Phenotypic variability in expressivity and deleterious capability of the homozygous UGT1A1*28 genotype
Raquel Rodríguez-López1*, Jorge Ferriz Vivancos1, Ana Cristina Requena Piedrabuena2, Irene Ferrer Bolufer1, Carola Guzmán Luján1, Otilia Zomeño Alcalá1, Joan Más Lloret1, Jennifer Valero García2, Ana de Blas Zapata3, Mireia Gil Raga4, Ana Vallejo Antolín5, Mercedes Latorre Sánchez5, Mª José Safont Aguilera4
1 Genetics Laboratory, Clinical Analysis Service, General Universitary Hospital Consortium of Valencia (CHGUV)
2 IMEGEN (HealthinCode), Valencia
3 Pediatric Service, CHGUV
4 Oncology Service, CHGUV
5 Digestive Service, CHGUV. SPAIN
*Corresponding Author: Raquel Rodríguez-López, MD, PhD, Genetics Laboratory, General Universitary Hospital of Valencia, Av. Tres Cruces, 2, 46014, Valencia, Spain. And Member of the Genetics Commission of the Spanish Society of Clinical Chemistry (SEQC).
Received: 15 December 2022; Accepted: 30 December 2022; Published: 04 January 2023
Article Information
Citation: Raquel Rodríguez-López, Jorge Ferriz Vivancos, Ana Cristina Requena Piedrabuena, Irene Ferrer Bolufer, Carola Guzmán Luján, Otilia Zomeño Alcalá, Joan Más Lloret, Jennifer Valero García, Ana de Blas Zapata, Mireia Gil Raga, Ana Vallejo Antolín, Mercedes Latorre Sánchez, Mª José Safont Aguilera. Phenotypic variability in expressivity and deleterious capability of the homozygous UGT1A1*28 genotype. Journal of Analytical Techniques and Research 5 (2023): 01-07.
View / Download Pdf Share at FacebookAbstract
Background :
The UGT1A1*28 polymorphism (rs3064744) is responsible for Gilbert's Syndrome (GS) and is characterized by allelic variability based on TA repeats in the TATA sequence of the UGT1A1 gene promoter, whose alleles 7TA and 8TA are associated with elevated bilirubin, as well as an increased risk of certain neoplasms and adverse reactions to the chemotherapy Irinotecan. In contrast, the existence of hyperbilirubinemia has been considered a preventive factor for certain cardiovascular and oncological pathologies. We analyzed the allelic distribution of the UGT1A1 allele in the Valencian population with hyperbilirubinemia and Valencian patients diagnosed with digestive neoplasms; we studied their biochemical profiles, specifically comparing individuals with the UGT1A1*28 genotype in homozygosis of both groups.
Methods :
We collected a sample of 1,358 individuals: 621 patients with suspected GS (cases) and 737 individuals affected by one oncological disease (possible reference population). After extracting DNA, samples were analyzed the number of TA repeats by both Sanger sequencing and fragment analysis technique. In addition, the clinical history and set of analytical parameters of 242 patients carrying the genotype UGT1A1*28 in homozygosis were reviewed, with special attention to the consecutive measurement of bilirubin levels.
Results :
The differences in the distribution of the UGT1A1*28 genotypes in both groups, heterozygous and homozygous, were statistically significant; the homozygous UGT1A1*28 genotype reached 55.4% among patients with suspected GS. Severe H-W E deviations were observed in this group (OR: 45.37), while in cancer patients they were distributed according to the H-W E balance. Bilirubin levels in the group of homozygous UGT1A1*28 individuals among oncology patients were lower, showing a higher risk (OR: 2.86) in males.
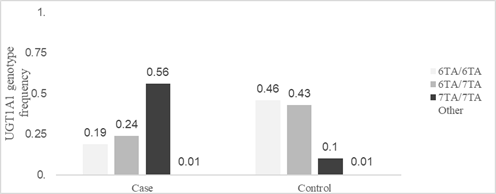
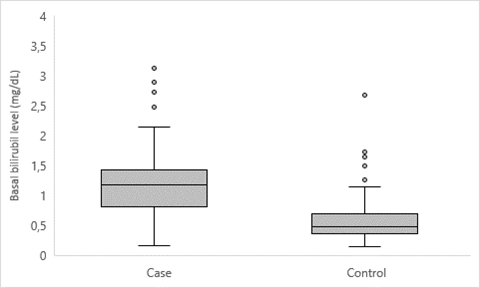
<p>Gilbert syndrome; Hyperbilirubinemia; pharmacogenetics; UGT1A1</p> Enzyme uridine diphosphate-glucuronosyl transferase 1A1 is an enzyme responsible for the glucuronidation of lipophilic molecules into water-soluble and excretable molecules. Diverses hormones and drugs can be the substrate of this enzyme, as well as bilirubin, a yellow bile pigment derived from the metabolism of hemoglobin [1]. This UDP-glucuronosyltransferase is encoded by the UGT1A1 gene and the presence of genetic variations cause alteration of the enzyme leading to reduced activity, with an increase in bilirubin that can act as an indirect antioxidant. These variations in UGT1A1 gene are the most common cause of mild hereditary intermittent unconjugated hyperbilirubinemia-Gilbert syndrome (GS) (OMIM#143500) [2], one of the four disorders associated with the UGT1A1 gene. In the Human Phenotype Ontology, GS is described as an autosomal recessive inherited disorder characterized by unconjugated hyperbilirubinemia, resulting in harmless intermittent jaundice [3]. The associated HPO terms are Jaundice (HP:0000952), Unconjugated hyperbilirubinemia (HP:0008282), and Dehydration (HP:0001944). Other authors consider GS as an autosomal dominant hereditary condition with incomplete penetrance [4]. The genetic variant code rs3064744 (allele UGT1A1*28) in the UGT1A1 gene describes the polymorphic insertion of thymine-adenine (TA) repeats in the promoter region (wild type consists of 6 TA repeats and UGT1A1*28 variant allele contains 7 TA repeats), associated with a reduction of the gene transcription even 67–82%, with subsequent reduction of enzyme activity by 20–30% [5]. Individuals with GS carry 7/7 homozygous genotype with two UGT1A1*28 alleles. GS is considered a benign condition that is present in 5-10% of the population, about which there is quite a bit of contradictory literature. On the one hand, bilirubin has been attributed anti-inflammatory and antioxidant capacities of its substrate [6]. SG has also been linked to the prevention of liver disease, diabetes, coronary heart disease and even obesity [7-9]. In addition, a relationship between GS and a lower incidence of cancer, specifically Hodgkin's lymphoma or endometrial cancer, has been observed [10]. On the other hand, alleles that induce GS have been related to increased risks of developing breast and colorectal cancer, due to decreased detoxification of carcinogens. Also very important are the new pharmacogenetic discoveries that associate severe reactions to antineoplastic drugs, such as Irinotecan, in patients with the homozygous 7TA allele UGT1A1 (or double heterozygous with the low-frequency 8TA allele; 7TA /8TA genotype [5]. Considers the cause that some xenobiotics share the same conjugation and excretion pathway as bilirubin [11]. The prevalence of GS varies worldwide, estimated at 5 to 16% of European blood donors, but as many cases are asymptomatic and undiagnosed, the true prevalence is probably much higher [12]. The heterozygosity rate for the (TA)7 allele in our control population was previously estimated to be around 32% and the rate for the TA7/7 genotype was 11.33% [13], almost double the 5% reported for the genotypes European control patients. This confirms the observed increasing gradient of allelic frequencies in populations from the North to the South, as well as the high rate of GS reported in the Spanish population, with no significant differences observed between the geographical extremes from the west to the east of the country. The objectives of this study were to describe the population distribution of the UGT1A1*28 (rs3064744) variant in a Valencian population and to verify whether the allelic frequencies among the group of patients with suspected GS differed from the cancer patients who act as a control population, in order to assess whether referred allele could confer increased susceptibility to suffering it. Finally, to compare the biochemical evolution between patients with genotype 7TA/7TA diagnosed with neoplasms and patients with genotype 7TA/7TA who had been diagnosed with GS. Patient characteristics Patients who attended the Hospital General Universitario de Valencia (Valencia, Spain) between 01/2014 and 07/2020 were included in this study. They were divided into: 1,621 individuals derived from the Service of Digestive for suspicion of GS, and a reference population of 737 cancer patients who had been referred to our laboratory for the indicated pharmacogenetic study, in order to predict possible increased susceptibility to Irinotecan toxicity; its characteristics make this series be considered representative of the general population. Later, to increase the potency of statistical analysis of biochemical parameters comparison 52 more oncological patients with 7TA homozygous UGT1A1 allele were included. All patients included belonged to the same population, with less than 3% coming from non-Caucasian ethnicity. Control group that we refer to in our previous article [13] came from the same health area where the cases included in this study came from. All patients gave their written informed consent for the acquisition of genetic data in the context of scientific studies, according to institutional guidelines. Genomic DNA extraction A peripheral blood sample (PB) of each patient included in this study, was collected in EDTA tubes. 200 microliters of PB samples collected, were extracted using a fully automated, spin-column-based nucleic acid extraction method with the automatic QIAcube Connect equipment (QIAGEN, Hilden, Germany). 50 µL of DNA were obtained and quantified with NanoDropTM spectrophotometer (ThermoFisher Scientific, Massachusetts, USA) and stored at -20ºC until all the genotyping assays were performed by the two techniques, Sanger sequencing and fragment analysis, with the aim of validating the fragment method, assessing the profitability and efficiency of both, finally choosing the one considered best for the characteristics of our laboratory and hospital. Sanger sequencing Custom primers were used to amplify and sequence the target region. The results were analyzed using SeqScapeTM Software V3.0 (ThermoFisher Scientific®). Fragment analysis Samples were analyzed in parallel with ImegenTM-Gilbert Plus commercial kit, provided by Health in Code S.L®. and used according to the manufacturer’s recommended protocol. This kit allows the detection of the mutant alleles A(TA)5TAA, A(TA)7TAA, and A(TA)8TAA, as well as the normal allele A(TA)6TAA of the UGT1A1 gene promoter by fragment analysis. The amplification of the interest region was performed by conventional PCR with SimpliAmp Thermal Cycler (ThermoFisher Scientific®). Then, the PCR products were separated by capillary electrophoresis and were detected due to 6-Carboxyfluorescein fluorescent dye (6-FAM) with a 3730xl DNA Analyzer (ThermoFisher Scientific®) using POP-7™ polymer. The analysis of the obtained results was carried out with GeneMapperTM Software 5 (ThermoFisher Scientific®). Biochemical analysis Clinical history of 242 patients with 7TA/7TA genotype was reviewed to obtain minimum, considered as basal, and maximum values of bilirubin to compare between patients with GS suspicion and oncological patients. Bilirubin was analyzed with an AU5400 analyzer (Beckman Coulter®) and a cut-off point of 1.2 mg/dL for bilirubin was established, considering >1.2mg/dL as hyperbilirubinemia. Statistical Analysis Statistical analysis was performed using Statistical Package for Social Sciences (SPSS®, version 11.5) and included both descriptive and analytical methods. Subjects were grouped and categorized according to the presence of three different genotypes (6/6 wild type, 6/7 heterozygote, 7/7 homozygote), depending on the number of TA repeats in the promoter region of UGT1A1 gene. Hardy-Weinberg law was applied by com-paring the allelic and genotypic frequencies in both population groups with SNPStats software, and odds ratio (OR) was calculated using X2 test. Both parametric (Student’s test) and non-parametric tests (c2, Kruskal Wallis, Fisher’s exact test) were used, depending on the normality of variables. Linear regression and beta coefficient were also computed for the analysis of association. Values at the p 0.05 level were considered statistically significant, the confidence interval (CI) was 95% and all performed tests were 2-tailed. No differences were found in the obtained genotypes by using both methods (Sanger sequencing and Fragment analysis). The correlation of the genotyping results of individuals included was absolute (100%) and no discrepancy was detected between the results obtained by using both techniques. The low-frequency alleles 5TA (one 5TA/6TA genotype) and 8TA (two 6TA/8TA genotypes and one 7TA/8TA genotype) were also detected with extensive precision. A total of 1,358 patients with a mean age of 57.6 years (range 1 to 93 years) and 63.3% male were analyzed. 621 with suspected GS (cases) and 737 affected by one of the oncological diseases (reference population): colon 51.6%, rectum 26.7%, pancreas 6.15%, gastric 3.21%, esophagus 2.8%, cholangiocarcinoma 2.65%, breast 1.24%, candidate to rs3064744 UGT1A1 polymorphism previously to treatment with Fluoropyrimidines and/or Irinotecan. The frequency of the UGT1A1*28 (TA)7/(TA)7 genotype in the reference series associated with the Valencian population was 9.8%. The prevalence of the hyperbilirubinemia risk genotype reached 55.4% among patients with suspected GS. Statistically significant differences were found between the genotypes of both groups (chi-square test, p<0.001) (Figure 1). Fig 1: Distribution of genotypes of the rs3064744 polymorphism in the association study Statistically significant differences were also found in terms of allelic frequencies. Cases: 6TA 0.317 and 7TA 0.683 and among cancer patients: 6TA 0.684 and 7TA 0.316. It was observed that the genotypes among the individuals of the Oncology Department were in Hardy-Weinberg equilibrium (H-W E) (OR: 2.27, p-value 0.13), but severe H-W E deviations were observed within the series of Digestive Service (OR: 45.37; p-value <0.001). The comparison of both populations showed a comparable allelic distribution (6TA/7TA genotype: OR: 1.08; 95% CI 0.72-1.64; 7TA/7TA genotype: OR: 0.98; 95% CI 0.53 -1.80). In the codominant model (6TA/7TA genotype) the ability of the heterozygous allele to cause bilirubin elevation was observed (OR: 1.66; 95% CI 1.05-2.62), however, the recessive model (7TA/7TA genotype) indicated a greater increased risk attributable to the genotype of the risk allele in homozygosity (OR: 14.01; 95% CI 8.63-22.74). In both cases, statistically significant values were reached, with the sample size of the analyzed series. The biochemical phenotype of 93 controls with 7TA/7TA genotype (mean age: 64.3 years) versus 149 cases (mean age: 38.3 years) with the same genotype, who had been referred for sustained hyperbilirubinemia from the Digestive Service, showed differences in the basal bilirubin level between both groups: 0,61 mg/dL in controls versus 1.24 mg/dL in cases (p-value <0.001) (Figure 2). No statistically significant differences were observed in terms of maximum bilirubin. It was observed that patients with suspected GS (cases) presented higher basal bilirubin values than controls (chi-square, p<0.001), with just 5.4% of controls with basal hyperbilirubinemia vs 48.6% of cases. Also, only 1 case (0.67%) never presented hyperbilirubinemia, while 19 controls (20.43%) never reached that condition (chi-square, p<0.001). There were no statistically significant differences in the sex distribution between carriers homozygous for the risk allele (p-value = 0.07). However, linear regression of bilirubin levels between both sexes and the presence or absence of basal hyperbilirubinemia showed that the male sex confers a higher risk (OR: 2.86) to higher bilirubin levels (p-value <0.01). Fig 2: Comparison of basal bilirubin levels for cases and controls The comparison between Sanger sequencing technologies versus fragment analysis to obtain the genotypes corresponding to the existing alleles of the UGT1A1*28 variant (genetic variant code rs3064744) located in the promoter of the UGT gene, strongly validated the second methodology. Although the economic ad-vantages were not a determining cause for prioritizing it over the Sanger methodology, the speed and greater simplicity of its execution were, translated into the time of the specialized molecular technical staff. Reducing the response time for the issuance of genotype reports in the field of pharmacogenetics to the minimum possible has been a priority in our laboratory, which we have achieved with the change of methodology to fragment analysis. The distribution of alleles and genotypes of the UGT1A1*28 variant among the individuals analyzed for suspected GS leaves no doubt about its association (p-value <0.001) with the widely accepted biochemical phenotype currently described in Human Phenotype Ontology. It was to be expected that severe deviations would be observed among the patients of the Digestive Service, given that it is an allele with a very high association with hyperbilirubinemia. This fact is the basis for the indication of the genetic study in these patients, trying to confirm the diagnosis of GS and as a differential diagnosis with other causes of hyperbilirubinemia. That the distribution of the genotypes of the UGT1A1*28 allele in the series of patients referred from the Oncology Service complied with the Hardy-Weinberg equilibrium was a strong indicator that: 1. Our association study ruled out that the rs3064744 polymorphism could be an allele risk of suffering from gastrointestinal tumors (which are the majority in the diagnoses of neoplasms diagnosed in these patients) and 2. this and, in general, the series of patients referred for genotyping of analyzed polymorphism with a pharmacogenetic objective, can be considered as a control population (ClinVar ID:549828; Allele ID:540561). The allelic frequency described in this group of patients was the same as that calculated in 150 individuals from the correctly selected control population [13], confirming both assumptions. The distribution of the risk allele UGT1A1*28 (TA)7 among cancer patients was 0.32, as previously calculated in the general Valencian population. The reported frequencies for the polymorphism in the databases revealed gradients from higher to lower, from more ancestral populations and more southerly latitudes to the north [14]. Our data suggest that these genotypic differences and the difficulties in establishing a confident association of an allele with low to moderate risk for a given relatively common disease may suggest spurious or biased associations. Theories supporting that this polymorphism could be a colorectal/digestive cancer risk allele were based on the fact that the UGT1A1 gene encodes an enzyme that has antioxidant activity [15]. Other articles show that these genotypic differences are not accidental and that they are justified by advantageous evolutionary effects that take advantage of the anti-inflammatory capacity of bilirubin to generate protective effects in certain populations [16-17]. As has been widely described, the codominant model (6TA/7TA genotype) showed sufficient capacity of the heterozygous allele to cause bilirubin elevation (OR: 1.66; 95% CI 1.05-2.62), while the recessive (genotype 7TA/7TA) indicated a much higher risk of the genotype of the risk allele in homozygosity (OR: 14.01; CI 95% 8.63-22.74) [4]. In both cases, statistically significant values were reached, with the sample size of the analyzed series, which also showed that the indication of the genetic study in the patients by the specialists in digestive pathology of our Hospital was very appropriate. Our study suggests that there is an evident diversity in the phenotypic expressivity of the risk allele, evaluated among individuals carrying the high-risk genotype (risk allele 7TA/7TA in homozygosis) and evidenced in the lack of diagnosis among the referred patients of the Oncology Service; in them the diagnostic suspicion of GS had never been raised. We show that such individuals had baseline bilirubin levels well below the considered lower limit of the range calculated as normal in our reference population (1.2 mg/dL). The literature has already suggested the existence of a percentage of 7TA/7TA individuals without hyperbilirubinemia (in our population 8.26%) [18-19], although it has only hinted that other compensatory molecular causes of the effect of the risk allele/genotype could coexist in these individuals [20]. Finally, it would be interesting to see if this group of patients has fewer side effects to treatments such as Irinotecan, than those associated with the higher risk genotype, such as severe degrees of diarrhea and neutropenia. The strategy would be to be able to verify if lower rates of glucuronidation are found in this group, which is the basis of the greater susceptibility to gastrointestinal and bone marrow toxicity induced by Irinotecan [21]. When comparing the biochemical phenotype between patients with genotype 7TA/7TA our study confirms that patients with suspected GS had higher minimum bilirubin values than controls. Therefore, it is concluded that the cases start from higher basal bilirubin values than the controls. The distribution of UGT1A1 (TA) n/n alleles or genotypes did not vary with sex among included individuals (p value = 0.07) [1]. However, the comparison of bilirubin levels between both sexes attributed the male sex to a greater expressiveness of the hyperbilirubinemia phenotype (OR: 2.86). The highest penetrance of the risk allele for the associated HPO terms jaundice and unconjugated hyperbilirubinemia was revealed when comparing the minimum values of total bilirubin from where the individuals started and these levels were significantly higher among men [22]. Evolutionary theories have explained the greater population distribution of certain risk alleles for certain pathologies, due to the advantage they entail for certain traits and/or conditions. Something similar has been suggested by verifying that higher frequencies of GS risk alleles and genotypes in certain populations generate slightly increased bilirubin concentrations that confer strong protective effects against diseases related to inflammation, such as cardiovascular diseases and the development of neoplasms. This protective relationship appears to be linear; it has been reported that each unit increase in systemic bilirubin concentrations has been associated with decreased cardiovascular risk and even cancer. These hypotheses are supported by evidence that plasma bilirubin concentrations below 0.58 mg/dL, although still within the current physiological range, seem to confer a greater risk to this group of diseases [22]. Perhaps this is the association that we are observing among patients with the 7TA/7TA genotype affected by neoplasms, in whom, despite their hereditary susceptibility, they start with minimum bilirubin levels below the average of individuals with said genotype. risk; this effect was even more pronounced in women, whose cancer patients still exhibit trough bilirubin levels below the lower end of the population range. In any case, the molecular basis of these variations between individuals with the same 7TA/7TA genotype in the promoter polymorphism in the UGT1A1 gene is still unknown. Statements & Declarations The authors declare that they received no funds, grants or other support during the preparation of this manuscript. The authors have no relevant financial or non-financial interests to disclose, and declare that they have no conflict of interest. The authors confirm that they have no interest or relationship with the certification or marketing of the Kit used for fragment analysis, their only interest being to validate the technique for its implementation in the methodology used in their care activity; the results obtained allowed Sanger technology to be replaced by fragment analysis, with the advantages described in the manuscript. The authors Raquel Rodríguez-López, Jorge Ferriz-Vivancos, and Irene Ferrer Bolufer contributed to the conception and design of the study. The patients Ana de Blas Zapata, Mireia Gil Raga, Ana Vallejo Antolín, Marcedes Latorre Sánchez and M José Safont Aguilera were recovering. Preparation of material Carola Guzmán Luján, Otilia Zomeño Alcalá, Ana Cristina Requena Piedrabuena and Jennifer Valero García, data collection Carola Guzmán Luján and Ana Cristina Requena Piedrabuena. The analyzes were carried out by Raquel Rodríguez-López, Jorge Ferriz-Vivancos and Joan Más Lloret. The first draft of the manuscript was written by Raquel Rodríguez-López and Jorge Ferriz-Vivancos. All authors commented on previous versions of the manuscript, read and approved the final manuscript. In any case, it is an observational study that collects a series of patients analyzed in our clinical care practice, so it does not require specific ethical approval. Informed consent was obtained from all individual participants included in the study. All the patients included in the study gave their informed consent prior to the genetic study, consensual and standardized for each genetic study carried out in our Hospital Center; Informed Consent document was approved by the Ethics Committee of our Hospital, in accordance with those used for care practice in our care setting. We thank the nursing and technical staff of the Clinical Analysis Service of the General Hospital of Valencia for their collaboration and good teamwork.Keywords
Article Details
INTRODUCTION
PATIENTS AND METHODS
RESULTS
DISCUSSION
Funding
Conflict of interests
Author contributions
Ethical approval
Consent to Participate
ACKNOWLEDGEMENTS
References
Article Views: 1643
Journal Statistics




Discover More: Recent Articles


