Metabolic Study of Resveratrol in Rat Biosamples by UPLC-Q-TOF/MS
Miao Qiaoa,b,#, Chenxi Guanga,b,#, Liqin Dingb, Xuliu Shia,b,c, Liwei Chaia,b, Xinchi Fenga,*
aSchool of Chinese Materia Medica, Tianjin University of Traditional Chinese Medicine, Tianjin, China
bTianjin State Key Laboratory of Modern Chinese Medicine, Tianjin University of Traditional Chinese Medicine, Tianjin, China
cCollege of Science and Technology, Hebei Agricultural University, Cangzhou, China
# These authors contributed equally to this work
*Corresponding Author (s): Xinchi Feng, School of Chinese Materia Medica, Tianjin University of Traditional Chinese Medicine, Tianjin, 301617, China
Received: 26 September 2020; Accepted: 06 October 2020; Published: 09 October 2020
Article Information
Citation:
Miao Qiao, Chenxi Guang, Liqin Ding, Xuliu Shi, Liwei Chai, Xinchi Feng. Metabolic Study of Resveratrol in Rat Biosamples by UPLC-Q-TOF/MS. Journal of Analytical Techniques and Research 2 (2020): 96-109.
View / Download Pdf Share at FacebookAbstract
Abstract
Resveratrol is a polyphenol derived from a variety of food and beverages, which has been reported to possess diverse pharmacological activities. An ultra-high performance liquid chromatography/quadrupole time-of-flight mass spectrometry (UPLC-Q-TOF/MS) method was established to figure out the metabolic profiles of resveratrol in rat biological samples after oral administration. According to the results, 21 resveratrol-related metabolites in rat biosamples were found after oral administration. As far as we know, 14 of these metabolites (M3 to M10, M13, M16, M17 and M21) have not been reported previously. Furthermore, glucuronide conjugation, sulfate conjugation, methylation, hydroxylation and reduction were the main metabolic pathways. Besides, the hydroxylated product was also detected in rat liver microsome. The results may be conducive to a better understanding the metabolic mechanism of resveratrol and provide helpful chemical information for further pharmacological and action mechanism studies of resveratrol.
Keywords
<p>Resveratrol; Metabolites; UPLC-Q-TOF/MS</p>
Article Details
1. Introduction
Resveratrol, a polyphenol compound occurred naturally in nuts, berries, and the skin of grapes [1]. Since 1940, resveratrol has caused considerable interest in the medical community due to its ability to protect against a variety of diseases, including diabetes, cardiovascular disease, cancers [2] and so on.
Although many studies showed the momentous role of resveratrol in disease prevention [3], only few studies deal with the bioavailability and metabolism of resveratrol were available [4]. Resveratrol is rapidly metabolized in the body and its bioavailability is low [5]. It is speculated that metabolites of resveratrol may play an important role in effectiveness. Accordingly, it is necessary to systematically study the metabolism of resveratrol in vivo.
Metabolic researches play a crucial role in drug discovery and development [6]. With the aid of the metabolynx™ software, UPLC-Q-TOF/MS has been widely applied in the screening and identifying of the unknown compounds or their metabolites in biological samples [7]. In this study, a quick and credible UPLC-Q-TOF/MS method was developed to profile and identify the major metabolites of resveratrol in rat urine, plasma, bile and feces after oral administration of resveratrol.
2. Materials and Methods
2.1 Chemicals and Materials
Resveratrol (HPLC purity >98%) was purchased from Tianjin Vientiane hengyuan Technology Co., Ltd. (Tianjin, China). Dihydroresveratrol (The purity was no less than 98% by HPLC-UV analysis) was synthesized by our laboratory. Rat liver microsome (10 mg/mL microsomal protein) was purchased from Research Institute for Liver Disease (Shanghai) Co., Ltd. Other chemical reagents used were in analytical grade and obtained commercially.
2.2 Animals
Male SD rats (200~220 g, SPF grade) were obtained from the national quality inspection center of experimental animal (Beijing, China; Certificate No. SCXK 2017-0005). All animals were housed under standard conditions for a week. The study strictly adheres to the Regulations of Experimental Animal Administration. Before drug administration, the rats were fed in metabolic cages for 12 h. Resveratrol was orally administered at a dose of 50 mg/kg with normal saline as the vehicle.
2.3 Sample Collection and Preparation
The urine and feces samples were collected within 0–12 h and 12–24 h after oral administration respectively. Blood samples (0.3 mL) were collected from the orbital vein into heparinized tubes at 0.083, 0.25, 0.5, 1, 2, 4, 6, 8, 12 and 24 h, separately after drug treatment. Bile samples were collected for 0-12 h and 12-24 h after administration separately. All the samples were stored at −80°C (0–1 week) and thawed immediately before analysis.
The bile sample (2 mL) or urine sample (5 mL) was loaded onto a C18 SPE (500 mg) column, and washed with 5 mL ultra-pure water and then 5 mL methanol. The methanol eluent was dried under nitrogen gas at room temperature. The feces sample (1g) was added into 10 mL methanol with ultrasonic treatment for 1 h and subsequently centrifuged for 10 min. The supernatant was separated and dried with nitrogen gas at room temperature. Plasma samples obtained from different time points were combined. An aliquot of 6 mL acetonitrile was added to 2 mL of plasma samples and vortex mixed for 3 min to precipitate proteins. Then, the precipitated protein was removed by centrifuging and the supernatant was dried under nitrogen gas at room temperature. Residue was dissolved with 0.3 mL of methanol-water (1:1, v/v), vortexed and thoroughly mixed, centrifuged at 17,460 Í g for 10 minutes, finally, 200 μL of the supernatant was taken out for analysis.
2.4 Microsomal Incubations and Sample Preparation
The incubation mixture contained rat liver microsomes (0.2 mg/mL), potassium phosphate buffer (100 mM, pH 7.4), MgCl2 (5 mM) and resveratrol (100 µM). The final volume of the incubation system was 200 µL. Resveratrol was previously dissolved in 60% methanol aqueous solution at concentration of 10 mM as stock solution. An aliquot of 2 µL stock solution was added into the microsomal incubation system to obtain a final concentration of resveratrol at 100 µM and the total amount of organic solvent in the system was not over 1%. Microsomal mixtures were incubated at 37°C in an intelligent temperature metal bath with and without combinations of 1 mM NADPH. After 60 minutes of incubation, the reaction was terminated by adding 0.6 mL of ice-methanol to the system, and then the sample was centrifuged for 10 minutes at 4°C, and 200 μL supernatant was taken out and then injected into a UPLC for analysis.
2.5 Chromatography and Mass Spectrometry Conditions
The UPLC-Q-TOF/MS system was conducted on an ACQUITYTM UPLC I-Class system and a Xevo G2-S Q-TOF mass spectrometer interface (Waters, Milford, USA). The chromatographic separation was carried out on an ACQUITY UPLC BEH C18 column (2.1Í100 mm, 1.7 μm). The eluent was directly introduced into the mass spectrometry. The mobile phase consisting of 0.1% formic acid aqueous solution (A) and 0.1% formic acid acetonitrile solution (B) was delivered using a linear gradient program showed below: 5–10% B from 0 to 2 min, 10–30% B from 2 to 6 min, 50–85% B from 12 to 18 min and 85–95% B from 18 to 20 min. The optimal conditions of analysis were employed showed below: capillary voltage was 3.0 kV; extraction cone voltage was 3.0 V; sampling cone voltage was 40 V; desolvation temperature was 400°C; source temperature was 100°C; cone gas flow was 50 L/h; desolvation gas flow was 600 L/h. In MSE mode, trap collision energy of low-energy function was set at 0 eV, while the ramp trap collision energy of high-energy function was set at 15–60 eV. In MS/MS mode, the samples were detected at the collision energy of 20 eV. Data in centroid mode were from 50 to 1000 Da. All data were collected and processed by Metabolynx XS software under the Masslynx V4.1 (Waters, Milford, USA) operating interface.
. Results
This paper reported the metabolites of resveratrol. Firstly, the full scan mass spectra of biological samples were obtained (Figure 1). Secondly, the molecular formula of resveratrol (C14H12O3) used as the parent compound was put into the Metabolynx XS software. The element composition and elemental limits of possible substituents were determined by potential metabolic reactions (sulfate conjugation, glucuronide conjugation, methylation and hydroxylated et al.). Thirdly, the metabolites were screened, identified and affirmed by comparing the retention times, MS2 fragments with those of the reference materials or reported in the published literatures. Additionally, Clog P (calculated logarithm of its partition coefficient between n-octanol and water) values for several isomers were obtained from ChemDraw (version 12.0) and applied as important parameters for the identification of metabolites as reported [8]. Finally, 21 major metabolites were identified in rat biological samples (Table 1). The proposed metabolic pathways of resveratrol were shown in Figure 2. According to our results, resveratrol was able to underwent reduction, hydroxylation and methylation metabolism in rats. Subsequently, glucuronidation and sulfation metabolism occurred and plenty of phase II metabolites were generated. Interestingly, no methylation metabolites of resveratrol were detected directly; however, metabolites underwent both methylation and other metabolic pathways (M3, M8, M10, M17, M20 and M21) were detected. Due to the fact that there are three hydroxyl groups in resveratrol, bis-glucuronide conjugations (M3, M5, M6 and M9) and metabolites underwent both sulfate and glucuronide conjugation (M4 and M7) were also identified.
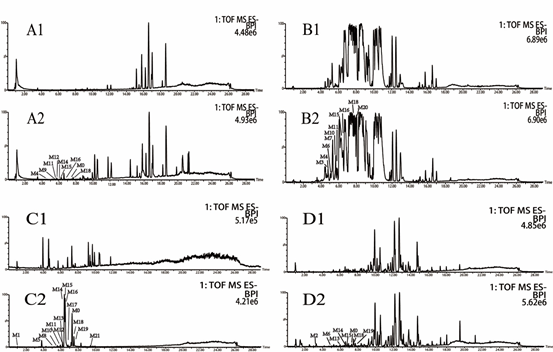
Figure 1: Total ion chromatograms of plasma (A), bile (B), urine (C) and feces (D) in rats after intragastric administration of resveratrol; and 1, blank samples, 2 resveratrol-related samples.
Table 1 : Chromatographic and mass data of resveratrol and its metabolites obtained by UPLC-Q-TOF/MS.
Footnote: P: Plasma; U: Urine; F: Feces; B: Bile. “+”, detected; “/” not detected; * : Compared with reference standards; ** : Detected in liver microsomes. # : Reported for the first time.
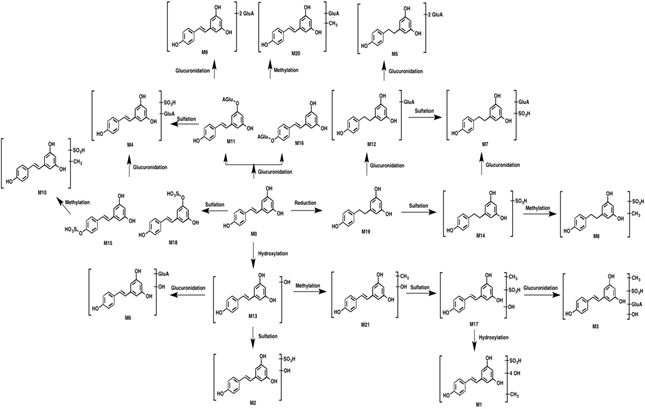
Figure 2: Proposed metabolic pathways of resveratrol.
3.1 Mass Fragmentation Behavior of Resveratrol and dihydroresveratrol
In order to fully understand the resveratrol metabolites, the MS2 fragmentation behaviors of the parent compound and reduction metabolites dihydroresveratrol were first studied in negative ion mode which was more suitable for resveratrol analysis (Figure 3A). Resveratrol was eluted at 7.27 min with a quasi-molecular ion [M-H]- at m/z 227.0713 (-2.2 ppm, C14H11O3), which yielded characteristic fragment ions at m/z 185.0590 and 143.0490. Dihydroresveratrol with the retention time of 7.50 min exhibited the [M-H]- at m/z 229.0865, which was 2 Da higher than that of resveratrol. MS/MS spectrum of dihydroresveratrol yielded fragment ions at m/z 123.0445. These results were in accordance with that reported in the published paper [9, 10]. The fragmentation pathways of resveratrol and dihydroresveratrol were showed in Figure 3A and Figure 3B. Since plenty of glucuronidation metabolites were detected, the proposed fragmentation pathways of glucuronidation metabolites were also summarized in Figure 3C. As we can see, in the negative ion mass spectra, fragment ions of m/z 175 and m/z 113 could be considered as characteristic ions of glucuronidation metabolites. Meanwhile, characteristic fragment ion of m/z 79.96 could be observed for sulfation metabolites in negative ion mass spectra. Taken together, characteristic product ions along with neutral losses (176 Da for glucuronic acid and 80 Da for SO3) provided a sound basis for the identification of the metabolites of resveratrol.
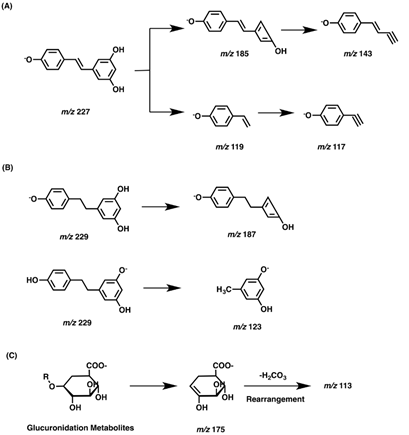
Figure 3: The proposed fragmentation pathways of resveratrol (A), dihydroresveratrol (B) and glucuronidation metabolites (C).
3.2 Metabolites of Resveratrol in Rat Biosamples
21 metabolites were identified. The extracted ion chromatograms (EICs) of the M0 and its metabolites were showed in Figure 4.
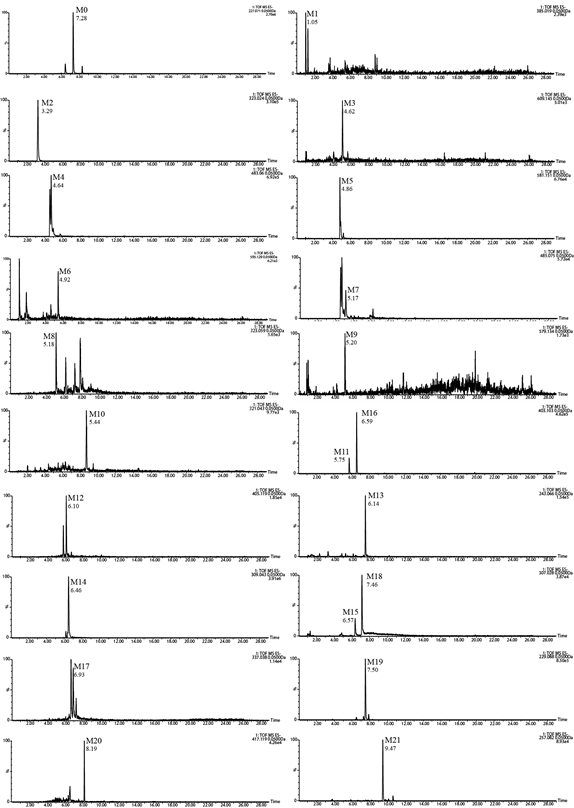
Figure 4: Extracted ion chromatograms (EICs) of resveratrol metabolites in rat biosamples.
3.2.1. Reduction metabolites
The retention time of M19 was 7.50 min and it exhibited the quasi-molecular ion at m/z 229.0873 (C14H14O3), which was 2 Da higher than that of resveratrol. Characteristic ions at m/z 143.0497 and 123.0445 suggested that M19 was dihydroresveratrol. By comparing the retention time and accurate mass spectrum with that of the authentic standard, M19 was identified as dihydroresveratrol.
3.2.2. Glucuronidation metabolites
M11 eluted at 5.75 min and the elemental formula was determined as C20H20O9 ([M-H]- m/z 403.1029). Fragment ion at m/z 227.0712 was yielded via the neutral loss of 176 Da (403.1029 - 227.0712= 176), indicating that M11 was the glucuronidated metabolite of resveratrol. Characteristic ions of glucuronic acid moiety at m/z 175.0264 and 113.0235 were also observed. Thus, M11 was determined to be glucuronide conjugate of resveratrol. Similarly, M16 was also identified as glucuronidated product of resveratrol and M9 was identified as bis-glucuronide conjugate of resveratrol. M11 and M16 was a pair of isomer and the Clog P values of M16 and M11 were 0.63 and 0.84. Therefore, M11 and M16 were tentatively identified separately according to the Clog P values.
M12 was detected at 6.10 min with the quasi-molecular ion at m/z 405.1188 (C20H22O9), which was 176 Da (405.1188 – 229.0873 = 176) higher than that of dihydroresveratrol. Characteristic fragment ions at m/z 175.0237 and 113.0237 indicated the existence of glucuronic acid moiety, thus, M12 was identified as glucuronidation metabolite of dihydroresveratrol. Similarly, M5 was determined to be bis-glucuronide conjugate of dihydroresveratrol.
3.2.3. Sulfation metabolites
M15 and M18 showed quasi-molecular ions at m/z 307.0275 (C14H14O6S), which was 80 Da higher than that of resveratrol. Characteristic ion at m/z 227.0707 was yielded via the neutral loss of SO3 (80 Da, 307.0275 – 227.0707 = 80). The neutral loss of SO3 together with characteristic ions at m/z 185.0602 and 143.0497 indicated that M15 and M18 were the sulfate conjugates of resveratrol and they were also identified separately according to the Clog P values. Similarly, the neutral loss of SO3 (80 Da, 309.0429 – 229.0862 = 80) and characteristic ions at m/z 229.0862 and 123.0442 indicated that M14 was the sulfate conjugate of dihydroresveratrol.
M4 was eluted at the retention time of 4.64 min and showed a quasi-molecular ion at m/z 483.0599 (C20H19O12S). Fragment ions displayed at m/z 403.1022 (C20H19O9), 307.0273 (C14H11O6S) were yielded via the neutral losses of SO3 and a glucuronic acid moiety, respectively. Therefore, M4 was determined of the glucuronide and sulfate conjugate of resveratrol. Similarly, characteristic ion at m/z 175.0240 was observed in the MS2 spectrum of M7, indicating that M7 was a glucuronidation metabolite. Additionally, M7 showed a quasi-molecular ion at m/z 485.0739 (C20H22O12S), which was 2 Da higher than that of M4, thus, M7 was identified as glucuronide and sulfate conjugate of dihydroresveratrol.
3.2.4. Hydroxylation metabolites
M13 eluted at 6.14 min and showed a quasi-molecular ion at m/z 243.0663 (C14H11O4), which was 16 Da higher than that of resveratrol. In the MS2 spectrum, M13 yielded the characteristic fragment ions at m/z 227.0708, 142.0659, 123.0442 and 79.9568. Characteristic fragment ion at m/z 227.0708 was generated via the loss of one atom of oxygen, indicating that M13 was the hydroxylation metabolite of resveratrol.
M2 was detected at 3.29 min with the quasi-molecular ion at m/z 323.0238 (C14H12O7S), which was 80 Da (323.0238 – 243.0663 = 80) higher than that of M13. Characteristic fragment ion at m/z 243.0659 was generated via the neutral loss of SO3, thus, M2 was identified as sulfation and hydroxylation metabolite of resveratrol. As for M6, it showed a quasi-molecular ion at m/z 595.1291 (C26H28O16), which was 2 × 176 Da (595.1291 – 243.0663 = 352 = 2 × 176) higher than that of M13. Additionally, characteristic fragment ion at m/z 419.0976 was generated via the neutral loss of a glucuronic acid moiety. Thus, M6 was determined to be bis-glucuronide conjugate of M13.
3.2.5. Methylation metabolites
Even though no methylation metabolite of resveratrol was detected directly in rat biosamples, metabolites underwent both methylation and other metabolic pathways were detected. Take M10 for example, M10 eluted at 5.44 min and showed a quasi-molecular ion at m/z 321.0437. Diagnostic ion at m/z 241.0872 was yielded via the neutral losses of SO3 and diagnostic ion at m/z 241.0872 was 14 Da (241.0872 – 227.0717 = 14) higher than that of resveratrol, indicated that M10 was methylation and sulfation metabolite of resveratrol.
Other metabolites including M3, M8, M17, M20 and M21 were presumed based on retention time, molecular weight, and characteristic fragment ions. It is worth mentioning that, among the 21 metabolites identified, only reduction metabolite was confirmed by comparison with reference standard. It would be difficult to unambiguously confirm the structures of other metabolites due to the fact that reference standards of them were commercially unavailable.
3.3 Metabolites of Resveratrol in Rat Liver Microsomal
Q-TOF-MS/MS was operated in a negative ion mode to obtain important information about resveratrol metabolites. After incubation of resveratrol with rat liver microsomal, metabolite M13, which attributed to the hydroxylation of resveratrol, was detected (Figure 5).
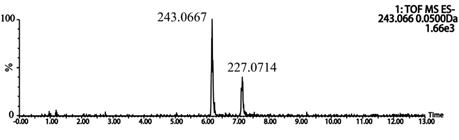
Figure 5: Extracted ion chromatograms (EICs) of resveratrol metabolites in rat liver microsomes.
4. Discussion
Generally speaking, the role of phase I metabolism in the body was to improve the water solubility of metabolites, reduce or abrogate their biological activities. The phase II metabolites can be detoxified through drug conjugation [11]. Some studies have shown that in the body, resveratrol was able to be transported across the intestinal epithelium and conjugated with either glucuronic acid or sulfate [12]. In our study, the number of phase II metabolites were significantly greater than the number of phase I metabolites. The most abundant metabolites were found in the urine, which suggested that the kidney pathway plays an important role in resveratrol excretion. Additionally, in our study, after resveratrol was cultured with rat liver microsomes, we only detected hydroxylated metabolites of resveratrol and no double bond reduction products were found. This may be due to the fact that the reduction products detected in vivo were produced in intestine rather than liver. It has been reported that the intestinal flora can hydrogenate the double bond of resveratrol to form dihydroresveratrol, which is then absorbed and metabolized [13].
5. Acknowledgment
This project was financially supported by the State Key Program of National Natural Science Foundation of China (81430095), the National Natural Science Foundation of China (81703776) and the National Science and Technology Major Project of China “Key New Drug Creation and Manufacturing Program” (Grants 2017ZX09301005).
Conflict of Interest:
The authors declare no conflicts of interest.
References
- Craveiro M, Cretenet G, Mongellaz C, et al. Resveratrol stimulates the metabolic reprogramming of human CD4+ T cells to enhance effector function. Science Signaling 10 (2017).
- Patel KR, Brown VA, Jones DJ, et al. Clinical pharmacology of resveratrol and its metabolites in colorectal cancer patients. Cancer Research 70 (2010): 7392-7399.
- Boocock DJ, Faust GE, Patel KR, et al. Phase I dose escalation 507 pharmacokinetic study in healthy volunteers of resveratrol, a potential cancer chemopreventive 508 agent. Cancer Epidemiol Biomarkers Prev 16 (2007): 1246-1252.
- Wang D, Hang T, Wu C, et al. Identification of the major metabolites of resveratrol in rat urine by HPLC-MS/MS. Journal of Chromatography B 829 (2005): 97-106.
- Walle T. Bioavailability of resveratrol. Annals of the New York Academy of Sciences 1215 (2011): 9-15.
- Tao JH, Zhao M, Jiang S, et al. UPLC-Q-TOF/MS-based metabolic profiling comparison of four major bioactive components in normal and CKD rat plasma, urine and feces following oral administration of Cornus officinalis Sieb and Rehmannia glutinosa Libosch herb couple extract. Journal of Pharmaceutical and Biomedical Analysis 161 (2018): 254-261.
- Fan Y, Han H, He C, et al. Identification of the metabolites of gigantol in rat urine by ultra-performance liquid chromatography combined with electrospray ionization quadrupole time-of-flight tandem mass spectrometry. Biomedical Chromatography 28 (2014): 1808-1815.
- Tian T, Jin Y, Ma Y, et al. Identification of metabolites of oridonin in rats with a single run on UPLC-Triple-TOF-MS/MS system based on multiple mass defect filter data acquisition and multiple data processing techniques. Journal of Chromatography B 1006 (2015): 80-92.
- Bode LM, Bunzel D, Huch M, et al. In vivo and in vitro metabolism of trans-resveratrol by human gut microbiota. The American Journal of Clinical Nutrition 97 (2013): 295-309.
- Huang Huilian, Yao Shun, Huang Xiaoying, et al. LC-MS-MS analysis of resveratrol metabolites in rat bile and urine. Chinese Journal of Experimental Pharmacology 20 (2014 ): pp.56-61.
- Zhu L, Sun S, Hu Y, et al. Metabolic study of paeoniflorin and total paeony glucosides from Paeoniae Radix Rubra in rats by high-performance liquid chromatography coupled with sequential mass spectrometry. Biomedical Chromatography 32 (2018): e4141.
- Walle T, Hsieh F, DeLegge MH, et al. High absorption but very low bioavailability of oral resveratrol in humans. Drug Metabolism and Disposition 32 (2004): 1377-1382.
- Juan ME, Alfaras I, Planas JM. Determination of dihydroresveratrol in rat plasma by HPLC. Journal of Agricultural and Food Chemistry 58 (2010): 7472-7475.


 Impact Factor: * 2.8
Impact Factor: * 2.8 Acceptance Rate: 77.30%
Acceptance Rate: 77.30%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 