Maternal Blood Lipid Biomarkers of Oligodendrocyte Pathology to Predict Fetal Alcohol Spectrum Disorders
Nune Darbinian1,*, Emily C Sparks1, Armine Darbinyan2, Nana Merabova1,3, Gabriel Tatevosian1, Ekaterina Vadachkoria1, Huaqing Zhao4, Shohreh Amini5, Laura Goetzl6, Michael E Selzer1,7,*
1Center for Neural Repair and Rehabilitation (Shriners Hospitals Pediatric Research Center), Lewis Katz School of Medicine, Temple University, Philadelphia, PA 19140, USA
2Department of Pathology, Yale University School of Medicine, New Haven, CT 06520, USA
3Medical College of Wisconsin-Prevea Health, Green Bay, WI 54304, USA
4Center for Biostatistics and Epidemiology, Department of Biomedical Education and Data Science, Lewis Katz School of Medicine at Temple University, Philadelphia, PA 19140, USA
5Department of Biology, College of Science and Technology, Temple University, Philadelphia, PA 19122, USA
6Department of Obstetrics & Gynecology, University of Texas, Houston, TX 77030, USA
7Department of Neurology, Lewis Katz School of Medicine at Temple University, Philadelphia, PA 19140 USA
*Corresponding Author’s: Dr. Michael Selzer, Dr. Nune Darbinian, Shriners Hospitals Pediatric Research Center,
Lewis Katz School of Medicine, Temple University, Room 682 MERB, 3500 N. Broad Street, Philadelphia, PA 19140, USA.
Received: 16 March 2023; Accepted: 20 March 2023; Published: 02 May 2023
Article Information
Citation:
Nune Darbinian, Emily C. Sparks, Armine Darbinyan, Nana Merabova, Gabriel Tatevosian, Ekaterina Vadachkoria, Huaqing Zhao, Shohreh Amini, Laura Goetzl, Michael E. Selzer. Maternal Blood Lipid Biomarkers of Oligodendrocyte Pathology to Predict Fetal Alcohol Spectrum Disorders. Obstetrics and Gynecology Research. 6 (2023): 127-138
View / Download Pdf Share at FacebookAbstract
Introduction:
Up to 9.9% of children have fetal alcohol spectrum disorders (FASD), the most frequent cause of intellectual disability in the US. FASD may involve abnormal brain development, including dysmyelination, suggesting abnormal development of oligodendrocytes (OLs), which make myelin and are rich in lipids. Indeed, low serum levels of omega-3 fatty acids (ω-3) have been reported in FASD. Free fatty acids bind to specific receptors (FFARs). We have isolated cell type-specific fetal brain-derived exosomes (FB-E) from maternal blood and sampled their contents to search for lipid-related biomarkers that predict FASD.
Methods:
Blood samples were collected from two groups of pregnant women: 1) those who consumed EtOH during pregnancy, and 2) non- EtOH using controls, under an IRB-approved protocol. Serum and OLderived exosomes (OL-Es) were used to assay myelin basic protein (MBP) and FFAR by ELISA and droplet digital PCR (ddPCR), respectively.
Results:
FFAR and MBP proteins were downregulated in the EtOH group compared to controls, and this difference was greatest in OL-Es from maternal blood compared maternal serum.
Conclusion:
MBP and FFAR levels were reduced in OL-Es from EtOHconsuming pregnant women. The data suggest potential therapeutic targets to predict which children are at risk for developing FASD and reduce dysmyelination in developing.
Keywords
<p>Biomarkers; Brain Development; FASD; Exosomes; Fatty Acid Receptors; Lipids; Oligodendrocytes</p>
Article Details
1. Introduction
There is evidence for a deficit of fatty acids in the blood of patients with FASD. Whether this is due to a genomic abnormality, or to diet or other environmental factor is not known. Nor is it known whether this is the only lipidomic abnormality, or even the most important one.
FASD and Oligodendrocyte (OL) Injury
Fetal exposure to ethanol (EtOH) during pregnancy is the leading cause of preventable cognitive impairment. Alterations of white matter integrity and subsequent white matter structural deficits are consistent findings in FASD, but knowledge regarding the molecular mechanisms underlying these abnormalities is incomplete. Ols are the last cells generated during development, with maturation, myelination and expression of MBP beginning during the second trimester, at 20 weeks gestational age (GA) in humans and continues postnatally [1]. While the alcohol-induced apoptosis of OLs was shown in the fetal macaque brain [2,3,4,5] less is known regarding human in vivo effects. Recently, we demonstrated that EtOH exposure was associated with reduced expression of markers of mature OLs in fetal brain in the mid-second trimester [6].
Oligodendrocyte Marker Studies
MBP is the second most abundant protein in central nervous system myelin. Since the 1980s, MBP has been regarded as a marker of brain tissue injury in trauma and disease [7,6,8]. Thus, the concentration of MBP in peripheral blood reflects the severity of the brain injury and correlates with treatment outcome. The expression of genes related to lipid metabolism in PBMC was studied recently in 54 healthy subjects [9]. This finding supports the use of PBMCs as a model system for investigating the role of dietary w-3 and w-6 fatty acids on expression of genes related to lipid metabolism. Studies on MBP and FFAR may explain how fatty acids influence lipid metabolism at a molecular level in humans in FASD, CP and Scoliosis. Although FFARs are involved in the regulation of energy metabolism in many neurological disorders, it is not known how specific or prevalent these findings are in CP patients who develop scoliosis.
Oligodendrocyte Alterations and Fatty Acids Deficiency in FASD
Researchers link alcohol exposure and fatty acids deficiency directly with brain OL injury during the pregnancy. Among the most important molecular pathways to study in this regard are those that are involved in lipid metabolism, i.e., the fatty acids and w-3 fatty acids pathways, including insulin and IGF, because these pathways are strongly implicated in OL dysregulation [10,11,12,13,14,15]. But it is not known how alcohol and FFA interact in determining the OL differentiation and myelination in FASD.
Lipid Metabolism and w-3 Fatty Acid (ω-3) Deficiency in FASD
There is evidence for an inflammatory contribution to FASD, and for low intake of ω-3 or a protective effect of dietary supplementation with polyunsaturated fatty acids (PUFAs; e.g., ω-3). Most of the clinical studies did not actually measure serum lipid levels but inferred low levels from response to supplementation or because of poor dietary intake.
Lipids and their Receptors in Neurodegenerative Disorders
Free fatty acids (FFAs) cross the placenta [16] and are key components of lipid membranes in brain and muscle development [17,18,19,20]. The essential fatty acids (EFAs), particularly ω-3, are important for brain development during fetal and postnatal periods [21,22] and may be critical in the pathogenesis of not only FAS [23,24], but also ADHD [25] depression and anxiety disorders [26]. Many of the effects of FFAs are mediated by binding to specific receptors (FFARs), which are GPRs that play essential roles as nutritional components and signaling molecules. FFARs have been identified by the GPR deorphanization strategy, derived from the human genome database [27]. Several FFARs have been identified as critical components in physiological and pathological processes, e.g., metabolism, inflammation, type 2 diabetes/obesity, and emotional behavior [28]. FFARs are categorized according to the chain length of FFA ligands that activate them. FFAR1 (GPR40) and FFAR4 (GPR120) are activated by long-chain saturated and unsaturated fatty acids, while FFAR2 (GPR43) and FFAR3 (GPR41) are activated by SCFAs, mainly acetate, butyrate, and propionate. GPR84 is activated by medium-chain FFAs. FFARs act as sensors for food derived FFAs and digestion products, and are involved in the regulation of energy metabolism, mediated by the secretion of insulin hormone, and by inflammatory responses related to insulin resistance.
FFAs are metabolized and synthesized as energy substrates, therefore, FFARs have been targeted in therapeutic strategies for the treatment of metabolic disorders. Injured OLs can release exosomes and abnormal lipids into the blood and contribute to the pathogenesis of FASD. Considering the previous data [6,29], it can be that increased release of soluble factors may be involved in the dysregulation of OL and neuronal growth and survival. We previously reported that changes in differentiation and chemokine secretion by OLs are associated with activation of apoptotic signaling in differentiated into OL rat O2A cells and neurons [8,6].
OL-derived exosomes (OL-Es) to study MBP and FFAR
OLs are damaged in FASD, and either fail to develop, or undergo excessive apoptosis. Failure to repair the damaged OLs in FAS hampers myelination and also leads to accumulation of neuronal damage. Because axonal demyelination is so important in the pathophysiology of FAS, brain-derived OL-exosomes (OL-Es) are an ideal tool for investigating the OL damage and lipid metabolism affected in FAS and have great translational potential. Direct fetal brain tissue examination is essentially impossible in ongoing human pregnancies, and non-invasive examination of the fetal brain has been limited to expensive and technically challenging in-utero functional and imaging studies. Maternal plasma miRNAs have been used to predict infant outcomes, and may be useful to classify difficult-to-diagnose FASD subpopulations [30,31,32]. In vivo non-invasive methods were developed recently to isolate fetal brain-derived CNS exosomes from maternal blood to study fetal injury [33,34,35,36] In prior publications, we measured OL markers in brain [8,6] and exosomal proteins [33,34,35,36] using human ELISAs. This strategy was refined further by sorting with the OL marker MBP, to study OL-derived fetal biomarkers of OL alterations and FFA deficiency, to determine whether their membranes show the OL abnormalities or changes identified in brain [37].
Llipid abnormalities associate with maternal EtOH use and reflect the lipid content of maternal blood.
Lipids, including w-3, are involved in the development and metabolism of OLs, and thus to myelination [38,39]. Fatty acids and their receptors play important roles not only in the initiation of immune-mediated demyelination, but also in remyelination and repair of lesions [40]. w-3 deficiency has been reported in FAS, but also independently in demyelinating disorders. Because OL injury is one of features of FASD patients, the w-3 or other FFA deficiency in FAS might be a good target in the treatment of FAS. Circulating FFAs in serum are associated with many chronic diseases. In a survey of 61 FFAs from an ethnically diverse population of 826 healthy young adults [41], plasma concentrations of some major FFAs averaged 12.0-186.9 μmol/L for α-linolenic acid, and 7.2-237.5 μmol/L for docosahexaenoic acid (DHA). Males had significantly higher plasma concentrations of γ-linolenic acid, and lower concentrations of LA and DHA than females. The expression of genes related to lipid metabolism in peripheral blood mononuclear cells (PBMC) was studied recently in 54 healthy subjects [42]. Among 285 genes related to cholesterol and triglyceride metabolism, 161 were expressed in the PBMCs, depending on the plasma fatty acid levels. This finding supports the use of PBMCs for investigating the role of dietary w-3 and w-6 fatty acids on expression of genes related to lipid metabolism. The plasma SFA to PUFA ratio seems to be very important.
The increased release of soluble factors may be involved in the dysregulation of OL and neuronal growth and survival. Changes in differentiation and chemokine secretion by OLs are associated with activation of apoptotic signaling in differentiated into OL rat O2A cells and neurons [8].
The abnormalities in OL markers identified in fetal brain are concentrated in oligodendrocyte-derived exosomes (OL-Es). Not only mRNA [43,44,45] but even double-stranded DNA can be found in exosomes [46,47], and exosomes secreted by OLs [48] contain major myelin and stress-protective proteins [49] lipids [50] and miRNAs [51].
We already determined the effect of prenatal alcohol exposure on human fetal OL apoptosis [6,29], and here we show effects of EtOH on levels of OL markers in OL-Es. Our exploration of possible effects of FFAR on fetal development is novel, since there have been only few human studies of this type.
2. Methods
2.1. Clinical Samples
. A comparison was performed between EtOH-using pregnant women and individually matched unexposed controls, in which none of the women used illicit drugs or CNS-active medications (n=20). The selection of cases and controls was driven by the availability of matching maternal blood samples and intact fetal brain tissues, and the availability of data for matching of sex, and gestational age (GA) [6,37,33,35]. Consenting mothers were enrolled between 11 and 21 weeks GA (Suppl. Table 1) under a protocol approved by our Institutional Review Board (IRB).
Cases (EtOH users) were matched to controls (non-users) by GA. All assays were performed in triplicates. Data from both sexes were combined.
|
Controls (n=20) |
EtOH Users (n=20) |
|
|
Age: maternal (years ±SD) |
24.15 ± 2.3 |
28.0 ± 2.7 |
|
GA (weeks ±SD) |
14.92 ± 1.58 |
15.22 ± 1.6 |
|
GA range (weeks) |
11.1 to 21 |
11.5 to 21 |
|
BMI: Body Mass Index (±SD) |
24.4 ± 3.3 |
26.1 ± 1.5 |
|
Hispanic Ethnicity (%) |
15 |
15 |
|
Race |
||
|
White (%) |
50 |
50 |
|
Black (%) |
50 |
50 |
|
Male Fetal Sex (%) |
50 |
50 |
Suppl. Table 1: Clinical characteristics of pregnant subjects used in all experiments.
Assessment of Alcohol Exposure in Pregnancy:
Maternal EtOH exposure was determined with a face-to-face questionnaire that also included questions regarding many types of drugs/medications used [6,37,29,33,35,36]. EtOH exposure was defined as current daily use, and samples were matched based on the last incidence of alcohol consumption, as indicated by the survey. Alcohol dose was calculated as the total number of drinks consumed in a week multiplied by the number of weeks of exposure. Alcohol exposure was assessed using a detailed questionnaire based on measures adapted from the NICHD PASS study [52]. EtOH consumption for each week since conception (2 weeks after last menstrual period) was self-reported using visual/photographic guides of different types of drinks to estimate actual EtOH dose. Women admitting to any EtOH use were classified as EtOH exposed [37].
Subject Recruitment:
Women reporting alcohol use (or no alcohol use) since conception were grouped in GA windows: 11-21 weeks (1st - late 2nd Trimester). GA was determined by a dating ultrasound performed immediately prior to recruitment; at the GAs planned, ultrasound can accurately determine GA ± 10 days (Spong et al, 2011). We collected blood sample of 20 EtOH-exposed and 20 case-matched control maternal blood samples.
2.2. RNA Preparation
RNA was isolated using the RNeasy kit (Qiagen, Valencia, CA) with on-column DNA digestion.
2.3. Droplet Digital PCR (ddPCR)
For absolute quantitation of mRNA copies, ddPCR was performed using the QX200 ddPCR system. Fifty ng of human fetal total RNA were used with the 1st Strand cDNA Synthesis Kit (Qiagen, Valencia, CA, USA). After reverse transcription, the cDNA (300 dilution) aliquots were added to BioRad master mix to conduct ddPCR (EvaGreen ddPCR Supermix, BioRad). The prepared ddPCR master mix for each sample (20-μl aliquots) was used for droplet formation. PCR conditions: Activation 95°C 5 min, PCR 45 cycles at 95°C 10 sec, 60°C 20 sec, 72°C 30 sec, melting curve (95–65°C), cool to 40°C 30 sec. The absolute quantity of DNA per sample (copies/µL) was calculated using QuantaSoft Analysis Pro Software (Bio-Rad) to analyze ddPCR data for technical errors (Poisson errors) (Darbinian et al, 2021a, 2023). The Poisson distribution relates the probability of a given number of events occurring independently in a sample when the average rate of occurrence is known and very low. Accurate Poisson analysis requires optimizing the ratio of the number of positive events (positive droplets) to the total number of independent events (the total number of droplets). A greater total number of droplets results in higher accuracy. With 20,000 droplets, the above ddPCR protocol yields a linear dynamic range of detection between 1 and 100,000 target mRNA copies/µL. The estimated error is negligible compared with other error sources, e.g., pipetting, sample processing, and biological variation. The ddPCR data were exported to Microsoft EXCEL for further statistical analysis.
2.4. Primers (IDT Inc.).
β-actin: S: 5’-CTACAATGAGCTGCG TGTGGC-3’,
AS: 5’-CAGGTCCAGACGCAGGATGGC-3’,
MBP: Myelin Basic Protein (human), S: 5’- ACTATCTCTTCCTCCCAGCTTAAAAA-3’,
AS: 5’-TCCGACTATAAATCGGCTCACA-3’,
2.5. Flow cytometry
Plasma samples were analyzed according to the previously published protocols with modifications [53,6,37]. In brief, cells were washed with cold phosphate-buffered saline (PBS) cocktail with 0.1% BSA and 1% protease inhibitors (Sigma). Cells were passed through 70 mM mesh, and 10,000 cells were placed onto 96-well plates and incubated with fluorescein isothiocyanate (FITC)-conjugated primary antibody for 1 hour. Myelin basic protein (MBP) was used as a late OL differentiation/myelination marker in the developing CNS, and FFAR was used as a marker of fatty acids receptors. Proportions were quantified using 5,000 cells and GUAVA FACS (Fluorescence-Activated Cell Sorting) software [6].
2.6. ELISA Quantification of Exosomal Proteins
MBP, FFAR and CD81 (American Research Products-Cusabio) were quantified by human-specific ELISAs according to suppliers’ directions.
2.7. Antibodies
Anti-human MBP (cat # AB5864), and anti-human FFAR were purchased from Millipore-Sigma (Bedford, MA USA).
2.8. Isolation of Fetal Brain-Derived Exosomes (FB-Es) or Fetal OL-Derived Exosomes (OL-Es) From Maternal Plasma, and ELISA Quantification of Exosomal Proteins
Human FB-Es were isolated as described previously [33,35,36,37]. Two hundred and fifty mL of plasma was incubated with 100 mL of thromboplastin-D (Fisher Scientific, Inc., Hanover Park, IL USA) and cocktails of protease and phosphatase inhibitors. After centrifugation, supernates were incubated with exosome precipitation solution (EXOQ; System Biosciences, Inc., Mountainview, CA), and the resultant suspensions centrifuged at 1,500g for 30 min at 4°C, and pellets resuspended in 400 mL of distilled water with protease and phosphatase inhibitor cocktail for immunochemical enrichment of exosomes. To isolate exosomes from fetal neural sources, total exosome suspensions were incubated for 90 min at 20°C with 50 mL of 3% bovine serum albumin (BSA) (Thermo Scientific, Inc., Waltham, MA) containing 2 mg of mouse monoclonal IgG1 antihuman contactin-2/TAG1 antibody (clone 372913, R&D Systems, Inc., Minneapolis, MN USA), or MBP antibody that had been biotinylated (EZLink sulfo-NHS-biotin System, Thermo Scientific, Inc., USA). Then, 10 mL of Streptavidin-Plus UltraLink resin (PierceThermo Scientific, Inc., Waltham, MA USA) in 40mL of 3% BSA were added, and the incubation continued for 60 min at 20°C. After centrifugation at 300g for 10 min at 4°C and removal of supernates, pellets were resuspended in 75 mL of 0.05 mol/L glycine-HCl (pH 3.0), incubated at 4°C for 10 min and recentrifuged at 4,000g for 10 min at 4°C. Each supernate was mixed in a new 1.5 mL Eppendorf tube with 5 mL of 1 mol/L Tris-HCl (pH 8.0) and 20 mL of 3% BSA, followed by addition of 0.40 mL of mammalian protein extraction reagent (M-PER; Thermo Scientific, Inc. Waltham, MA USA) containing protease and phosphatase inhibitors, prior to storage at -80°C. For exosome counts, immunoprecipitated pellets were resuspended in 0.25 mL of 0.05 mol/L glycine-HCl (pH = 3.0) at 4°C, with pH adjusted to 7.0 with 1 mol/L Tris-HCl (pH 8.6). Exosome suspensions were diluted 1:200 to permit counting in the range of 1-5 x 108/mL, with an NS500 nanoparticle tracking system (NanoSight, Amesbury, U.K.).
Isolation of OL-exosomes
Serum from EtOH used cases were precipitated in ExoQuick and OL-Es were isolated using biotinylated anti-MBP antibody. Nanoparticle-tracking analysis of exosomes revealed a mean particle diameter of 134 nm ± 46.6 nm and a mode is 109.8 nm. OL-E MBP protein levels were quantified by ELISA (normalized to exosome marker CD81). MBP, FFAR, and the tetraspanin exosome marker human CD81 (all from American Research Products, Waltham MA-Cusabio, USA), were quantified by human-specific ELISAs according to the supplier’s instructions. The mean value for all determinations of CD81 in each assay group was set at 1.00, and the relative values for each sample were used to normalize their recovery.
2.9. Statistical Analysis.
Statistical analysis was described previously [6,37]. In brief, analysis was performed using SPSS (IBM Corp., released 2017. IBM SPSS Statistics for Windows, Version 25.0. Armonk, NY). All data are represented as the mean ± standard error for all performed repetitions. Means were analyzed by a one-way ANOVA, with Bonferroni correction. Statistical significance was defined as p < 0.05. Sample numbers are indicated in the figure legends. Data from ddPCR, which measures absolute quantities of DNA per sample (copies/µL), were processed using QuantaSoft Analysis Pro Software (Bio-Rad) to analyze for technical errors (Poisson errors). Data from ddPCR were exported to Microsoft EXCEL for further statistical analysis.
2.10. Ethics. Human Subjects
Consenting mothers were enrolled between 11 - 21weeks gestation, under a protocol approved by our Institutional Review Board (IRB). This protocol involved no invasive procedures other than routine care. Maternal EtOH exposure was determined with a face-to-face questionnaire that also included questions regarding many types of drugs/medications used [33,35,36,6,37]. The questionnaire was adapted from that designed to identify and quantify maternal EtOH exposure in the NIH/NIAAA Prenatal Alcohol and SIDS and Stillbirth (PASS) study [52].
All procedures involving collection and processing of blood and tissues were done according to NIH Guidelines through a trained Study Coordinator. All investigators were trained annually to complete Citi Program - Human Subject training, Biohazard Waste Safety Training and Blood–Borne Pathogens Training, and all other required training.
- Eligibility Criteria: The blood samples were obtained according to NIH Guidelines through a trained Study Coordinator. Samples were collected regardless of sex, ethnic background, and race.
- Treatment Plan: Each patient was asked to sign a separate consent form for research on blood and tissue samples. Blood obtained was processed for collection of serum and plasma. No invasive procedures were performed on the mother, other than those used in her routine medical care. Fetal tissues were processed for RNA or protein isolation.
- Risk and Benefits: There are very small risks of loss of privacy as with any research study where protected health information is viewed. The samples were depersonalized before they were sent to the lab for analysis. There were no additional risks of blood sampling as this was only performed in patients with clinically indicated venous access. There was little anticipated risk from obtaining approximately 2-3 cc of blood, but a well-trained Study Coordinator collected all samples.
There was no direct benefit to the research subjects from participation, but there is significant potential benefit for the future FAS subjects and the general population. This research represents a reasonable opportunity to further the understanding, prevention, or alleviation of a serious problem affecting the health or welfare of FAS patients.
Informed Consent: Consent forms were maintained by the Study Coordinator and were not sent to the investigator with the samples. The de-identified log sheets and IRB protocol were sent by the Study Coordinator to Principal Investigator with each blood and tissue sample. This sheet contains an assigned accession number, the age, sex, ethnicity, and race of the patient. Except for an assigned accession number, no identification was kept on the blood and tissue samples.
Results
3.1. Down-regulation of the MBP protein expression by prenatal EtOH exposure
Our previous data in primary OL cultures and fetal neural exosomes from EtOH-exposed maternal blood, demonstrate an association between injury (EtOH exposure) and OL markers [6,54,37,33,35,36]. Therefore, we hypothesize that OL and other CNS damage that we saw in fetal brain, can also be studied using maternal blood, and that OL-Es will enable us to study the OL dysfunction that is correlated with fatty acid deficiency in FAS patients. If the abnormalities seen in fetal brain reflect disordered myelination, they might be more concentrated in OL-Es from plasma, compared to total/maternal plasma. Therefore, we isolated OL-Es from plasma to detect similar pattern of reduced MBP in OL-Es from EtOH-exposed plasma compared to control OL-Exs. Interestingly, OL-Es from plasma of pregnant women contained more exosomes compared to non-pregnant women, suggesting that majority of OL-Es from pregnant women had fetal origin (Figure 1A, 1B). MBP protein was downregulated in EtOH-exposed OL-Exs (Figure 1A) or in plasma (Figure 1B).
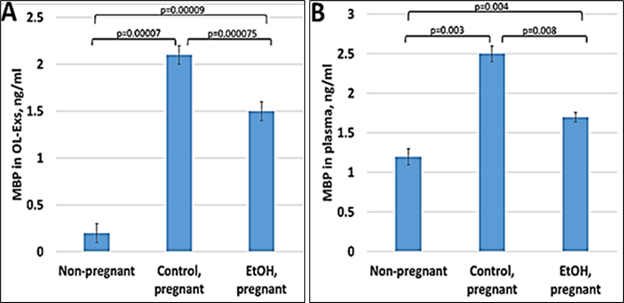
Figure 1: Downregulation of the MBP in EtOH cases. A. Downregulation of MBP protein levels in EtOH-exposed OL-Es or in plasma from the same cases (B). Plasma from 20 patients with or without EtOH, and 3 non-pregnant women, were studied by ELISA for MBP protein. Downregulation was greatest in the cases with EtOH exposure (graphs show means from triplicate assays +/- SD). Downregulation was statistically (p<0.05). For absolute quantitation of MBP by ELISA, values are shown in ng/mL (normalized to CD81).
3.2. Down-regulation of the OL marker, MBP mRNA by EtOH in OL-Es from patients with EtOH exposure.
OL-Exs and plasma from EtOH-exposed, were studied by ddPCR for MBP mRNA and downregulation of MBP was found in OL-Exs from EtOH group (Figure 2A). Similar pattern of MBP decrease was found for RNA levels in plasma (Figure 2B).
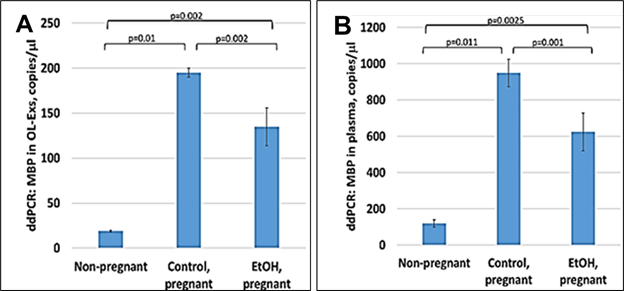
Figure 2: Downregulation of the MBP mRNA in EtOH-exposed cases. A. Downregulation of MBP mRNA levels in EtOH-exposed OL-Es or in plasma from the same cases (B). Plasma from 20 patients with or without EtOH, and 3 non-pregnant women, were studied by ddPCR for MBP mRNA. Downregulation was greatest in the cases with EtOH exposure (graphs show means from triplicate assays +/- SD). Downregulation was statistically significant (p<0.05). For absolute quantitation of MBP by ddPCR, values are shown in copies/ml.
3.3. Severe down-regulation of the FFAR by prenatal EtOH.
Because FFAR deficiency can be common among patients with FASD, the previously-reported average deficiencies of w-3 or other FFA in FASD might reflect only those changes for FFARs. To determine if this is so, in the same samples used for MBP studies, FFAR analysis were performed and downregulation of FFAR protein by maternal EtOH use was found in plasma for FFAR protein (Figure 3B and Figure 3C). FFAR was strongly downregulated in OL-Es, compared to plasma (Figure 3A). Thus, FFAR expression was affected in the cases with EtOH exposure, and EtOH cases had lower levels of FFAR (Figure 3A-C) than controls. Flow cytometry showed that the proportion of blood cells expressing FFAR protein was reduced (46% in controls vs 38% in EtOH cases), indicating that FFARs are affected most in EtOH cases.
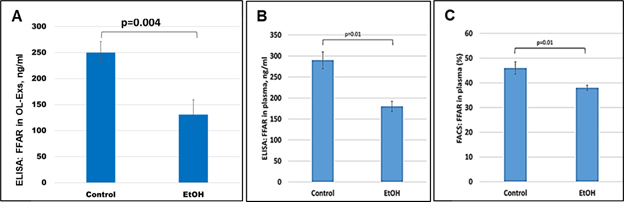
Figure 3: Severe downregulation of the FFAR by maternal EtOH use. OL-Es from the plasmas were measured for FFAR protein by ELISA (A). The same plasma from pregnant women who drank EtOH or did not use EtOH were measured (in triplicates) for FFAR by ELISA (B) and flow cytometry (C). Downregulation was greatest in the cases with EtOH exposure (graphs show means from triplicate assays +/- SD). Downregulation was statistically significant (p<0.05). For absolute quantitation of MBP by ELISA, values are shown in ng/mL (normalized to CD81).
3.4. Down-regulation of the FFAR mRNA by prenatal EtOH exposure in EtOH cases.
FFAR gene expression was studied by ddPCR. FFAR was downregulated in OL-Es (Figure 4) in the cases with EtOH exposure. Thus, EtOH cases had lower levels of FFAR than controls. The results of this study could allow us to predict which children are at risk for developing demyelination and FASD. This could lead to clinical trials of dietary fatty acid supplements or w-3 to prevent the development of FASD or other demyelinating disorders.
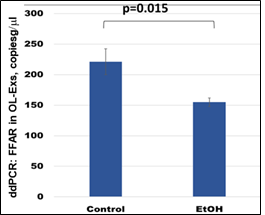
Figure 4: Downregulation of the FFAR mRNA in EtOH-exposed cases. OL-Es from the same plasmas were measured for FFAR mRNA by ddPCR. Downregulation was greatest in the cases with EtOH exposure (graphs show means from triplicate assays +/- SD). Downregulation was statistically significant (p<0.05). For absolute quantitation of FFAR in OL-Exs by ddPCR, values are shown in copies/ml.
4. Discussion
The number of children diagnosed with FASD is a small proportion of those prenatally exposed to alcohol, but the frequency with which FASD is diagnosed has been increasing. Moreover, exposure to doses lower than those that result in the most severe manifestation of prenatal exposure to alcohol, FASD, still carries risks for non-trivial developmental abnormalities. Unfortunately, FAS remains underdiagnosed. Recent random testing of schoolchildren by public health facilities resulted in the establishment of newly diagnosed cases of FASD, microcephaly, autism, and cerebral palsy, all associated with prenatal alcohol exposure. Thus, our findings may have implications that go beyond FASD. In clinical practice, the diagnosis of FASD is not made prenatally. The present study demonstrates that the blood exosomal test can diagnose FASD very early during fetal development, and is highly correlated with molecular markers (e.g., low levels of MBP mRNA and protein, and low levels of FFAR) in fetal cell type-specific exosomes. These can be isolated noninvasively from maternal blood samples, making them potentially valuable as a source of biomarkers for FASD. The biomarkers could have mechanistic relevance, and thus inform research aimed at devising therapeutic interventions, e.g., antiapoptotic drugs, or molecular approaches to enhance MBP or FFAR transcription, by targeting the affected biomarkers, or their promotors and suppressors.
We have isolated fetal cell type-specific exosomes from the blood of pregnant women who consumed EtOH, and these fetal OL-Es also showed low MBP expression, which is significant because FAS includes dysmyelination. Thus, we may be able to predict which at-risk fetuses will develop FASD, and thus be able to develop early neuroprotective interventions to prevent FASD and possibly other developmental disorders.
We have previously established primary cultures of neurons, OPC, and OL for use in molecular studies [55,8,54,56]. Our data in primary OL cultures and fetal neural exosomes from EtOH-exposed maternal blood [6,37,33,35,36] demonstrate an association between injury (EtOH exposure) and OL markers. Therefore, we hypothesize that OL and other CNS damage that we saw in fetal brain, can also be studied using maternal blood, and that OL-Es will enable us to study the OL dysfunction that is correlated with fatty acid deficiency in FAS patients. If the abnormalities seen in fetal brain reflect disordered myelination, they might be more concentrated in OL-Es from plasma, compared to total/maternal plasma. Therefore, we isolated OL-Es from plasma to detect similar pattern of reduced MBP in OL-Es from EtOH-exposed plasma compared to control OL-Es. Interestingly, OL-Es from plasma of pregnant women contained more exosomes compared to non-pregnant women, suggesting that majority of OL-Es from pregnant women had fetal origin (Figures 1 & 2).
The results of these studies have the potential to be extended to non-invasive diagnostic analyses of fetal development in future, that could predict the emergence of FAS, and might lead to clinical trials of readily available nutritional supplements to prevent the emergence of FASD prenatally, and possibly other neurological disorders, particularly those with prominent demyelination. A future long-term follow-up study could determine whether decreased lipid biomarkers predict eventual development of FASD in utero, and whether FASD can be prevented by early lipidomic analysis in blood and OL- or other neural cell-specific exosomes.
We demonstrated that the blood exosomal test can diagnose FASD by assaying molecular markers (e.g., low levels of MBP mRNA and protein, and low levels of FFAR) in cell type-specific exosomes. These can be isolated noninvasively from blood samples, making them potentially valuable as a source of biomarkers for FASD. The biomarkers could have mechanistic relevance, and thus inform research aimed at devising therapeutic interventions, e.g., antiapoptotic drugs, or molecular approaches to enhance MBP or FFAR transcription, by targeting the affected biomarkers, or their promotors and suppressors.
Since we determined that MBP and FFAR mRNA and protein levels were reduced in the plasma of EtOH cases, and the same pattern of downregulation was seen in OL-E isolated from the plasma, thus, we may be able to use MBP and FFAR abnormalities to predict which fetuses with EtOH exposure will develop FASD, and to institute dietary therapies, to prevent it.
As far as we know, these are the first EtOH-exposed cases that have been studied for OL marker, MBP, and FFAR using OL-derived exosomes. We must emphasize the potential value of OL-Es in the identification of lipid deficiencies in CNS because of its advantages of sensitivity, accuracy, and noninvasiveness.
Finally, our previous data in primary OL cultures and fetal neural exosomes from EtOH-exposed maternal blood, demonstrate an association between injury (EtOH exposure) and OL markers. Thus, OL and other CNS damage that we saw in fetal brain, can also be studied using maternal blood, and that OL-Es will enable us to study the OL dysfunction that is correlated with fatty acid deficiency in FAS patients. If the abnormalities seen in fetal brain reflect disordered myelination, they might be more concentrated in OL-Es from plasma, compared to total/maternal plasma. Therefore, we isolated OL-Es from plasma to detect similar pattern of reduced MBP in OL-Exs from EtOH-exposed plasma compared to control OL-Es. Importantly, OL-Es from plasma of pregnant women contained more exosomes compared to non-pregnant women, suggesting that majority of OL-Es from pregnant women had fetal origin.
5. Conclusions
Our studies investigated involvement of FFAR and MBP in EtOH cases. Recent publications demonstrate FFAR involvement in lipid metabolism, thus, the results of these studies have the potential to be extended to non-invasive diagnostic analyses of fetal development that could predict the emergence of FASD and might lead to clinical trials of readily available nutritional supplements to prevent the emergence of FASD prenatally. In addition, the role of w-3 in FASD can be also investigated in future studies.
Our data on blood from 20 patients with EtOH use, and 20 healthy controls, showing that gene and protein expression for MBP (Figures 1&2) or FFARs (Figures 3&4) was lower in cases with EtOH exposure than in controls. RNA was tested by ddPCR for OL marker MBP (Figures 2A, 2B), and FFAR gene expression (Figures 4). Levels of MBP gene expression were reduced in both EtOH OL-Exs (Figure 2A) and in EtOH serum (Figures 2B). Downregulation of MBP protein was greatest also in EtOH cases (Figures 1A, 1B). FFAR gene expression (Figure 4) and protein levels were reduced in EtOH cases (Figures 3A, 3B, 3C). Flow cytometry for FFAR showed that the proportion of blood cells expressing FFAR protein was reduced in EtOH serum, but most in EtOH OL-Exs cases, again indicating that FFARs are affected most in EtOH cases (Figure 3). Thus, EtOH group had lowest levels of OL MBP and FFAR.
We a) isolated OL-derived exosomes from the plasma samples, using OL-specific MBP, b) assayed MBP, a biomarker of OL injury, by ELISA in OL-Exs and plasma; c) confirmed MBP changes in OL-Es by flow cytometry; d) analyzed gene expression of MBP by ddPCR using OL-exosomal RNA and plasma RNA, e) assayed FFAR by ELISA and FACS in plasma from mothers in two clinical group, EtOH and unexposed controls. Maternal blood and OL-Es matched to fetal brain tissues studied previously [6,37].
These results might predict which fetuses are at risk for developing FASD, and lead to therapeutic trials of fatty acids and w-3 for FASD, and for other neurological disorders that involve demyelination. Number of samples in further studies will be increased and stratified by alcohol dose, gender, and race/ethnicity. The results of this study, using a unique human biobank of EtOH-exposed maternal blood, established under an NICHD-funded project with approved by Temple University IRB protocol, could lead to further analyses to determine whether the lipidomic abnormalities have genetic causes, and to clinical trials aimed at dietary correction of the lipid abnormalities (e.g., w-3 supplementation).
Ethics Statement
All procedures involving collection and processing of blood and tissues were done according to NIH Guidelines through a trained Study Coordinator. All investigators were trained annually to complete Citi Program - Human Subject training, Biohazard Waste Safety Training and Blood–Borne Pathogens Training, and all other required training. Written informed consent has been obtained from the parents of patient(s) for studies, and de-identified samples were used for this publication. Informed Consent forms were maintained by the Study Coordinator. The de-identified log sheets contain an assigned accession number, the age, sex, ethnicity, and race of the patient. Except for an assigned accession number, no identification was kept on the blood samples.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Temple University for studies involving humans.
Declaration of Interest
The authors declare no competing financial interests.
Contributors
Dr. Nune Darbinian and Dr. Michael Selzer are corresponding co-authors:
Dr. Darbinian: designing of the experiments, developing exosome studies, managing the project, supervising all experimental processes, writing of the first draft, participation in reviewing and editing the manuscript, and supporting the project financially.
Ms. Emily C. Sparks: experimental approach, RNA studies, editing a manuscript.
Dr. Armine Darbinyan: conceptualization, methodology, data analysis, writing, reviewing, and editing, visualization.
Dr. Nana Merabova: flow cytometry analysis, supervising students, participated in exosome isolation, and editing of the manuscript.
Dr. Gabriel Tatevosian: statistical support.
Ms. Ekaterina Vadachkoria, MD: discussion and carrying out data collection.
Dr. Shohreh Amini: reviewing and editing of the manuscript.
Dr. Goetzl: reviewing, and editing of the manuscript, developing exosome studies, and supporting the project financially.
Dr. Selzer: designing of the study and interpretating the data, provided overall scientific expertise, participating in the writing of the first draft of the manuscript, reviewing and editing the final manuscript, and supporting the project financially.
All authors read and approved the last version of the manuscript.
Acknowledgements
We thank study coordinators, Mrs. Gabrielle Rebillard and Tamara Tatevosian-Geller (MPH) for their assistance in the collecting of blood samples. We also wish to thank Karen Tamul and Richard Demarco (Luminex) for important suggestions in using of GUAVA FACS software. This work was supported by NIH grant R01HD069238, and Gates Foundation grant OPP1119489 to Dr. Laura Goetzl, by NIH grants R01NS97846, R01NS097846-02S1 and R01NS092876 awarded to Dr. Michael Selzer; Shriners research grant SHC-85400 awarded to Dr. Michael Selzer; and USA Pennsylvania State Health Department grant Project 10: 420491-04400-02 to Dr. Nune Darbinian.
Funding:
NIH grant R01HD069238 and Gates Foundation grant OPP1119489 to Dr. Laura Goetzl; NIH grant R01NS97846 and R01NS097846-02S1 to Dr. Michael Selzer; NIH grant R01NS092876 to Dr. Michael Selzer; Pennsylvania State Department grant Project 10: 420491-04400-02 to Dr. Nune Darbinian.
References
- Jiang X, Nardelli J. Cellular and molecular introduction to brain development. Neurobiol Dis 92 (2016): 3-17.
- Creeley CE, Dikranian KT, Johnson SA, et al., Alcohol-induced apoptosis of oligodendrocytes in the fetal macaque brain. Acta Neuropathologica Communications 1 (2013): 1-11.
- Kane CJ, Drew PD. Inflammatory responses to alcohol in the CNS: Nuclear receptors as potential therapeutics for alcohol-induced neuropathologies. J. Leukoc Biol 100 (2016): 951-959.
- Sowell ER, Mattson SN, Kan E, et al. Abnormal cortical thickness and brain-behavior correlation patterns in individuals with heavy prenatal alcohol exposure. Cereb Cortex 18 (2008): 136-144.
- Newville J, Valenzuela CF, Li L, et al. Acute oligodendrocyte loss with persistent white matter injury in a third trimester equivalent mouse model of fetal alcohol spectrum disorder. Glia 65 (2017): 1317-1332.
- Darbinian N, Darbinyan A, Merabova N, et al. Ethanol-mediated alterations in oligodendrocyte differentiation in the developing brain. Neurobiol Dis 148 (2021): 105181.
- Wasik N, Sokól B, Holysz M, et al. Serum myelin basic protein as a marker of brain injury in aneurysmal subarachnoid haemorrhage. Acta Neurochir 162 (2020): 545-552.
- Darbinyan A, Kaminski R, White MK, et al. Polyomavirus JC infection inhibits differentiation of oligodendrocyte progenitor cells. J Neurosci Res 91 (2013): 116-127.
- Larsen SV, Holven, KB, Ottestad I. Plasma fatty acid levels and gene expression related to lipid metabolism in peripheral blood mononuclear cells: a cross-sectional study in healthy subjects. Genes Nutr 13 (2018): 1-11.
- Sowell KD, Roberta RH, Janet YUA, et al. Altered Maternal Plasma Fatty Acid Composition by Alcohol Consumption and Smoking during Pregnancy and Associations with Fetal Alcohol Spectrum Disorders. Journal of the American College of Nutrition 39 (2020): 249-260.
- Lee JH, Lee YA, Oh KH, et al. Effects of dietary folic acid on the expression of myelin basic protein in the brain and spinal cord of pregnant and lactating rats. Ann Nutr Metab 56 (2010): 83-90.
- Vidakovic AJ, Jaddoe VW, Voortman T, et al. Maternal plasma polyunsaturated fatty acid levels during pregnancy and childhood lipid and insulin levels. Nutr Metab Cardiovasc Dis 27 (2017): 78-85.
- Grimes SB, Wild R. Effect of Pregnancy on Lipid Metabolism and Lipoprotein Levels. In: Feingold KR, Anawalt B, Boyce A, et al. editors. Endotext [Internet]. South Dartmouth (MA): MDText.com, Inc. 2000 (2018).
- Wiznitzer A, Mayer A, Novack V, et al. Association of lipid levels during gestation with preeclampsia and gestational diabetes mellitus: a population-based study. Am J Obstet Gynecol 201 (2009): 1-8.
- Crocker I, Nigel L, Ian D, et al. Significance of Fatty Acids in Pregnancy-Induced Immunosuppression. Clinical and Diagnostic Laboratory Immunology 6 (1999): 587-593.
- Elphick MC, Hull D. Transfer of fatty acid across the cat placenta. J Dev Physiol 6 (1984): 517-525.
- Roux JF, Takeda Y, Grigorian A. Lipid concentration and composition in human fetal tissue during development. Pediatrics 48 (1971): 540-546.
- Svennerholm L, Vanier MT. The distribution of lipids in the human nervous system. II. Lipid composition of human fetal and infant brain. Brain Res 47 (1972): 457-468.
- Mansson J E, Vanier M T, Svennerholm L. Changes in the fatty acid and sphingosine composition of the major gangliosides of human brain with age. J Neurochem 30 (1978): 273-275.
- Prinetti A, Chigorno V, Prioni S, et al. Changes in the lipid turnover, composition, and organization, as sphingolipid-enriched membrane Limited to 3 Pages - Developmental domains, in rat cerebellar granule cells developing in vitro. J Biol Chem 276 (2001): 21136- 21145.
- Crawford MA. Essential fatty acids and neurodevelopmental disorder. Adv Exp Med Biol 318 (1992): 307-314.
- Mead JF, Dhopeshwarkar GA. Types of fatty acids in brain lipids, their derivation and function. In: lipids, malnutrition & the developing brain. Ciba Found Symp (1971): 59-72.
- Burdge GC, Postle AD. Effect of maternal ethanol consumption during pregnancy on the phospholipid molecular species composition of fetal guinea-pig brain, liver and plasma. Biochim Biophys Acta 1256 (1995): 346-352.
- Burdge GC, Delange E, Dubois L, et al. Effect of reduced maternal protein intake in pregnancy in the rat on the fatty acid composition of brain, liver, plasma, heart and lung phospholipids of the offspring after weaning. Br J Nutr 90 (2003): 345-352.
- Rojas MAE, Padilla VE, Ortuno SD. Prenatal Alcohol Exposure in Rodents As a Promising Model for the Study of ADHD Molecular Basis. Front Neurosci 10 (2016): 565.
- Muller CP, Reichel M, Muhle C, et al. Brain membrane lipids in major depression and anxiety disorders. Biochim Biophys Acta 1851 (2015): 1052-1065.
- Hara T, Kimura I, Inoue D, et al. Free fatty acid receptors and their role in regulation of energy metabolism. Rev Physiol Biochem Pharmacol 164 (2013): 77-116.
- Kimura I, Ichimura A, Ohue-Kitano R, et al. Free Fatty Acid Receptors in Health and Disease. Physiological Reviews 100 (2020): 171-210.
- Darbinian N. Selzer M. Oligodendrocyte Pathology in the Developing Brain in Fetal Alcohol Syndrome. Neural Regeneration Research 17 (2022): 497-502.
- Balaraman S, Lunde ER, Sawant O, et al. Maternal and neonatal plasma microRNA biomarkers for fetal alcohol exposure in an ovine model. Alcohol Clin Exp Res 38 (2014): 1390-1400.
- Balaraman S, Schafer JJ, Tseng AM, et al. Plasma miRNA Profiles in Pregnant Women Predict Infant Outcomes following Prenatal Alcohol Exposure 11 (2016): e0165081.
- Tseng AM, Chung DD, Pinson MR, et al. Ethanol Exposure Increases miR-140 in Extracellular Vesicles: Implications for Fetal Neural Stem Cell Proliferation and Maturation. Alcohol Clin Exp Res 43 (2019): 1414-1426.
- Goetzl L, Darbinian N, Goetzl EJ. Novel window on early human neurodevelopment via fetal exosomes in maternal blood. Annals of Clinical and Translational Neurology 3 (2022): 381-385.
- Goetzl L, Merabova N, Darbinian N, et al. Diagnostic Potential of Neural Exosome Cargo as Biomarkers for Acute Brain Injury. Ann Clin Transl Neurol 5 (2018): 4-10.
- Goetzl L, Darbinian N, Merabova N. Noninvasive Assessment of Fetal Central Nervous System Insult: Potential Application to Prenatal Diagnosis. Prenat Diagn. Prenat Diagn 39 (2019): 609-615.
- Goetzl L, Thompson FT, Darbinian N, et al. Novel Biomarkers to Assess In-Utero Effects of Maternal Opioid Use: First Steps towards Understanding Short and Long Term Neurodevelopmental Sequelae. Genes Brain Behav 18 (2019): 1-20.
- Darbinian N, Darbinyan A, Sinard J, et al. Molecular Markers in Maternal Blood Exosomes Allow Early Detection of Fetal Alcohol Spectrum Disorders. International Journal of Molecular Sciences 24 (2023): 135.
- Williams BY, Boularan C, Vural A, et al. Omega-3 free fatty acids suppress macrophage inflammasome activation by inhibiting NF-kappaB activation and enhancing autophagy 9 (2014): e97957.
- Bernardo A, Giammarco ML, De Nuccio C, et al. Docosahexaenoic acid promotes oligodendrocyte differentiation via PPAR-gamma signalling and prevents tumor necrosis factor-alpha-dependent maturational arrest. Biochim Biophys Acta Mol Cell Biol Lipids 1862 (2017): 1013-1023.
- Siegert E, Paul F, Rothe M, et al. The effect of omega-3 fatty acids on central nervous system remyelination in fat-1 mice. BMC Neurosci 18 (2017): 1-9.
- Abdelmagid SA, Clarke SE, Nielsen DE, et al. Comprehensive profiling of plasma fatty acid concentrations in young healthy Canadian adults.10 (2015): 1-16.
- Larsen SV, Holven KB, and Ottestad I. Plasma fatty acid levels and gene expression related to lipid metabolism in peripheral blood mononuclear cells: a cross-sectional study in healthy subjects. Genes Nutr 13 (2018): 1-11.
- Batagov AO, Kurochkin IV. Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3′-untranslated regions. Biol Direct 8 (2013): 1-8.
- Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9 (2007): 654-659.
- Skog J, Würdinger T, Van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10 (2008): 1-16.
- Thakur BK et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 24 (2014): 766-769.
- Witwer K W et al. Updating the MISEV minimal requirements for extracellular vesicle studies: building bridges to reproducibility. J Extracell Vesicles 6 (2017): 1396823.
- Fruhbeis C, Frohlich D, Kramer AEM. Emerging roles of exosomes in neuron-glia communication. Front Physiol 3 (2012): 1-7.
- Kramer AEM, Bretz N, Tenzer S, et al. Oligodendrocytes secrete exosomes containing major myelin and stressprotective proteins: Trophic support for axons? Proteomics Clin Appl 1 (2007): 1446-1461.
- Skotland T, Sandvig K, Llorente A (2017). Lipids in exosomes: Current knowledge and the way forward. Prog Lipid Res 66 (2017): 30-41.
- Ebrahimkhani S, Vafaee F, Young PE, et al. Exosomal microRNA signatures in multiple sclerosis reflect disease status. Sci Rep 7 (2017): 14293.
- Dukes KA, Burd L, Elliott AJ, et al. The Safe Passage Study: Design, Methods, Recruitment, and Follow-Up Approach. Paediatric and Perinatal Epidemiology 8 (2014): 455-465.
- Khoenkhoen S, Ádori M, Pedersen GK, et al. Flow Cytometry-Based Protocols for the Analysis of Human Plasma Cell Differentiation. Front Immunol 11 (2020): 571321.
- Darbinian N, Darbinyan A, Merabova N, et al. Fetal Brain Injury Models of Fetal Alcohol Syndrome (FAS): Examination of Neuronal Morphologic Condition Using Sholl Assay. Methods Molecular Biology 2311 (2021): 195-201.
- Darbinyan A, Kaminski R, White M K, et al. Isolation and propagation of primary human and rodent embryonic neural progenitor cells and cortical neurons. Methods Molecular Biology 2311 (2021).
- Amini S, Merabova N, Khalili K, et al. p38SJ, a novel DINGG protein protects neuronal cells from alcohol induced injury and death. J Cell Physiol 221 (2009): 499-504.


 Impact Factor: * 3.2
Impact Factor: * 3.2 Acceptance Rate: 76.63%
Acceptance Rate: 76.63%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 