Development of a Diagnostic Prototype for the Detection of Salmonella by the LAMP Method and Investigation of Diagnostic Value of this Prototype
Gulnur SERDAR1*, Oktay GENC2
1Samsun veterinary control institute, Turkey
2Ondokuz Mayİs University, Turkey
* Corresponding Author: Gulnur SERDAR, Samsun veterinary control institute, Turkey.
Received: 17 September 2024; Accepted: 23 September 2024; Published: 30 September 2024
Article Information
Citation: Gulnur SERDAR, Oktay GENC. Development of a Diagnostic Prototype for the Detection of Salmonella by the LAMP Method and Investigation of Diagnostic Value of this Prototype. Archives of Veterinary Science and Medicine 7 (2024): 27-38.
View / Download Pdf Share at FacebookAbstract
Salmonellosis is a contagious disease of humans and animals caused by Salmonella bacteria. The disease affects all animals. But young, pregnant, and nursing animals are more at risk. The main symptom is enteritis. But septicemia, abortion, arthritis, and respiratory diseases can also happen. The LAMP method stands out in the diagnosis of Salmonella. It is easy to use, gives quick results, and does not need an expert. LAMP relies on the isothermal strand-displacement activity of the Bsm (Bacillus subtilis) or Bst (Bacillus stereothermophilus) polymerase enzyme. The method is simple. The reaction components stay at the same temperature. All amplification and fixing occur in one step. In this study, the invasive A (invA) gene, which is widely used for identification of Salmonella at the genus level, was preferred. The LAMP reaction was done at 65ºC for 60 minutes. The results were seen by electrophoresis and with SYBR Green I dye. The LAMP test's detection limit was 3.66x102 cfu/ml for S. Enteritidis. The PCR and RT-PCR tests' limits were 3.66x104 cfu/ml and 3.66x103 cfu/ml. The specificity of the LAMP test was determined as 100%.
Keywords
<p>LAMP; Molecular detection; PCR; Salmonellosis</p>
Article Details
Introduction
Salmonellosis is an infectious disease in humans and animals caused by Salmonella bacteria. They usually cause subclinical infections. They also contaminate the environment by scattering with the feces of carrier animals. The disease can affect all types of pets. Young, pregnant, and nursing animals are most at risk. Salmonellosis can be diagnosed by isolating the agent from necropsy materials, feces, rectal swabs, environmental samples, and food [1]. various tests can definitively identify suspected Salmonella isolates. These include biochemical, serological, and molecular tests. Other methods like Polymerase Chain Reaction (PCR) and PCR based tests, such as MLST and MLVA with phage typing, can also detect the agent [1,2]. In addition, Loop Mediated Isothermal Amplification (LAMP) method has been developed as an alternative to PCR for pathogen detection in clinical samples and food matrices. These developments have directed attention to LAMP tests for rapid diagnosis in recent years [3]. The LAMP technique is a method characterized by its simplicity, in which the reaction components are subjected to isothermal conditions and all amplification and fixation are performed in a single step [4,5]. LAMP needs less special gear than traditional PCR. So, it is easy for labs in poor countries to use. The method is based on the isothermal strand-displacement activity of the Bacillus subtilis (Bsm) or Bacillus stereothermophilus (Bst) polymerase enzyme. This enzyme, when used with four target-specific primers, allows amplification of a specific region in a DNA template at one temperature. This process produces more product than an equivalent PCR [4,6]. In this study, it was aimed to detect diagnostic performance of Salmonella by LAMP method which is fast, easy and possible to use in field conditions instead of nucleic acid amplification based methods (PCR, RT-PCR).
Material and Methods
Materials
Reference Strains
Bacterial strains were used for the detection of sensitivity and specificity of the test in the study. The reference strains are Salmonella Thyphimirium (NCTC74) and Salmonella Enteritidis (NC4444) isolates. They were obtained from Ondokuz Mayıs University, Department of Microbiology. They were checked with Eschericia coli (ATCC35218), Yersinia pseudotuberculosis (ATCC29833), Pseudomonas aeruginosa (ATCC27853), Klebsiella pneumonia (NCTC13465), Enterococcus faecalis (ATCC29212), and Staphylococcus aureus (ATCC29213). S. Gallinarum and S. Pullorum field strains were obtained from Samsun Veterinary Control Institute Poultry Diseases Laboratory.
Primers
The invA gene was chosen for the genus level identification of Salmonella. Optimization studies were performed with the primers suggested by Yang et al. (2018) for the LAMP test. Primers designed by Yang et al.[3] are shown in Table 1.
|
Target / Reference |
Primer |
Sequence (5'-3') |
|
Salmonella spp. Yang et al. (2018) |
FIP |
FIP (F1c+F2): GCGCGGCATCCGCATCAATA-TCTGGATGGTATGCCCGG |
|
BIP |
BIP (F1c+F2): GCGAACGGCGAAGCGTACTG-TCGCACCGTCAAAGGAAC |
|
|
F3 |
F3: GAACGTGTCGCGGAAGTC |
|
|
B3 |
B3: CGGCAATAGCGTCACCTT |
|
|
LF |
LF (5’-3’): TCAAATCGGCATCAATACTCATCTG |
|
|
LB |
LB (5’-3’):AAAGGGAAAGCCAGCTTTACG |
F3: Forward inner primer; B3: Backward inner primer; FIP: Forward inner primer; BIP: Backward inner primer; LF: Loop forward primer; LB: Loop Backward primer
Table 1: Primers used for S. Enteritidis LAMP optimization.
External primers recommended by Yang et al. [3] were also used for both PCR and real-time PCR tests (Table 1).
qRT-PCR
iTaq universal SYBR®Green reaction mix (2x) (SİGMA, S9430-5ml) was used for qRT-PCR applications.
Conventional PCR
Conventional PCR mix was performed with Taq DNA polymerase (5u/µl), MgCl2 (25µM), 10XTaq (Mg-free) reaction buffer, dNTP (deoxynucleotide triphosphate) (2mM, 100 pmol Forward Primer and 100 pmol Reverse Primer mix.
Method
Revival of Bacterial Standard Strains
- Thyphimurium and S. Enteritidis reference strains were inoculated on blood agar to determine test detection limits in LAMP, PCR and real time PCR tests. Having been considered microscopic morphology and Gram staining of culture of S.Enteritidis and S. Typhimurium, dilutions were prepared according to McFarland standard.
DNA Extraction
For DNA extraction, Salmonella cultures were boiled in a dry block heater at 100°C for 10 minutes and then centrifuged at 10000 rpm for 10 minutes. The supernatant was collected and stored at -20°C for use in PCR, RT-PCR and LAMP.
Conventional PCR
Conventional PCR optimization was performed with primers designed by Yang et al. [3]. After optimization, the detection limit was determined. In addition, to detect the specificity of the test, DNA extracts of Eschericia coli (ATCC35218), Yersinia pseudotuberculosis (ATCC29833), Pseudomonas aeruginosa (ATCC27853), Klebsiella pneumonia (NCTC13465), Enterococcus faecalis (ATCC29212) and Staphylococcus aureus (ATCC29213) reference strains were prepared.
|
PCR Components |
Reaction Volume |
Final Concentration |
|
|
10X Standard Taq (Mg-free) Reaction Buffer |
2,5 µl |
Mix |
1X |
|
25 µM MgCl2 |
2 µl |
1,5-2 mM |
|
|
2 mM dNTPs |
2,5 µl |
200 µM |
|
|
100 pmol Forward (F) Primer |
1 µl |
20pmol |
|
|
100 pmol Reverse (R) Primer |
1 µl |
20pmol |
|
|
Taq DNA Polimerase 5u/µl |
0,5 µl |
1 unite |
|
|
Distilled water |
20 µl |
||
|
Template DNA |
3 µl |
< 1µg |
|
Table 2: Conventional PCR Components and Concentrations.
In order to prepare PCR ingredients, reaction mixtures were prepared as indicated in Table 2 and the test was performed on the following conditions in Table 3.
|
Stage |
Time |
Temperature |
Explanation |
|
Initial PCR activation |
95°C |
10 minute |
Taq DNA polymerase activation |
|
Cyclus |
|||
|
Denaturation |
95°C |
15 second |
Total 35 cycles |
|
Annealing |
57°C |
1 min 30 second |
|
|
Extending |
72°C |
1 min 40 second |
|
Table 3: Conventional PCR Protocol.
Analysis of PCR Products
The amplified products were examined by 1% agarose gel electrophoresis. The PCR product of each sample was stained with 1µl dye (6X Loading Dye; Thermo Scientific). The amplicons were visualized and photographed with the help of a UV-transilluminator (Fig. 1 and 2).
Real Time (RT)-PCR Application
Detection limits of target DNAs were determined by qRT-PCR. For this purpose, 10-fold dilutions of the reference Salmonella Enteritidis serotype were prepared by calculating the colony count and threshold cycle (Ct) value in the last dilution was determined as minimum cfu.
LAMP Optimization
Primers in the study were applied to the Yang et al. [3] (Table 1). The primer mix ratios used for preperation of the mastermix in LAMP method are indicated in Table 4. Primer concentration was optimized in a total of 25 ul volume. In the reaction, a primer mix for S. Enteritis was prepared to a total volume of 1.1 ul with the proportion respectively 1:2:8 (F3/B3, LF/LB, FIP/BIP) (Table 4).
|
Primers |
Volume (µl) |
Preparation |
|
FIP |
0.4 |
The specified volumes were taken from each primer (100µmol/l) and the LAMP was optimized so that the primer mixture was 1,1µl per reaction. |
|
BIP |
0.4 |
|
|
F3 |
0.05 |
|
|
B3 |
0.05 |
|
|
LoopF |
0.1 |
|
|
LoopB |
0.1 |
|
|
Total |
1.1µl |
Table 4: LAMP Primary Mix Ratios.
LAMP Mastermix Preparation
Preparation of mastermix for LAMP test reported by Parida et al. [7,8] were modified. The invA gene was chosen for the genus level identification of Salmonella. In order to determine the end-products obtained as a result of LAMP analysis, electrophoresis and fluorometric SYBR Green I dye were used. The components used in the LAMP reaction and their concentrations are shown in Table 5.
|
Components |
Volume (25µl/Reaction) |
|
1X DNA Buffer |
2.5µl |
|
100mM MgSO4 |
1.5µl (6µM) |
|
10mM dNTPs |
3.5µl (1.4µM) |
|
Primer mix |
2.5µl(1.6µM FIP/BIP,0.2µM F3/B3,0.4µM LF/LB) |
|
Betaine (5M) |
5 µl (0.5M) |
|
Bst DNA polymerase (8,000 U/ml) |
1µl |
|
Template DNA |
5µl |
|
Distiled water |
made up to 25µl with distilled water. |
Table 5: LAMP protocol, components and concentrations.
Results
Confirmation of Salmonella by Conventional PCR
PCR sensitivity for Salmonella reference strains (S. Typhimurium and S. Enteritidis) and clinical S. Pullorum and S. Gallinarum strains was 100% (Figure 1).
In conventional PCR test, the detection limit of Salmonella was determined as 3.66x104 seen in Fig 2. This value was determined as 5.37x104 for S. Typhimurium.
Figure 2: Determination of the detection limit of the Conventional PCR test in the identification of S. Enteritidis. (cfu/ml) M: marker 1: S. Enteritidis 3.66x107 2: S. Enteritidis 3.66x106 3: S. Enteritidis 3.66x105 4: S. Enteritidis 3.66x104 5: S. Enteritidis 3.66x103 6: S. Enteritidis 3.66x102 7: S. Enteritidis 3.66x101
Detection limit with Real Time (RT)-PCR
Test optimizations were performed with real-time PCR based on the cfu with reference strains (S. Typhimurium and S. Enteritidis). For this purpose, the colony count of the reference S. Enteritidis serotype was determined as 3.66x1010 cfu/ml by the colony counting method. DNAs were prepared from the cultures of which 10-fold dilutions were prepared, and target genes were amplified by qRT-PCR as template DNA. The concentration of 3.66x103 determined in the qRT-PCR shown in Table 7 was calculated as the detection limit of the test.
LAMP Optimization
In the study, LAMP optimization was performed by Yang et al. [3]. Mastermix prepared for LAMP optimization is based on the invA gene and S. Enteritidis was the strain amplified in a thermalcycler at 65°C for 60 minutes. Finally, the master mix was kept at 80°C for 2 minutes to stop the reaction. The results are shown in Fig 3.
After the amplification process, the master-mix was treated with 0,1% diluted SYBR Green I and the result in Fig 4 was obtained. With SYBR Green I, if the glow color remained orange, which was the original color of the dye, the result was considered negative. When the SYBR Green I dye changed to fluorescent green light, the result was interpreted as positive.
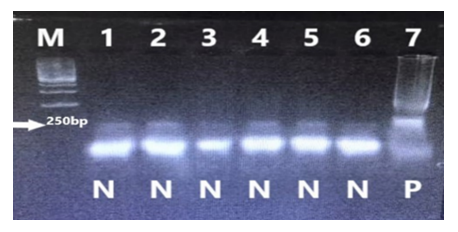
To determine the specificity of the LAMP test, 5 different microorganisms (Escherichia coli, Yersinia pseudotuberculosis, Pseudomonas aeruginosa, Klebsiella pneumonia, Enterococcus faecalis) were tested. The results are shown in Fig. 5 and 6.
Figure 6: Determination of the specificity of the LAMP test for S. Enteritidis identification with SYBR Green I. 1: No DNA 2: Escherichia coli (ATCC35218) 3: Yersinia pseudotuberculosis (ATCC29833) 4: Pseudomonas aeruginosa (ATCC27853) 5: Klebsiella pneumonia (NCTC13465) 6: Enterococcus faecalis (ATCC29212) 7: Salmonella Enteritidis (NC4444)
Analytical sensitivity of the LAMP test was evaluated by diluting of SE by log10. The sensitivity of the LAMP test was determined as 3.66x102 compared to PCR and RT-PCR. The results are shown in Figures 7 and 8.
Figure 7: Determination of the detection limit of S. Enteritidis by making a 1/10 subdilution with the LAMP test (cfu/ml) M: marker 1: S. Enteritidis 3.66x107 2:S. Enteritidis 3.66x106 3:S. Enteritidis 3.66x105 4: S. Enteritidis 3.66x104 5: S. Enteritidis 3.66x103 6: S. Enteritidis 3.66x102 7: S. Enteritidis 3.66x101
Figure 8: Determining the detection limit of the S. Enteritidis LAMP test result with SYBR Green I by making a 1/10 subdilution. 1: S. Enteritidis 3.66x107 2: S. Enteritidis 3.66x106 3: S. Enteritidis 3.66x105 4: S. Enteritidis 3.66x104 5: S. Enteritidis 3.66x103 6: S. Enteritidis 3.66x102 7: S. Enteritidis 3.66x101
In this study, the presence of Salmonella spp. was detected with specific DNA sequences of the invA gene by conventional PCR, RT-PCR and LAMP methods. Although the sensitivity and specificity of the test were evaluated with a limited number of reference strains, the presence of Salmonella spp. invA gene was detected with high specificity and sensitivity.
Discussion
Salmonellosis is a zoonotic infection that causes intestinal and extraintestinal infections in many animal species, including poultry, wild and mammals, reptiles, and cold-blooded animals. For the direct diagnosis of infection, molecular methods such as PCR, RT-PCR, PCR-ELISA are used together or independently with culture, which is the gold standard method, to increase the accuracy of diagnosis. LAMP is an innovative gene amplification technique alternative to PCR, consisting of the use of 4-6 different primers at constant temperature (isothermal reaction), with the help of the strand displacement polymerase enzyme [9]. LAMP for the diagnosis of salmonellosis was first used by Notomi et al. [4], and in subsequent periods, many studies have been carried out in various fields for the detection of microorganisms with different applications such as qLAMP, RT-LAMP, multiplex LAMP [10]. In this research, studies were carried out based on the invA gene. It has been reported in various studies and the detection limit of the test was >2.2-4.8-6.0 cfu/ml depending on bacterial concentration and 13.5fg-1pg-1.4pg/ul depending on DNA concentration [11-14]. In this study, the detection limit of the test was determined as 102 cfu/ml. Fig. 7 shows that this unit is 3.66x102 cfu/ml for S. Enteritidis. These results show that the detection limit is similar or even more sensitive than other studies. In conventional PCR and Real time PCR comparisons, it was determined that 3.66x104 cfu/ml and 3.66x103 cfu/ml were detected, respectively. These results show that LAMP is more sensitive than other methods. Similar results were obtained in both conventional and qRT-PCR studies. However, detection limits may vary under field conditions [12-16]. The specificity of the test can be determined by measuring the test results in cases where the material contaminated with other than Salmonella strains or serotypes of microorganisms and found in similar environments. LAMP results showed that the specificity of the test was similar to conventional PCR. Hara-Kudo et al. [11], in a study in which they evaluated 39 Salmonella serotypes (32 Salmonella supsp. enterica, 7 Salmonella enterica subsp. arizonae) and 62 non-Salmonella bacterial strains, found that the sensitivity of the test was higher than the classical PCR test (>2.2 cfu/test tube), while its specificity was higher. They determined that it was similar to the classical PCR method.
In this study, 4 reference Salmonella strains and reference bacterial strains of E.coli, Y.pseudotuberculosis, P. aeruginosa, K.pneumonia and E.faecalis with higher cross-reaction potential were used to determine the specificity of the LAMP test. It was shown that all strains examined gave negative results with the LAMP test, both by electrophoresis (Fig. 7) and SYBR Green 1 dye (Fig. 8). In the research, sensitivity and specificity were determined as 100% to the mastermix composition. In this study, primers reported by Yang et al. [3] were obtained and studies were carried out. Test optimizations were carried out by preparing the mastermix composition at the concentrations specified in Table 6. LAMP test results were evaluated by electrophoresis and visually. With SYBR Green I, the reaction was interpreted as negative when the color was orange, which is the original color of the dye, and when the fluorescence gave green light, the reaction was interpreted as positive (Fig. 6). Mastermixes prepared for LAMP optimization based on the invA gene of S. Enteritidis were amplified in a thermalcycler at 65°C for 60 minutes. Finally, it was kept at 80°C for 2 minutes to stop the reaction. Although the test seems easy in principle, for LAMP optimization; there are many critical parameters such as primer selection, concentration, reaction components, reaction temperature and time, and detection method. In our study, the optimal temperature and duration were determined as 60 minutes at 65ºC. These results are consistent with Mei et al. [10] and Zhang et al. [17]. Some reported that they optimized the LAMP test for similar periods of time between 60-65ºC, up to 60 minutes, and some within 30 minutes [18]. The most important feature in completing these tests in a short time is the use of loop primers as the third pair of primers in addition to the 2 pairs of primers. In addition to these factors, many parameters such as primer selection, researchers' DNA extraction methods, time and temperature preference play an important role in the specificity of the test [19]. To detect Salmonella by LAMP method Ou et al. [12] tested contaminated and uncontaminated clinical samples to evaluate the sensitivity and specificity of the method. They applied LAMP with the optimal reaction temperature of 60-65°C and the optimal reaction time of 25-30 minutes, and as a result, they determined that the sensitivity of the method was 1.4 pg/μL with real-time fluorescence and visual observation. In another study, Wang et al. [14] developed the LAMP method for the rapid detection of Salmonella in foods and designed primers targeting the invA gene of Salmonella. As a result, they showed that Salmonella strains were successfully amplified with the developed LAMP assay and that this method was 1000 times more sensitive than conventional PCR with an analytical sensitivity of 8x102 copies per μL of sample. They directly visualized the results by adding calcein and MnCl2 to the LAMP reaction tube and observed that the amplified products positively turned green after a 2-minute incubation. In a study conducted in Turkey in 2018, Yüksel [16] compared the classical cultural method, LAMP and Real time PCR in the diagnosis of Salmonella spp. and Campylobacter spp. As a result, they determined that the LAMP test is compatible with these methods and can be an alternative to real time PCR thanks to its features such as being fast, specific, sensitive and low-cost. The importance of new methods, which are as reliable as the cultural methods considered to be the gold standard today, save both time and economy, and have high sensitivity and specificity, is gradually increasing. For this reason, the use of molecular methods such as classical conventional PCR, real time PCR and LAMP is gaining more importance. Considering the advantages of rapid amplification, simple use, and easy detection, LAMP has potential applications for clinical diagnosis as well as monitoring of infectious diseases, especially in developing countries where resources are more limited, without requiring advanced equipment or qualified personnel. In this study, specific DNA sequences of the invA gene and the presence of Salmonella spp. were tried to be detected by classical conventional PCR, RT-PCR and LAMP methods. Although the sensitivity and specificity of the test were evaluated with a limited number of reference strains in the this study, the presence of the Salmonella spp. invA gene was detected with high specificity and sensitivity.
References
- Salmonellosis.https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.09.08_ SALMONELLOSIS.pdf 2016.
- Pijnacker R, van den Beld M, van der Zwaluw K, et al. Comparing Multiple Locus Variable-Number Tandem Repeat Analyses with Whole-Genome Sequencing as Typing Method for Salmonella Enteritidis Surveillance in The Netherlands, January 2019 to March 2020. Microbiol Spectr 10 (2022): e01375-22.
- Yang Q, Domesle KJ, Ge B. Loop-Mediated Isothermal Amplification for Salmonella Detection in Food and Feed:Current Applications and Future Directions. Foodborne Pathogens and Disease 15 (2018): 309-331.
- Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Research 28 (2000): e63.
- Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Molecular Cell Probes 16 (2002a): 223-229.
- Nagamine K, Kuzuhara Y, Notomi T. Isolation of single-stranded DNA from loop-mediated isothermal amplification products. Biochemical and Biophysical Research Communities 290 (2002b): 1195-1198.
- Parida M, Posadas G, Inoue S, et al. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. Journal of Clinical Microbiology 42 (2004): 257-263.
- Parida M, Horioke K, Ishida H, et al. Rapid detection and differentiation of dengue virüs serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. Journal of Clinical Microbiology 43 (2005): 2895-2903.
- Montrasio C. Development of a Software Application for Loop-Mediated Isothermal Amplification (LAMP) Primer Design. UNIVERSITY OF MILAN - BICOCCA Department of Biotechnology and Biosciences PhD in Industrial Biotechnology. Doctoral Thesıs (2016).
- Mei X, Zhai X, Lei C, et al. Development and application of a visual loop-mediated isothermal amplification combined with lateral flow dipstick (LAMP-LFD) method for rapid detection of Salmonella strains in food samples. Science Direct 2019: 9-19.
- Hara-Kudo Y, Manabu Yoshino M, Kojima T, et al. Loop-mediated isothermal amplification for the rapid detection of Salmonella. FEMS Microbiology Letters 253 (2005): 155-161.
- Ou H, Wang Y, Gao J, et al. Rapid detection of Salmonella based on loop-mediated isothermal amplification. Annals of Palliative Medicine 10 (2021): 6850-6858.
- Tang T, Cheng A, Wang M, et al. Development and clinical verification of a loop-mediated isothermal amplification method for detection of Salmonella species in suspect infected ducks. Poultry Science 9 (2012): 979-986.
- Wang L, Shi L, Alam MJ, et al. Specific and rapid detection of foodborne Salmonella by loop-mediated isothermal amplification method. Food Research International 41 (2008): 69-74.
- Kreitlow A, Becker A, Schotte U, et al. Evaluation of different target genes for the detection of Salmonella sp. by loop-mediated isothermal amplification. Letters in Applied Microbiology 72 (2021): 420-426.
- Yüksel, M. Tavuk Etlerinde Salmonella spp. ve Campylobacter spp.’nin Standart Kültürel Yöntem, İlmiğe Dayalı İzotermal Amplifikasyon (LAMP) Ve Real-Tıme PCR İle Belirlenmesi Ve Doğrulanması. Basılmamış Doktora Tezi. Atatürk Üniversitesi Fen Bilimleri Enstitüsü Gıda Mühendisliği Anabilim Dalı, Erzurum (2018).
- Zhang L, Shi Y, Chen C, et al. Visual and rapid detection of Klebsiella pneumoniae by magnetic immunocapture-loop-mediated isothermal amplification assay. Jundishapur J. Microbiol 12 (2019): e90016.
- Tomlinson J. In-Field Diagnostics Using Loop-Mediated Isothermal Amplification. Phytoplasma 938 (2019): 291-300.
- Soraka M, Wasowicz B, Rymaszewska A. Loop-Mediated Isothermal Amplification (LAMP): The Better Sibling of PCR? Cells 10 (2021): 1931.








 Impact Factor: * 1.1
Impact Factor: * 1.1 Acceptance Rate: 80.20%
Acceptance Rate: 80.20%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 