carcinogenesis and anti-immunodeficiency tools were presented in the red squares as the AI virtual reality.
4.3 The etiology analysis by miRNA biomarkers as an early HCC diagnostic tool
In HCC cancer stages, therapeutic targets were different among HBV- and HCV-related HCCs, and conventional HCC. HBV-related HCC was induced by downregulation of the hub miRNAs, miR-26a-5p, miR-27a-5p, miR-125b-5p and let-7c-5p via augmentation of HMGA2, ZEB2 and TP53. Tumor suppressor, TP53 upregulation is implicated in HBV HBx protein overexpression and the HBx upregulates HMGA2. Therefore, HBV-related HCC is deeply involved into HBV genome integration into the host cell genome. Downregulation of miR-23b-3p and upregulation of miR-494-3p and miR-486-5p tuned upregulation of MET and reduction of PTEN in the case of HCV-related HCC. HCV core protein inhibits PTEN. As PTEN upregulation suppresses HCV replication, PTEN downregulation is also implicated in HCV production. About conventional HCC, upregulation of miR-106b-5p resulted suppression of tumor suppressors, PTEN and RUNX3. Thus, three different HCC etiological phenotypes were preciously distinguished by miRNA biomarker panels of HCCs. The further progressed classification and etiology simulation by the miRNA panel would be necessary to confirm the precious medicine among cancer stages and individuals in HCC.
4.4 Meet the quantum miRNA immunity with Covid-19
We have already discussed about the deep relation of the immune and cancer in the retrovirus-related integrating site BIC/miR-155 in the human genome [41, 180]. Although oncomir miR-155 is the seed paralogue of KSHV-miR-K12-11 and spumavirus miR-S4, the relation between host immune system and carcinogenesis upon host and viral miRNAs has not yet been cleared. Here, using AI computer virtual reality, miRNA biomarker of HCC was showed as the etiology of carcinogenesis in the liver by the quantum miRNA analysis upon a proof of the concept and it was found that there is the new programmed host defense system ‘the quantum miRNA immunity’ against virus infection, which might be quite difficult to prove in the in vitro and in vivo experiments, and clinical investigations. The quantum miRNA immunity against viruses would be presented in common use. Under Covid-19 pandemic outbreak as the RNA storm [Fujii, 2017], biomarker panels of Covid-19 infection and Covid-19-associated pneumonia should be investigated soon. The quantum miRNA algorithm with AI machine learning could reveal both the etiology and the target of Covid-19 infection and Covid-19-associated pneumonia. The hidden quantum miRNA immunity against Covid-19 would be cleared under asymptomatic infection of Covid-19 [181]. Further investigations would be required for clinical use of miRNA biomarker and the clinical etiology analysis with AI.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
References
- Sherman M. Risk of hepatocellular carcinoma in hepatitis B and prevention through treatment. Cleve Clin J Med 76 (2009): S6-S9.
- Lin CL, Kao JH. Review article: the prevention of hepatitis B-related hepatocellular carcinoma. Aliment Pharmacol Ther 48 (2018): 5-14.
- Axley P, Ahmed Z, Ravi S, et al. Hepatitis C virus and hepatocellular carcinoma: a narrative review. J Clin Transl Heaptol 6 (2018): 79-84.
- Petruzziello A. Epidemiology of hepatitis B virus (HBV) and hepatitis C virus (HCV) related hepatocellular carcinoma. Open Virol J 12 (2018): 26-32.
- Yan JD, Roberts LR. Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol 7 (2010): 448-458.
- Grimm D, Thimme R, Blum H. HBV life cycle and novel drug target. Hepatol Int 5 (2011): 644-653.
- Kew MC. Hepatitis B virus x protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. J Gastroenterol Hepatol 26 (2011): 144-152.
- Noh R, Lee DH, Kwon BW, et al. Clinical impact of viral load on the development of hepatocellular carcinoma and liver-related mortality in patients with hepatitis C virus infection. Gastroenterol Res Pract 2016 (2016): 7476231.
- Duvoux C, Pawlotsky JM, Bastie A, et al. Low HCV replication levels in end-stage hepatitis C virus-related liver disease. J Hepatol 31 (1999): 593-597.
- Yates SC, Hafez M, Beld M, et al. Hepatocellular carcinoma in Egyptians with and without a history of hepatitis B virus infection: association with hepatitis C virus (HCV) infection but not with (HCV) RNA level. Am J Trop Med Hyg 60 (1999): 714-720.
- Sobesky R, Feray C, Rimlinger F, et al. Distinct hepatitis C virus core and F protein quasispecies in tumoral and nontumoral hepatocytes isolated via microdissection. Hepatology 46 (2007): 1704-1712.
- Tunissiolli NM, Castanhole-Nunes MMU, Biselli-Chicote PM, et al. Hepatoccelular carcinoma: a comprehensive review of biomarkers, clinical aspects, and therapy. Asian Pac J Cancer Prev 18 (2017): 863-872.
- El-Ahwany E, Nagy F, Zoheiry M, et al. Circulating miRNAs as predictor markers for activation of hepatic stellate cells and progression of HCV-induced liver fibrosis. Elect Phys 8 (2016): 1804-1810.
- Schuppan D, Afdhal NH. Liver cirrhosis. Lancet 371 (2008): 838-851.
- Ramachandran P, Dobie R, Wilson-Kanamori JR, et al. Resolving the fibrotic niche of human liver cirrhosis using single-cell level. Nature 575 (2019): 512-518.
- Dobie R, Wilson-Kanamori JR, Henderson BEP, et al. Single-cell transcriptomics uncovers zonation of function in the mesenchyme during liver fibrosis. Cell Rep 29 (2019): 1832-1847.
- Yan JD, Kim WR, Coelho R, et al. Cirrhosis is present in most patients with hepatitis B and hepatocellular carcinoma. Clin Gastroenterol Hepatol 9 (2011): 64-70.
- Chen JD, Yang HI, Iloeje UH, et al. Carrier of inactive hepatitis B virus are still at risk for hepatocellular carcinoma and liver-related death. Gastroentrology 138 (2010): 1747-1754.
- Xu J, Shi J, Wang YP, et al. Milder liver cirrhosis and loss of serum HBeAg do not imply lower risk for hepatocellular carcinoma development in HBV-related cirrhosis. Med Sci Monit 15 (2009): CR274-279.
- Yan HI, Lu SN, Liaw YF, et al. Hepatitis B e antigen and the risk of hepatocellular carcinoma. New Engl J Med 347 (2002): 168-174.
- Liu J, Hu HH, Chang CL, et al. Association between high levels of hepatitis B core antibody and seroclearance of hepatitis B e antigen in individuals with chronic hepatitis B virus infection. Clin Gastroenterol Hepatol 17 (2019): 1413-1415.
- Tseng TC, Liu CJ, Yang HC, et al. High levels of hepatitis B surface antigen increase risk of hepatocellular carcinoma in patients with low HBV load. Gastroenterology 142 (2012): 1140-1149.
- Sinn DH, Lee J, Goo J, et al. Hepatocellular carcinoma risk in chronic hepatitis B virus-infected compensated cirrhosis patients with low viral load. Hepatology 62 (2015): 694-701.
- Kawanaka M, Nishino K, Nakamura J, et al. Quantitative levels of hepatitis B virus DNA and surface antigen and the risk of hepatocellular carcinoma in patients with hepatitis B receiving long-term nucleos(t)ide analogue therapy. Liver Cancer 3 (2014): 41-52.
- Qu LS, Liu JX, Zhu J, et al. Risk factors for prognosis of hepatocellular carcinoma after curative resection in patients with low hepatitis B viral load. Annals Hepatol 16 (2017): 412-420.
- Plissonnier ML, Herzog K, Levrero M, et al. Non-coding RNAs and hepatitis C virus-induced hepatocellular carcinoma. Viruses 10 (2018): 591.
- Ormeci N, Gülsen MT, Sezgin O, et al. Treatment of HCV infection with direct-acting antiviral agents. Real life experiences from the Euro-Asian region. Turk. J Gastroenterol 31 (2020): 148-155.
- Morgan RL, Baak B, Smith BD, et al. Eradication of hepatitis C virus infection and the development of hepatocellular carcinoma. Ann Inter Med 158 (2013): 329-337.
- Shiomai A, de Jong YP, Rice CM. Virus associated malignancies: the rile of viral hepatitis in hepatocellular carcinoma. Semin Cancer Biol 26 (2014): 78-88.
- Sartorius K, Makarova J, Sartorius B, et al. The regulatory role of microRNA in hepatitis-B virus-associated hepatocellular carcinoma (HBV-HCC) pathogenesis. Cells 8 (2019): 1504.
- Shen S, Lin Y, Yuan X, et al. Biomarker microRNAs for diagnosis, prognosis and treatment of hepatocellular carcinoma: a functional survey and comparison. Sci Rep 6 (2016): 38311.
- Fiorino S, Bacchi-Reggiani ML, Visani M, et al. MicroRNAs as possible biomarkers for diagnosis and prognosis of hepatitis B- and C-related-hepatocellular-carcinoma. World J Gastroenterol 22 (2016): 3907-3936.
- Zhang Q, Xu M, Qu Y, et al. Analysis of the differential expression of circulating microRNAs during the progression of hepatic fibrosis in patients with chronic hepatitis B virus infection. Mol Med Rep 12 (2015): 5647-5654.
- Khatun M, Ray RB. Mechanisms underlying hepatitis C virus-associated hepatic fibrosis. Cells 8 (2019): 1249.
- Fujii YR. The quantum language of the microRNA gene and anti-cancer: with a dynamic computer simulation of human breast cancer drug resistance. Integr Mol Med 5 (2018): 1-13.
- Fujii YR. Cancer simulation from stage minus one by quantum microRNA language: lung, colorectal and pancreatic cancers. Med One 4 (2019): e190023.
- Fujii YR. Quantum microRNA network analysis in gastric and esophageal cancers: xenotropic plant microRNAs cure from cancerous paradox via Helicobacter pylori infection. Gastroentrrol Hepatol Endosc 4 (2019): 1-18.
- Fujii YR. The RNA gene information: retroelement-microRNA entangling as the RNA quantum code. Methods Mol Biol 936 (2013): 47-67.
- Yoshikawa M, Osone T, Fujii YR. MicroRNA memory I: the positive correlation between synergistic effects of microRNAs in cancer and a novel quantum scoring system. J Adv Med Phar Sci 5 (2016): 1-16.
- Osone T, Yoshikawa M, Fujii YR. MicroRNA memory II: a novel scoring integration model for prediction of human disease by microRNA/microRNA quantum multi-interaction. J Adv Med Phar Sci 5 (2016): 1-18.
- Fujii YR. The microRNA 2000: from HIV-1 to healthcare. Scientific Research Publishing Inc (2017).
- Fujii YR. Quantum language of microRNA: application for new cancer therapeutic targets. Methods Mol Biol 1733 (2018): 145-157.
- Yoshikawa M, Fujii YR. Human ribosomal RNA-derived resident microRNAs as the transmitter of information upon the cytoplasmic cancer stress. Biomed Res Int 2016 (2016): 7562085.
- Chahal J, Gebert LFR, Gan HH, et al. miR-122 and Ago interactions with the HCV genome alter the structure of the viral 5’ terminus. Nucleic Acids Res 47 (2019): 5307-5324.
- Xu K, Lin J, Zandi R, et al. MicroRNA-mediated target mRNA cleavage and 3’-uridylation in human cells. Sci Rep 6 (2016): 30242.
- Yang X, Li H, Sun H, et al. Hepatitis B virus-encoded microRNA controls viral replication. J Virol 91 (2017): e01919-16.
- Li LM, Hu ZB, Zhou ZX, et al. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res 70 (2010): 9798-9807.
- Jin X, Cai C, Qiu Y. Diagnostic value of circulating microRNAs in Hepatitis B virus-related hepatocellular carcinoma: a systematic review and meta-analysis. J Cancer 10 (2019): 4758-4764.
- Yamada H, Suzuki K, Ichino N, et al. Association between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin Chim Acta 424 (2013): 99-103.
- Guicciardi ME, Gores GJ. Apoptosis: a mechanism of acute and chronic liver injury. Gut 54 (2005): 1024-1033.
- Terradillos O, de La Coste A, Pollicino T, et al. The hepatitis B virus X protein abrogates Bcl-2-mediated protection against Fas apoptosis in the liver. Oncogene 21 (2002): 377-386.
- Pan J, Duan IX, Sun BS, et al. Hepatitis B virus X protein protects against anti-Fas-mediated apoptosis in human liver cells by inducing NF-kB. J Gen Virol 82 (2001): 171-182.
- Li J, He J, Fu Y, et al. Hepatitis B virus X protein inhibits apoptosis by modulating endoplasmic reticulum stress response. Oncogene 8 (2017): 96027-96034.
- Kong F, You H, Zhao J, et al. The enhanced expression of death receptor 5 (DR5) mediated by HBV X protein through NF-kappaB pathway is associated with cell apoptosis induced by (TNF-α related apoptosis inducing ligand) TRAIL in hepatoma cells. Virol J 12 (2015): 192.
- Richman RA, Claus TH, Pilkis SJ, et al. Hormonal stimulation of DNA synthesis in primary culture of adult rat hepatocyte. Proc Natl Acad Sci USA 73 (1976): 3589-3593.
- Huang P, Xu X, Wang L, et al. The role of EGF-EGFR signaling pathway in hepatocellular carcinoma inflammatory microenvironment. J Cell Mol Med 18 (2014): 218-230.
- Zhao X, Sun L, Mu T, et al. An HBV-encoded miRNA activates innate immunity to restrict HBV replication. J Mol Cell Biol 2019 (2019): 104.
- Chen YJ, Chien PH, Chen WS, et al. Hepatitis B virus-encoded X protein downregulates EGFR expression via inducing microRNA-7 in hepatocellular carcinoma cells. Evid. Based Complement. Alternat Med 2013 (2013): 682380.
- Fuchs BC, Hoshida Y, Fujii T, et al. EGFR inhibition attenuates liver fibrosis and development of hepatocellular carcinoma. Hepatology 59 (2014): 1577-1590.
- Iwamoto M, Saso W, Sugiyama R, et al. Epidermal growth factor receptor is a host-entry cofactor triggering hepatitis B virus internalization. Proc Natl Acad Sci USA 116 (2018): 8487-8492.
- Wu J, Zhang W, Xu A, et al. Association of epidermal growth factor and epidermal growth factor receptor polymorphisms with the risk of hepatitis B virus-related hepatocellular carcinoma in the population of north China. Genet Test Mol Biomarkers 17 (2013): 595-600.
- Tsujinaka S, Soda K, Kano Y, et al. Spermine accelerates hypoxia-initiated cancer cell migration. Int. J. Oncol 38 (2011): 305-312.
- Svensson KJ, Welch JE, Kucharzewska P, et al. Hypoxia-mediated induction of the polyamine system provides opportunities for tumor growth inhibition by combined targeting of vascular endothelial growth factor and ornithine decarboxylase. Cancer Res 68 (2008): 9291-9301.
- Kim YH, Coon A, Baker AF, et al. Antitumor agent PX-2 inhibits HIF-1α protein levels through an Nrf2/PMF-1-mediated increase in spermidine/spermine acetyl transferase. Cancer Chemother Pharmacol 68 (2011): 405-413.
- Fuhrmann V, Jäger B, Zubkova A, et al. Hypoxic hepatitis-epidemiology, pathophysiology and clinical management. Wien Klin Wochenschr 122 (2010): 129-139.
- Jeong W, Jung Y, Kim H, et al. Thioredoxin-related protein 14, a new member of the thioredoxin family with disulfide reductase activity: implication in the redox regulation of TNF-alpha signaling. Free Radic Biol Med 47 (2009): 1294-1303.
- Nogami S, Satoh S, Nakano M, et al. Taxilin; a novel syntaxin-binding protein that is involved in Ca2+-dependent endocytosis in neuroendocrine cells. Genes Cells 8 (2003): 17-28.
- Horii Y, Nogami S, Kawano Y, et al. Interaction of α-taxilin localized on interacellular components with the microtubule cytoskeleton. Cell Struct Func, 37 (2012): 111-126.
- Hoffmann J, Boehm C, Himmeisbach K, et al. Identification of α-taxilin as an essential factor for the life cycle of hepatitis B virus. J Hepatol 59 (2013): 934-941.
- Ford R, Tamayo A, Martin B, et al. Identification of B-cell growth factors (interleukin-14; high molecular weight-B-cell growth factors) in effusion fluids from patients with aggressive B-cell lymphomas. Blood 86 (1995): 283-293.
- Lou X, Hou Y, Liang D. Effects of hepatitis B virus X protein on human T cell cytokines. Can J Microbiol 59 (2013): 620-626.
- Henke JI, Goergen D, Zheng J, et al. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J 27 (2008): 3300-3310.
- Sedano CD, Sarnow P. Interaction of host cell microRNAs with the HCV RNA genome during infection of liver cells. Semin Liver Dis 35 (2015): 75-80.
- Cheng JC, Yeh YJ, Tseng CP, et al. Let-7b is a novel regulator of hepatitis C virus replication. Cell Mol Life Sci 69 (2012): 2621-2633.
- Matsuura K, Aizawa N, Enomoto H, et al. Circulating let-7 levels in serum correlate with the severity of hepatic fibrosis in chronic hepatitis C. Open Forum Infect Dis 268 (2018): 1-6.
- Nadeem AED, Thomas P, Ulf ML, et al. Cell culture-derived HCV cannot infect synovial fibroblast. Sci Rep 5 (2015): 18043.
- Ono C, Fukuhara T, Motooka D, et al. Characterization of miR-122-independent propagation of HCV. PLoS Pathog 13 (2017): e1006374.
- Pang PS, Pham EA, Elazar M, et al. Structural map of a microRNA-122: HCV complex. J Virol 86 (2012): 1250-1254.
- Bernier A, Sagan SM. Beyond sites 1 and 2, miR-122 target sites in the HCV genome have negligible contributions to HCV RNA accumulation in cell culture. J Gen Virol 100 (2019): 217-226.
- Fujii YR. RNA wave for the HIV therapy: foods, stem cells and the RNA information gene. World J AIDS 3 (2013): 131-146.
- Zhang S, Ouyang X, Jiang X, et al. Dysregulation serum microRNA expression profile and potential biomarkers in hepatitis C virus-infected patients. Int J Med Sci 12 (2015): 590-598.
- Seeff LB. Natural history of hepatitis C. Hepatology 26 (1997): 21S-28S.
- Xie KL, Zhang YC, Liu J, et al. MicroRNAs associated with HBV and infection and HBV-related HCC. Theranostics 4 (2014): 1176-1192.
- Gupta P, Carins MJ, Saksena NK. Regulation of gene expression by microRNA in HCV infection and HCV-mediated hepatocellular carcinoma. Virol J 11 (2014): 64.
- Li Q, Zhang YY, Chiu S, et al. Integrative functional genomics of hepatitis C virus infection identifies host dependencies in complete viral replication cycle. PLoS Pathog 10 (2014): e1004163.
- Qadri I, Choudhury M, Rahman SM, et al. Increased phophoenolpyruvate carboxykinase gene expression and steatosis during hepatitis C virus subgenome replication. J Biol Chem 287 (2012): 37340-37351.
- Yao W, Cai H, Li X, et al. Endoplasmic reticulum stress links hepatitis C virus RNA replication to wild-type PGC-1α/liver-specific PGC-1α upregulation. J Virol 88 (2014): 8361-8374.
- Argemi J, Latasa MU, Atkinson SR, et al. Defective HNF4alpha-dependent gene expression as a driver of hepatocellular failure in alcoholic hepatitis. Nat Commun 10 (2019): 3126.
- Li X, Jiang H, Qu L, et al. Hepatocyte nuclear factor 4α and downstream secreted phospholipase A2 GXIIB regulate production of infectious hepatitis C virus. J Virol 88 (2014): 612-629.
- Fahmy AM, Labonté P. The autophagy elongation complex (ATG5-12/16L1) positively regulates HCV replication and is required for wild-type membranous web formation. Sci. Rep 7 (2017): 40351.
- Tsai PH, Wang ML, Chang JH, et al. Dual delivery of HNF4α and cisplatin by mesoporous silica nanoparticles inhibits cancer pluripotency and tumorigenicity in hepatoma-derived CD133-expressing stem cells. ACS Appl Mater Interfaces 11 (2019): 19808-19818.
- Aydin Y, Kurt R, Song K, et al. Hepatic stress response in HCV infection promotes STAT3-mediated inhibition of HNF4A-miR-122 feedback loop in liver fibrosis and cancer progression. Cancers 11 (2019): 1407.
- Bao L, Chandra PK, Moroz K, et al. Impaired autophagy response in human hepatocellular carcinoma. Exp Mol Pathol 96 (2014): 149-154.
- Sasaki R, Sur S, Cheng Q, et al. Repression of microRNA-30e by hepatitis C virus enhances fatty acid synthesis. Hepatol Commun 3 (2019): 943-953.
- Jin T, Guo F, Wang Y, et al. High-resolution crystal structure of human Dim2/TXNL4B. Acta Cryst F69 (2013): 223-227.
- Turunen J, Niemelä EH, Verma B, et al. The significant other: splicing by the minor spliceosome. WIREs RNA 4 (2013): 61-76.
- Paek KY, Kim CS, Park SM, et al. RNA-binding protein hnRNP D modulates internal ribosome entry site-dependent translation of hepatitis C virus RNA. J Virol 82 (2008): 12082-12093.
- Webster NJ. Alternative RNA splicing in the pathogenesis of liver disease. Front Endocrinol 8 (2017): 133.
- Kubo M, Ihn H, Kuwana M, et al. Anti-U5 snRNP antibody as a possible serological marker for scleroderma-polymyositis overlap. Rheumatology 41 (2002): 531-534.
- Devadas K, Biswas S, Haleyurgirisetty M, et al. Identification of host micro RNAs that differentiate HIV-1 and HIV-2 infection using genome expression profiling techniques. Viruses 8 (2016): E121.
- Biswas S, Haleyurgirisetty M, Lee S, et al. Development and validation of plasma miRNA biomarker signature panel for the detection of early HIV-1 infection. EBioMedicine 43 (2019): 307-316.
- Su B, Fu Y, Liu Y, et al. Potential application of microRNA profiling to the diagnosis and prognosis of HIV-1 infection. Front MicroBiol 9 (2018): 385.
- Pauls E, Ruiz A, Badia R, et al. Cell cycle control and HIV-1 susceptibility are linked by CDK6-dependent CDK2 phosphorylation of SAMHD1 in myeloid and lymphoid cells. J Immunol 193 (2014): 1988-1997.
- Zack JA. The role of the cell cycle in HIV-1 infection. Adv Exp Med Biol 374 (1995): 27-31.
- Davy C, Doorbar J. G2/M cell cycle arrest in the life cycle of viruses. Virology 368 (2007): 219-226.
- Ruiz A, Pauls E, Badia R, et al. Cyclin D3-dependent control of the dNTP pool and HIV-1 replication in human macrophages. Cell Cycle 14 (2015): 1657-1665.
- Ammosova T, Berro R, Jerebtsova M, et al. Phosphorylation of HIV-1 Tat by CDK2 in HIV-1 transcription. Retrovirology 3 (2006): 78.
- Badia R, Pujantell M, Riveira-Muoñz E, et al. The G1/S specific cyclin D2 is a regulator of HIV-1 restriction in non-proiferating cells. PLoS Pathog 12 (2016): e1005829.
- Guendel I, Agbottah ET, Kehn-Hall K, et al. Inhibition of human immunodeficiency virus type-1 by cdk inhibitors. AIDS Res Ther 7 (2010): 7.
- Aillet F, Masutani H, Elbim C, et al. Human immunodeficiency virus induces a dual regulation of Bcl-2, resulting in persistent infection of CD4+ T- or monocyte cell lines. J Virol 72 (1998): 9698-9705.
- Yamamoto T, Omoto S, Mizuguchi M, et al. Double-stranded nef RNA interferes with human immunodeficiency virus type 1 replication. Microbiol Immunol 46 (2002): 809-817.
- Omoto S, Ito M, Tsutsumi Y, et al. HIV-1 nef suppression by virally encoded microRNA. Retrovirology 1 (2004): 44.
- Brisibe EA, Okada N, Mizukami H, et al. RNA interference: potentials for the prevention of HIV infections and the challenges ahead. Trends Biotechnol 21 (2003): 306-311.
- Gekonge B, Raymond AD, Yin X, et al. Retinoblastoma protein induction by HIV viremia or CCR5 in monocytes exposed to HIV-1 mediates protection from activation-induced apoptosis: ex vivo and in vitro study. J Leuko Biol 92 (2012): 397-405.
- Patro SC, Pal S, Bi Y, et al. Shift in monocyte apoptosis with increasing viral load and change in apoptosis-related ISG/Bcl2 family gene expression in chronically HIV-1-infected subjects. J Virol 89 (2015): 799-810.
- Mondai S, Farberov L, Herzig E, et al. HIV-1 infection increases microRNAs that inhibit Dicer1, HRB and HIV-EP2, thereby reducing viral replication. PLoS One 14 (2019): e0211111.
- Klockow LC, Sharifi HJ, Wen X, et al. The HIV-1 protein Vpr targets the endoribonuclease Dicer for proteasomal degeneration to boost macrophage infection. Virology 444 (2013): 191-202.
- Fujii YR. Symphony of AIDS: an miRNA-based therapy. In R. K. Gaur and J. J. Rossi, Eds., Regulation of Gene Expression by small RNAs. CRC Press (2009).
- Roy S, Benz F, Luedde T, et al. The role of miRNAs in the regulation of inflammatory processes during hepatofibrogenesis. Hepatobiliary Surg Nutr 4 (2015): 24-33.
- Lambrecht J, Verhulst S, Reynaert H, et al. The miRFIB-Score: a serological miRNA-based scoring algorithm for the diagnosis of significant liver fibrosis. Cells 8 (2019): 1003.
- Chen SC, Hu TH, Huang CC, et al. Hepatoma-derived growth factor/nucleolin axis as a novel oncogenic pathway in liver carcinogenesis. Oncotarget 6 (2015): 16253-16270.
- Guo Y, Xiao L, Sun L, et al. Wnt/beta-catenin signaling: a promising new target for fibrosis diseases. Physiol Res 61 (2012): 337-346.
- Ge WS, Wang YJ, Wu JX, et al. β-catenin is overexpressed in hepatic fibrosis and blockage of Wnt/β-catenin signaling inhibits hepatic stellate cell activation. Mol Med Rep 9 (2014): 2145-2151.
- Behari J. The Wnt/β-catenin signaling pathway in liver biology and disease. Expert Rev Gastroenterol Hepatol 4 (2010): 745-756.
- Ihara A, Koizumi H, Hashizume R, et al. Expression of epithelial cadherin and α- and β-catenins in nontumoral livers and hepatocellular carcinomas. Hepatology 23 (1996): 1441-1447.
- Verna EC. Management of advanced fibrosis in the context of hepatitis C virus infection. Top Antivir Med 25 (2017): 7-11.
- Zhou T, Sun Y, Li M, et al. Enhancer of zeste homolog 2-catalysed H3K27 trimethylation plays a key role in acute-on-chronic liver failure via TNF-mediated pathway. Cell Death Dis 9 (2018): 590.
- Tsou PS, Campbell P, Amin MA, et al. Inhibition of EZH2 prevents fibrosis and restores normal angiogenesis in scleroderma. Proc Natl Acad Sci USA 116 (2019): 3695-3702.
- Knipe RS, Tager AM, Liao JK. The Rho kinases: critical mediators of multiple profibrotic processes and rational targets for new therapies for pulmonary fibrosis. Pharmacol Rev 67 (2015): 103-116.
- Li M, Zhou W, Yuan R, et al. ROCK2 promotes HCC proliferation by CEBPD inhibition through phosphor-GSK3β/β-catenin signaling. FEBS Let 589 (2015): 1018-1025.
- Sun S, Li Y, Han S, et al. A comprehensive genome-wide profiling comparison between HBV and HCV infected hepatocellular carcinoma. BMC Med Genomics 12 (2019): 147.
- George A, Panda S, Kudmulwar D, et al. Hepatitis C virus NS5A binds to the mRNA cap-binding eukaryotic translation initiation 4F (eIF4F) complex and up-regulates host translation initiation machinery through eIF4E-binding protein 1 inactivation. J Biol Chem 287 (2012): 5042-5058.
- Zhang C, Huys A, Thibault PA, et al. Requirements of human Dicer and TRBP in microRNA-122 regulation of HCV translation and RNA abundance. Virology 433 (2012): 479-488.
- De Vito C, Riggi N, Cornaz S, et al. A TARBP2-dependent miRNA expression profile underlies cancer stem cell properties and provides candidate therapeutic reagents in Ewing sarcoma. Cancer Cell 21 (2012): 807-821.
- Lai HH, Li CW, Hong CC, et al. TARBP2-mediated destabilization of Nanog overcomes sorafenib resistance in hepatocellular carcinoma. Mol Oncol 13 (2019): 928-945.
- Kamal FA, Travers JG, Schafer AE, et al. G protein-coupled receptor-G-protein βγ-subunit signaling mediates renal dysfunction and fibrosis in heart failure. J Am Soc Nephrol 28 (2017): 197-208.
- Paudyal P, Xie Q, Vaddi PK, et al. Inhibition of G protein βγ signaling blocks prostate cancer progression and enhances the efficacy of paclitaxel. Oncotarget 8 (2017): 36067-36081.
- Zelsel MB, Baumert TF. Clinical development of hepatitis C virus host-targeting agents. Lancet 389 (2017): 674-675.
- Gebert LFR, Rebhan MAE, Crivelli SEM, et al. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res 42 (2014): 609-621.
- Drury RE, O’Connor D, Pollard AJ. The clinical application of microRNAs in infectious disease. Front Immunol 8 (2017): 1182.
- Zhou J, Yu L, Gao X, et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol 29 (2011): 4781-4788.
- Chen S, Chen H, Gao S, et al. Differential expression of plasma microRNA-125b in hepatitis B virus-related liver diseases and diagnostic potential for hepatitis B virus-induced hepatocellular carcinoma. Hepatol Res 47 (2017): 312-320.
- Xiong F, Ma H, Qu Y, et al. Profiles of serum miR-99a, let-7c and miR-125b in hepatitis B virus (HBV)-associated chronic hepatitis, liver cirrhosis and hepatocellular carcinoma. Int J Clin Exp Pathol 9 (2016): 7087-7095.
- Luo Y, Li W, Liao H. HMGA2 induces epithelial-to-mesenthymal transition in human hepatocellular carcinoma cells. Onc Let 5 (2013): 1353-1356.
- Zha Y, Yao Q, Liu JS, et al. Hepatitis B virus X protein promotes epithelial-mesenchymal-transition and metastasis in hepatocellular carcinoma cell line HCCLM3 by targeting HMGA2. Onc Let 16 (2018): 5709-5714.
- Li O, Li Z, Tang Q, et al. Long stress induced non-coding transcripts 5 (LSINCT5) promotes hepatocellular carcinoma progression through interaction with high-mobility group AT-hook 2 and miR-4516. Med Sci Monit 24 (2018): 8510-8523.
- Jiang H, Li Y, Li J, et al. Long noncoding RNA LSINCT5 promotes endometrial carcinoma cell proliferation, cell cycle, and invasion by promoting the WNT/β-catenin signaling pathway via HMGA2. Ther Adv Med Oncol 11 (2019): 1-13.
- Ou W, Lv J, Zou X, et al. Propofol inhibits hepatocellular carcinoma growth and invasion through the HMGA2-mediated Wnt/β-catenin pathway. Exp Ther Med 13 (2017): 2501-2506.
- He Q, Li W, Ren J, et al. ZEB2 inhibits HBV transcription and replication by targeting its core promoter. Oncotarget 7 (2016): 16003-16011.
- Gao HB, Gao FZ, Chen XF. MiRNA-1179 suppresses the metastasis of hepatocellular carcinoma by interacting with ZEB2. Eur Rev Med Pharmacol Sci 23 (2019): 5149-5157.
- Yeom S, Jeong H, Kim SS, et al. Hepatitis B virus X protein activates proteasomal activator 28 gamma expression via upregulation of P53 levels to stimulate virus replication. J Gen Virol 99 (2018): 655-666.
- Li J, Jin B, Wang T, et al. Serum microRNA expression profiling identifies serum biomarkers for HCV-related hepatocellular carcinoma. Cancer Biomark 26 (2019): 501-512.
- Shehab-Eldeen S, Nada A, Abou-Elela D, et al. Daignoatic performance of microRNA-122 and microRNA-224 in hepatitis C virus-induced hepatocellular carcinoma (HCC). Asian Pac J Cancer Prev 20 (2019): 2515-2522.
- Sun Q, Li J, Jin B, et al. Evaluation of miR-331-3p and miR-23b-3p as serum biomarkers for hepatitis C virus. Clin Res Hepatol Gastroenterol 44 (2019): 21-28.
- Weis A, Marquart L, Calvopina DA, et al. Serum mciroRNAs as biomarkers in hepatitis C: Preliminary evidence of a microRNA panel for the diagnosis of hepatocellular carcinoma. Int J Mol Sci 20 (2019): 864.
- Rahman MA, Kyriazanos ID, Ono T, et al. Impact of PTEN expression on the outcome of hepatitis C virus-positive cirrhotic hepatocellular carcinoma patients: possible relationship with cox II and inducible nitric oxide synthase. Int J Cancer 100 (2002): 152-157.
- Zhang Y, Li RQ, Feng XD, et al. Down-regulation of PTEN by HCV core protein through activating nuclear factor-kB. Int J Clin Exp Pathol 7 (2014): 7351-7359.
- Wu Q, Li Z, Mellor P, et al. The role of PTEN-HCV core interaction in hepatic C virus replication. Sci Rep 7 (2017): 3695.
- Goyal L, Muzumdar MD, Zhu AX. Targeting the HGF/c-MET pathway in hepatocellular carcinoma. Clin Cancer Res 19 (2013): 2310-2318.
- Zhang SZ, Pan FY, Xu JF, et al. Knockdown of c-Met by adenovirus-delivered small interfering RNA inhibits hepatocellular carcinoma growth in vitro and in vivo. Mol Cancer Ther 4 (2005): 1577-1584.
- You H, Ding W, Dang H, et al. c-Met represents a potential therapeutic target for personalized treatment in hepatocellular carcinoma. Hepatology 54 (2011): 879-889.
- Bang YJ, Su WC, Schuler M, et al. Phase 1 study of capmatinib in MET-positive solid tumor patients: dose escalation and expansion of selected cohorts. Cancer Sci 111 (2019): 536-547.
- Qin S, Chan SL, Sukeepaisarnjaroen W, et al. A phase II study of the efficacy and safty of the MET inhibitor capmatinib [INC280] in patients with advanced hepatocellular carcinoma. Ther Adv Med Oncol 11 (2019): 1-12.
- Fornari F, Ferracin M, Trerè D, et al. Circulating microRNAs, miR-939, miR-595, miR-519d and miR-494, identify cirrhotic patients with HCC. PLoS One 10 (2015): e0141448.
- Zhang Y, Li T, Qiu Y, et al. Serum microRNA panel for early diagnosis of the onset of hepatocellular carcinoma. Medicine 96 (2017): 2.
- Liu HN, Wu H, Chen YJ, et al. Serum microRNA signatures and metabolomics have high diagnostic value in hepatocellular carcinoma. Oncotarget 8 (2017): 108810-108824.
- Jiang Y, He J, Li Y, et al. The diagnostic value of microRNAs as a biomarker for hepatocellular carcinoma: a meta-analysis. BioMed Res Int 219 (2019): 517904.
- Nishina SI, Shirahata H, Nakanishi Y, et al. Restored expression of the tumor suppressor gene RUNX3 reduces cancer stem cells in hepatocellular carcinoma by suppressing Jagged 1-Notch signaling. Oncol Rep 26 (2011): 523-531.
- Shi BM, Lu W, Ji K, et al. Study on the value of serum miR-106b for early diagnosis of hepatocellular carcinoma. World J Gastroenterol 23 (2017): 3713-3720.
- Yen CS, Su ZR, Lee YP, et al. miR-106b promotes cancer progression in hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol 22 (2016): 5183-5192.
- Yau WL, Lam CSC, Ng L, et al. Over-expression of miR-106b promotes cell migration and metastasis in hepatocellular carcinoma by activating epithelial-mesenchymal transition process. PLoS One 8 (2013): e57882.
- Gu H, Gu S, Zhang X, et al. miR-106b-5p promotes aggressive progression of hepatocellular carcinoma via targeting RUNX3. Cancer Med 8 (2019): 6756-6767.
- Kanwal F, El-Serag HB, Ross D. Surveillance for hepatocellular carcinoma: can we focus on the mission? Clin Gastroenterol Hepatol 13 (2015): 805-807.
- Mikami S, Tateishi R, Hagiwara S, et al. Tumor markers are more useful in patients undergoing surveillance for hepatocellular carcinoma with unreliable results by ultrasonography. Hepatol Res 45 (2015): 4150422.
- Girardi E, López P, Pfeffer S. On the importance of host microRNAs during viral infection. Front. Genet 9 (2018): 439.
- Fujii Y, Higashi H, Ikuta K, et al. Specificities of human heterophilic Hanganutziu-Deicher (H-D) antigen-active glycosphingolipids. Mol Immunol 19 (1982): 87-94.
- Tashiro M, Fujii Y, Nakamura N, et al. Cell-mediated immunity induced in mice after vaccination with a protease activation mutant, TR-2 of Sendai virus. J Virol 62 (1987): 2490-2497.
- Ressel S, Rosca A, Gordon K, et al. Extracellular RNA in viral-host interactions: thinking outside the cell. WIREs RNA 10 (2019): e1535.
- Cullen BR. How do viruses avoid inhibition by endogenous cellular microRNAs? PLoS Pathog 9 (2013): e1003694.
- Fujii YR. Oncoviruses and pathogenic microRNAs in humans. Open Virol J 3 (2009): 37-51.
- Bai Y, Yao L, Wei T. Presumed asymptomatic carrier transmission of COVID-19. JAMA 323 (2020): 1406-1407.
Article Views: 850
Journal Statistics





Discover More: Recent Articles