Characterization of Two Novel EF-Hand Proteins Identifies a Clade of Putative Ca2+-Binding Protein Specific to the Ambulacraria
Arisnel Soto-Acabá1, Pablo A. Ortiz-Pineda2, Joshua G. Medina-Feliciano1, Joseph Salem-Hernández3, José E. García-Arrarás1, *
1Department of Biology, University of Puerto Rico, Río Piedras Campus
2Laboratorio de Biología Molecular y Genómica. Fundación InnovaGen. Popayán. Colombia.
3School of Medicine, University of Puerto Rico, Medical Sciences Campus
*Corresponding author: José E. García-Arrarás, Department of Biology, University of Puerto Rico, Río Piedras Campus, USA.
Received: 22 December 2021; Accepted: 13 January 2022; Published: 03 February 2022
Article Information
Citation:
Arisnel Soto-Acabá, Pablo A. Ortiz-Pineda, Joshua G. Medina-Feliciano, Joseph Salem-Hernández, José E. García-Arrarás. Characterization of Two Novel EF-Hand Proteins Identifies a Clade of Putative Ca2+-Binding Protein Specific to the Ambulacraria. Journal of Bioinformatics and Systems Biology 5 (2022): 1-25.
View / Download Pdf Share at FacebookAbstract
In recent years, transcriptomic databases have become one of the main sources for protein discovery. In our studies of nervous system and digestive tract regeneration in echinoderms, we have identified several transcripts that have attracted our attention. One of these molecules corresponds to a previously unidentified transcript (Orpin) from the sea cucumber Holothuria glaberrima that appeared to be upregulated during intestinal regeneration. We have now identified a second highly similar sequence and analyzed the predicted proteins using bioinformatics tools. Both sequences have EF-hand motifs characteristic of calcium-binding proteins (CaBPs) and N-terminal signal peptides. Sequence comparison analyses such as multiple sequence alignments and phylogenetic analyses only showed significant similarity to sequences from other echinoderms or from hemichordates. Semi-quantitative RT-PCR analyses revealed that transcripts from these sequences are expressed in various tissues including muscle, haemal system, gonads, and mesentery. However, contrary to previous reports, there was no significant differential expression in regenerating tissues. Nonetheless, the identification of unique features in the predicted proteins and their presence in the holothurian draft genome suggest that these might comprise a novel subfamily of EF-hand containing proteins specific to the Ambulacraria clade.
Keywords
<p>Ambulacraria</p>
Article Details
1. Introduction
Modern genome and transcriptome studies allow for the identification and discovery of hitherto unknown sequences that code for different types of proteins. This discovery process has been possible due to the ease by which DNA and/or RNA sequences are obtained, even from non-model organisms that make available millions of sequences for comparative analyses. Our group has focused on transcriptomes obtained from normal and regenerating tissues of an echinoderm, the sea cucumber Holothuria glaberrima [1-4]. Studies in this model have been done to explore gene expression of intestinal and nervous systems in an attempt to expand our knowledge of the Echinodermata, a phylum which lies on the evolutionary branch of chordates [5, 6]. In this effort, we have constructed several transcriptomic libraries using high throughput sequence analyses, including EST (expressed sequence tag) analyses [3], 454 and Illumina sequencing [7, 8]. Moreover, we have performed differential gene expression studies, particularly microarrays and transcriptomic comparisons between normal and regenerating tissues. The results from these experiments have been a large number of differentially expressed genes associated with the regenerating tissues. Out of these hundreds of genes, we have focused on the study of unknown sequences that show increased expression during regeneration. One of these molecules corresponds to a previously unidentified transcript from H. glaberrima that was shown to be upregulated during the initial stages of intestine regeneration by microarray analyses [3,8]. The sequence was annotated to public databases as Orpin (GU191018.1, ACZ73832.1) on 12-13-2009.
We now provide a full report on the putative Orpin sequence including the prediction of an N-terminal signal peptide, which is characteristic of secreted proteins. Moreover, we have discovered an additional Orpin isoform in H. glaberrima and provide a full description of both Orpin isoforms. Both sequences are newly discovered putative EF-hand coding proteins with structural characteristics that are evolutionarily related to this group of proteins. We have also probed other available databases and have found previously undescribed sequences whose similarities suggest they are part of the Orpin family, a protein family that appears to be restricted to the Ambulacraria clade.
2. Materials and Methods
2.1 Ethics Statement
This research deals only with invertebrate animals, thus the University of Puerto Rico IACUC waives ethical approval of research performed on invertebrates. Animals were sacrificed by immersion in ice cold water for 29-30 min and then sectioning the anterior part of the animal close to the oral nerve ring, which accounts for the main component of the nervous system.
2.2 Animals
Adult specimens (10-15 cm in length) of the sea cucumber H. glaberrima were collected in coastal areas of northeastern Puerto Rico and kept in indoor in aerated seawater aquaria at room temperature (RT: 22°C ± 2°C). Evisceration was induced by 0.35 M KCl injections (3-5 mL) into the coelomic cavity [1]. Eviscerated animals were let to regenerate for 3, 5, 7, 10, and 14 days before the dissection and tissue extraction. For the dissection, organisms were anesthetized by placement in ice-cold water for 1 h. [1,3,9]. Dissected tissues were rinsed in ice-cold filtered seawater and processed for RNA isolation.
2.3 RNA Extraction and cDNA Synthesis
RNA extraction was performed on tissue extracts of normal and 3, 5, 7, 10, and 14 dpe animals. Extracted tissues included gonads, mesentery, haemal system, respiratory tree, longitudinal muscle, and radial nerve cords. After dissection, tissues were placed in 1 mL of TRIzol reagent (Invitrogen), homogenized with a Power Gen Model 125 Homogenizer (Thermo Scientific) and incubated 30 min on ice. These samples were mixed vigorously with 200 μL of chloroform and incubated 10 min at RT. After centrifuged at 12,000 rpm at 4°C, the aqueous RNA phase was separated, mixed with 70% ethanol, and transferred to an RNeasy Mini Kit column (QIAGEN) for deoxyribonuclease (DNase) treatment (QIAGEN). Total RNA was extracted following the manufacturer’s protocol. The concentration and purity of the total RNA was measured using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific). The cDNA was synthesized from 1 μg of the total RNA using the ImProm-II Reverse Transcription System (Promega) and oligo (dT) 23 primers.
2.4 Semi-quantitative RT-PCR
RT-PCR reactions were performed using cDNAs prepared from extracted RNA. These reactions were set up in a reaction volume of 25 μL with the final concentration of the PCR primers of 100 nM. Specific primers for the most variable regions between Orpin A and Orpin B sequences were designed using OligoAnalizer tools from the Integrated DNA Technology webpage (www.idtdna.com). The primers used were: Orpin B forward: 5’-ACAGGGAGTACAAACAGTCGTCAA-3’ and Orpin B reverse: 5’-CTATTTACTCTGCAACTGACACTTTCT-3’; Orpin A forward: 5’-ACTTCTGCAGAATCAGTTGTTAAGA-3’ and Orpin A reverse: 5’-TTCAGTGGAGTCGCCAAC-3’. RT-PCR reactions were performed on three independent RNA samples purified from each of the regeneration stages (previously mentioned) as well as from the normal intestines. The PCR amplification was done by an initial denaturation step of 94°C (45 s), a primer annealing step of 50.2°C (45 s), and an extension step of 72°C (45 s) with a final additional 72°C (10 min) for 28 cycles for Orpin A, 26 cycles for Orpin B, and 26 cycles for NADH, as the amplification parameters for each pair of primers. All samples were analyzed in triplicate. Additional tissues were amplified for 35 cycles (2-4 replicates). The relative expression of Orpin A and Orpin B was normalized relative to the expression of the housekeeping gene NADH dehydrogenase subunit 5 using ImageJ software [10] from the optical density values from electrophoresed sample bands on 1% agarose gels, using a Molecular Imager ChemiDoc XRS+ (BioRad). The primers used for the NADH sequence amplification were: forward: 5’-CGGCTACTTCTGCGTTCTTC-3’ and reverse: 5’-ATAGGCGCTGTCTCACTGGT-3’. The Orpin A and Orpin B sequences were confirmed by sequencing excised electrophoresed sample bands at the Sequencing and Genotyping Facility (UPR-RP).
2.5 Bioinformatics Analyses
Homolog sequences were identified and retrieved from the NCBI GeneBank protein database [11] using the original Orpin sequence previously identified [8] as a query. BLASTp [12, 13] were performed against the public non-redundant protein database in GeneBank. Conserved domain identification and UTR analysis were performed using CDD [14], RegRNA [15], UTRScan [16] and PSIPRED [17,18], ScanProsite [19], InterProScan 5 [20,21], Phobos [22], SignalP 5.0 [23], and Phobius [24] on Geneious 11.1.5 software (https://www.geneious.com). Sequence alignments were carried out with MUSCLE [25] (10 iterations) and the Blosum62 matrix and edited with Geneious software 11.1.5 (https://www.geneious.com). Note: It is possible that there are N-terminal sequencing artifacts on two annotated sequences from A. japonicus sequences (ARI48335.1 and PIK49419.1). If we delete the residues from the predicted cytoplasmic N-terminal region from the ARI48335.1 sequence and from PIK49419.1 up until their next methionine, they also show a predicted signal peptide of 21 residues each.
2.6 Phylogenetic Analysis
EF-Hand proteins and other similar sequences were retrieved from literature and protein database as mentioned in results section and the multiple sequence alignment was performed on MAFFT v7.309 [26] with BLOSUM62 scoring matrix, gap open penalty of 1.57, and offset value of 0.123. For the tree building, the Maximum-Likelihood analysis was done using JTT model of sequence evolution with 1000 bootstraps using PhyML 3.0 [27] plugin using Geneious 11.1.5 software (https://www.geneious.com). The corresponding sequences are included in S1 Table. The tree was edited for better visualization and colors in iTOL v4 online tool [28]. The (frog) X. laevis, (mouse), M. musculus, and (human) H. sapiens calcineurin A sequences were selected as outgroups and does not contain EF-Hand motifs. In addition, the Orpin homologs from A. japonicus ARI48335.1 and PIK49419.1 were edited for the analyses by deleting the N-terminal residues down to the second predicted methionine for the reason mentioned above.
2.7 Genome Analysis
Genome loci of H. glaberrima Orpin transcripts were annotated manually by BLASTn alignments of the sequences against the draft genome (NCBI ID: PRJNA497079) [29]. To narrow our search, we initially aligned the identified Orpin transcripts to the genome to determine the scaffolds in which they were present and then aligned them to these scaffolds (22267 and 51815) with BLASTn. Alignments and annotation were carried out manually identifying each exon of both genes. Then, TopHat (v2.1.1) with default parameters was utilized to confirm the previously identified gene junctions. For the TopHat analysis we utilized raw RNA-seq reads (NCBI SRA BioProject: PRJNA660762) from previous H. glaberrima studies. Identified junctions that were not congruent with our current search of these Orpin transcripts were not considered. OrpinA & B sequence alignments were performed using MAFFT (v7.312).
2.8 Statistical Analyses
Statistical significance of the resulting data was evaluated through one-way ANOVA using the JMP®, Version 12. SAS Institute Inc, Cary, NC, 1989-2019. The multiple comparison procedure and statistical test Tukey-Kramer HSD (honestly significant difference) was used to determine significant differences between means from optical densities determined by Image J software as mentioned before [10]. The Tukey-Kramer results are displayed as small circles for high number of data points and large circles for low number of data points. The large red circle shows significant differences to small grey circles sample means. All values were reported as the mean ± standard mean error, including mean diamond with confidence interval ([1 − alpha] x 100), and outlier box plot from a quantiles report. While a P < .05 and P < .001 were considered to indicate statistical significance difference between groups.
3. Results
3.1 Identification of the Original Orpin (Orpin A) Sequence and Characterization of a Second Orpin Isoform (Orpin B)
The original report [3] described a contig sequence (4766-1) which was later annotated as Orpin. This contig was used as a template to identify the remaining nucleotides upstream from the open reading frame (ORF) region through RACE-PCR analysis [30]. The Orpin sequence is composed of 106 nucleotides from the 5’ UTR and 291 nucleotides from the 3’ UTR with a 366 nucleotide ORF (plus stop codon) that encodes a putative 122 amino acid peptide followed by a stop codon (Figs 1 and 2). The nucleotide composition of this gene sequence was validated by sequencing the RT-PCR products amplified from a normal intestine tissue cDNA sample (Fig 2). At the time it was annotated in the NCBI database (ACZ73832.1; 12/13/2009), there was no match with other sequences. Two similar sequences from the hemichordate Saccoglossus kowalevskii were later added as Orpin-like sequences (XP_006824981.1 and XP_002736736.1).
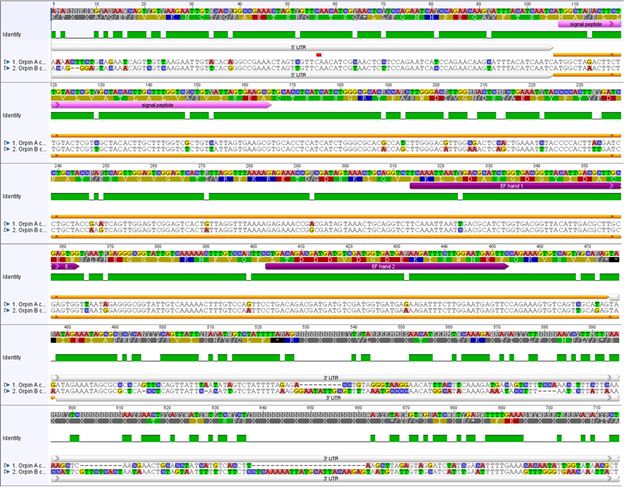
Figure 1: Orpin A and Orpin B are isoforms. Differences between sequences are highlighted. White bars: 5’ UTR and 3’ UTR regions of both sequences; pink bar: predicted signal peptides; orange bar: ORF regions; purple bars: predicted EF-hand motifs; green: conservation level; top sequences: nucleotide and amino acid consensus sequences. Differences between nucleotide sequences are highlighted. It is shown a significant difference, especially between both 3’ UTR sequences. Analysis was done using the MAFFT plugin in Geneious 11.1.5.

Figure 2: Primers for sq-RT-PCR of Orpin A. Orpin A UTRs regions (blue boxes) and coding region (green box) of the Orpin A gene. Primer sequences designed to specifically amplify Orpin A (light red letters). Primer sequences used for the identification of the original Orpin sequence in previous reports (green letters). These primers were designed prior to identification Orpin isoform.
After performing further in-depth analyses of the available transcriptome libraries from regenerating and non-regenerating intestine and regenerating and non-regenerating radial nerve, we discovered an additional highly similar sequence that was identified as a putative Orpin isoform. This new putative protein shared 90% identity and 98% similarity with the original Orpin sequence but displayed different UTR’s from the original sequence. We refer to this sequence as Orpin B to differentiate it from the original Orpin which we refer from now on as Orpin A. The sequence corresponding to Orpin B was also validated through RT-PCR amplification and sequencing (Fig 3 and S1 Fig). Orpin B mRNA sequence is composed of 103 nucleotides from the 5’ UTR and 364 nucleotides from the 3’ UTR (Figs 1 and 3). Its ORF is 369 nucleotides (plus stop codon) long and encodes a putative 123 amino acid protein.

Figure 3: Primers for sq-RT-PCR of Orpin B. Orpin B UTR sequences (blue boxes) and coding region (green box). Primer sequences designed to specifically amplify Orpin B (blue letters).
Sequence comparisons among the two Orpins from H. glaberrima and the two Orpin-like sequences from S. kowalevskii show that the latter shared 46-50% identity and 76-77% similarity with the Orpin A (Fig 4). Similarly, Orpin B translated amino acid sequence shared 46-50% identity and 66-67% similarity with the sequences from S. kowalevskii (Fig 4). Furthermore, we identified three additional putative Orpin homologs from another sea cucumber species, Apostichopus japonicus, one from the starfish Acanthaster planci, and two from the sea urchin Strongylocentrotus purpuratus with expected values (E-value < 0.001 and total scores > 47.8). All Orpin-like sequences contain one domain that is predicted to be a calcium-binding domain composed of two EF-hand motifs at their carboxy-terminal (Figs 5 and 6).
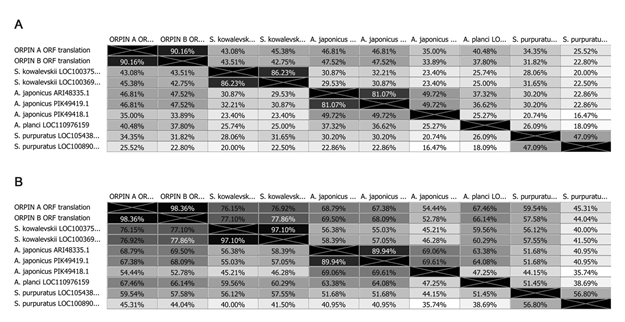
Figure 4: Orpin homologs pairwise sequence divergence. Translated amino acid sequences comparison by (A) identity% and (B) similarity%. The alignments were done using Muscle with 50 iterations using Geneious 11.1.5.
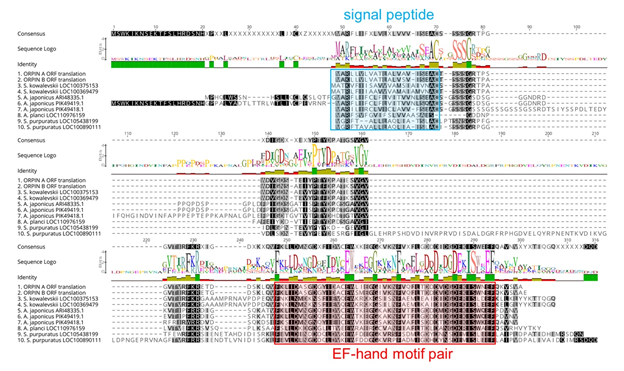
Figure 5: Orpin homologs alignment. The most conserved residues are indicated by letters in black boxes, green identity regions, and large cartoon letters at the sequence Logo. The exception is PIK49419.1 because 20 amino acid residues from the N-terminal portion are not compared to other sequences. We can see the additional N-terminal regions from A. japonicus sequences ARI48335.1 and PIK49419.1 that did not match to the other homologs. Blue box: signal peptide prediction; red box: EF-Hand motif pair prediction. This alignment was done by Muscle plugin with 50 iterations using Geneious 11.1.5.
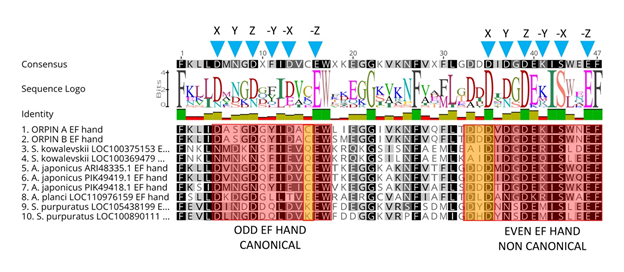
Figure 6: Orpin homologs EF-hand motifs alignment. The predicted odd EF-hands match with the canonical EF-hand pattern and the predicted even EF-hands were identified as non-canonical motifs (14 residues vs 12 residues) (red boxes). The non-canonical motifs are similar to vertebrates S100s. Predicted calcium coordinating residues from the identified EF-hands patterns are indicated by blue triangles. Holothurian Orpins contain a Cys residue at the −Z−1 position of the predicted calcium-binding loop (left orange box). The characteristic residues from Orpin residues are highlighted by orange boxes. Alignment was done using the MAFFT plugin in Geneious 11.1.5.
3.2 Domain analyses
Orpin A and Orpin B amino acid sequences were analyzed using different bioinformatics tools (refer to methodology) for evidence that could point towards a possible function. After evaluating these sequences for domain composition using InterProScan and NCBI’s CDD, and Phobos [14, 20-22], we identified that both sequences contain putative calcium-binding domain regions. Both Orpin isoforms shared identical calcium-binding loops residue composition. The key residue positions that participate in calcium chelation within these loops are conserved when compared with other known EF-hand proteins. The X, Y, Z, −X, −Z positions from each loop of the EF-hands are Asp, Asp, Asp, Asp, Glu (“odd loop”) and Asp, Asp, Asp, Ser, Glu (“even loop”), respectively (Fig 6). The only difference is located downstream to the “odd” loop. There are two consecutive amino acids, Leu87 and Ile88, immediately after the Trp86 (−Z+1) of the first calcium-binding loop from Orpin A which are changed to Ser87 and Met88 in Orpin B. Interestingly, Orpin A and Orpin B included a Cys residue at −Z−1 position which is particular to both sea cucumber sequence homologs and is an unusual feature in EF-hand proteins. When we compared H. glaberrima Orpin EF-hand sequences to the other identified putative homologs from S. kowalevskii, A. japonicus, A. planci, and S. purpuratus, we found that additional positions are highly conserved as well. All Orpin sequences share conserved positions at X−4 (Phe), −Z (Glu), −Z+1 (Trp) and −Z+6 (Gly) positions from the “odd” EF-hands, and X−8 (Phe), X (Asp), Z (Asp), −X−1 (Ile), −X (Ser), −Z (Glu) and −Z+1 (Phe) positions from the “even” EF-hands. Alternatively, there are residues particular to the EF-hands from H. glaberrima Orpin isoforms, such as Ala at X+1, Ser at Y, Ala at −X+1 from the odd EF-hand, and Val at X+1 and Asn at −X+2 from the even EF-hand. Moreover, there are residues that are particular to the holothurians such as Lys at X−3, Cys at −Z−1, Lys at −Z+9 from the odd EF-hand, and Lys at −Y from the even EF-hand (Fig 6).
Additional bioinformatics analyses revealed the presence of a signal peptide in the N-terminal of both isoforms (Fig 7). These signal peptides are 20 amino acids long each and are mainly composed of hydrophobic residues. The predicted signal peptides of Orpin A and Orpin B are nearly identical, with the exception of two residues at positions 3 (Arg/Lys) and 15 (Ala/Ser). Furthermore, InterProScan and Phobius identified the same region of 20 residues as a possible transmembrane region. In both cases, a high probability of a cleavage site was identified at the Cys21 residue of each isoform. If the signal peptide is eliminated, the remaining sequence is predicted to be localized outside the cytoplasm (Fig 7). This strongly suggests that these peptides could be secreted to the extracellular space and not targeted to the membrane of other cell organelles.
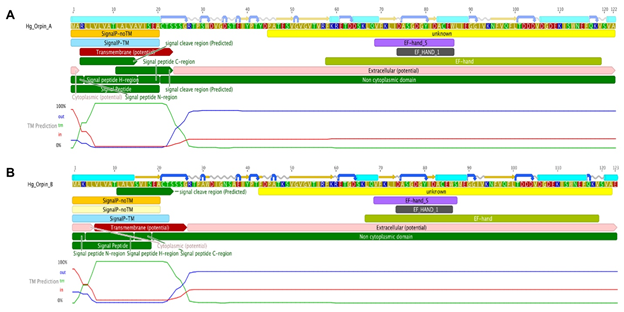
Figure 7: Orpin A and Orpin B bioinformatics characterization. (A) Orpin A and (B) Orpin B have predicted signal peptides at their transmembrane N-terminal regions including cleavage sites. Also, the two isoforms have predicted EF-hand motifs in their non-cytoplasmic regions. These were predicted by various bioinformatics plugin tools using Geneious 11.1.5.
The average length for the predicted signal peptides of the Orpin homologs is 20-22 residues based on Signal P and Phobius predictions [23,24,31], from the initial Met residue. The predicted signal peptides from the two S. kowalevskii sequences are longer (22 residues) than the other Orpins. The predicted signal sequence of a sea urchin homolog (XP_011664021.1) is the shortest (18) of the Orpins (Fig 5). The UTR’s of the H. glaberrima Orpin sequences were also analyzed. Even though the 5’ UTR’s from Orpin A and Orpin B are 80.2% identical, there are 20 nucleotide differences between them, mainly SNPs. A ribosome binding site within the 5’ UTR of each isoform sequence was identified. In contrast, the retrieved 3’ UTR’s of both Orpin isoforms were completely different. Polyadenylation sites were identified in both Orpin A [8] and Orpin B downstream to their corresponding stop codons. Interestingly, these analyses revealed the presence of two putative Musashi binding elements (MBEs) within the 3’UTR of Orpin A. Even though the available retrieved 3’ UTR from Orpin B is longer than its paralog, no MBEs were identified within this sequence. Surprisingly, two putative MBEs were also found within the coding sequence of each Orpin isotype.
3.3 Orpin phylogenetic analysis
In order to determine the relationship of the different Orpin homologs among themselves and with other EF-hand proteins, a phylogenetic tree was constructed with the PhyML program using a MAFFT alignment as input [26, 27]. Orpin A and Orpin B amino acid sequences were used as probes to identify the closest sequences through BLAST searches against the public databases. In addition, representative sequences from different EF-Hand subfamilies of various organisms were obtained from the scientific literature and available databanks. These sequences included members from the following protein families: S100s, calcineurin, recoverin, calbindin, parvalbumin, oncomodulin, osteonectin, SPARC, troponin C, calmodulin, centrin, Spec, and recoverin (S1 Table). Thus, these sequences were used for the final alignment to generate the phylogenetic tree.
The results from this analysis cluster Orpin and the identified hypothetical homologs from S. kowalevskii (acorn worm), A. japonicus (sea cucumber), A. planci (sea star), and S. purpuratus (sea urchin) together with a bootstrap value of 92, separately from other subfamilies of EF-Hand proteins (Fig 8). The other EF-Hand protein sequences cluster together as individual groups. The Orpin-like cluster was the most distant group after the outgroup sequences of mouse and frog calcineurin A, which do not contain EF-Hand motifs, suggesting that Orpins have evolved separately and are not direct homologs of EF-Hand proteins from other species. As expected, H. glaberrima Orpins were close to the other sea cucumber A. japonicus Orpin-like sequences. The most distant Orpin homologs were those from sea urchin S. purpuratus. The closest protein cluster was the osteonectins, BM-40, or SPARC proteins, which comprise a group of secreted CaBP modulators with a single pair of EF-Hand motifs. After these, the other group of proteins that appeared close by were the S100s, which also are small secreted proteins with two EF-hand motifs. The tree also showed the other outgroup EF-hand lacking protein, calcineurin A from humans, was placed separately from the other EF-Hand proteins of a high number of motifs (3 to 6 EF-Hands).
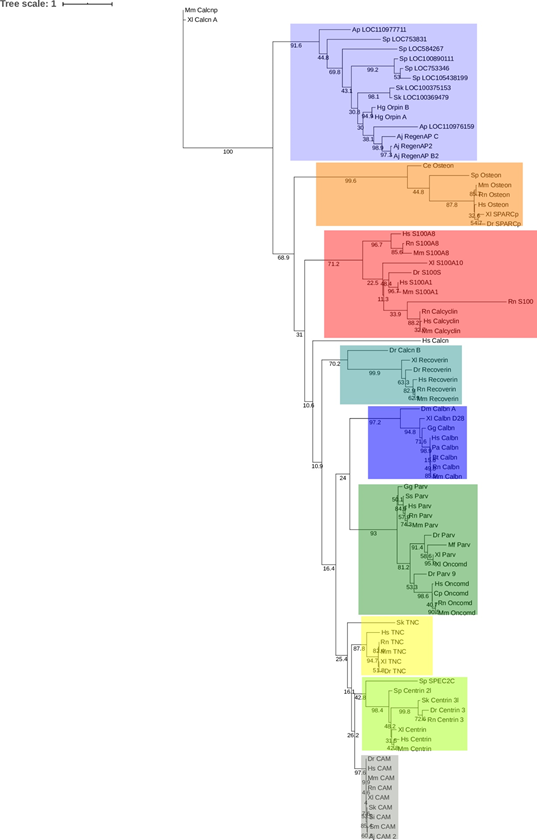
Figure 8: Orpin isoforms are specific to the Ambulacraria clade. EF-hand protein representative sequences from different subfamilies were aligned to build a phylogenetic tree. Orpin homologs were clustered together as a group, separated to the other EF-hand proteins. The tree was made using the PhyML plugin ran through Geneious 11.1.5. The parameters used for this analysis were JTT model of amino acid substitution and 1000 bootstraps. Scale bar: 1. Protein sequences accession numbers are included in S1 Table.
3.3 Orpin gene is expressed in several tissues of H. glaberrima
In order to determine the distribution of Orpin expression, mRNA was obtained from different tissues or organs of normal (non-regenerating) H. glaberrima specimens and processed for PCR analysis. The tissues and organs selected were: small intestine, large intestine, mesentery, radial nerve complex, longitudinal body wall muscle, gonads, and respiratory tree. Primers were designed for the specific detection of Orpin A and Orpin B mRNA sequences (Figs 2 and 3). Transcript levels were evaluated relative to the expression of NADH subunit 5, a constitutively expressed housekeeping gene. The results showed that Orpin A and Orpin B shared similar tissue specificity (Figs 9 and 10). Transcripts were detected in the gonads, muscle, mesentery, and haemal system but not in the respiratory tree nor in the nerve. Tissue expression varies significantly, with higher expression levels in the mesentery followed by the expression in muscle and gonads where it is slightly higher than in other tissues. Interestingly, a faint second lighter band was detected from Orpin B samples from gonads and muscle tissues.
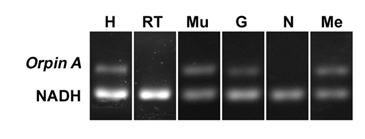
Figure 9: Orpin A expression in different tissues. Composite image from RT PCR amplification of Orpin A from H. glaberrima tissues. Orpin A expression (top band) was detected in haemal system (H), muscle (Mu), gonads (G), and mesentery (Me) relative to the expression of NADH. Orpin A was detected neither in the nerve (N) nor in the respiratory tree (RT). The image is a composite from different gels and is divided by a white line.
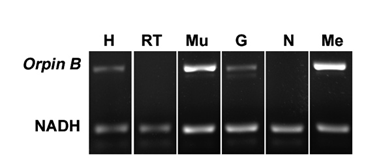
Figure 10: Orpin B expression in different tissues. Composite image from RT PCR amplification of Orpin B from H. glaberrima tissues. Orpin B expression (top band) was detected in haemal system (H), muscle (Mu), gonads (G), and mesentery (Me) relative to expression of NADH. Orpin B was detected neither in the nerve (N) nor in the respiratory tree (RT). The faint band below the NADH band corresponded to primer dimers. The image is a composite from different gels and is divided by a white line.
3.4 Orpin expression during intestinal regeneration in the sea cucumber H. glaberrima
Previous results from our laboratory have shown that Orpin was differentially expressed in regenerating intestinal tissues when compared to normal intestinal tissues [3, 8]. In order to validate the upregulation of this novel sequence during regenerative processes, Orpin transcript levels were measured during different stages of intestine regeneration. In contrast to previous experiments where no particular effort was made to separate the intestine of normal animals from the attached mesenteries, in the present experiments we measured separately the intestine (a mixed portion from the small intestine and from the large intestine) and the mesentery that attaches the intestine to the body wall, for the normal (non-regenerating) samples. Orpin transcript levels were measured relative to the housekeeping gene NADH subunit 5. The gene expression levels were monitored using semi-quantitative RT-PCR of tissue extracts from 3 days post evisceration (dpe), 5 dpe, 7 dpe, and 10 dpe along with tissues from non-eviscerated intestine and mesentery.
Previously it was found that the expression levels of Orpin A increased after 3 days of intestine regeneration and then gradually returned to the basal levels at 14 days of regeneration. In contrast, to those findings [3,8], there was no statistically detected difference found between the transcript expression of Orpin A from normal intestine samples and those from any of the studied regenerative days (Data not shown). This was also true for Orpin B (Data not shown). However a high differential expression was detected between tissues from Orpin A from normal mesentery and tissues from 7-10 dpe sample group, with a P < .05 (P=.002) (Fig 11). Orpin B exhibited a high differential expression between tissues from normal mesentery and tissues from 3-5 dpe with a P < 0.05 (P=.02), and 7-10 dpe with a P < .001 (Fig 12).
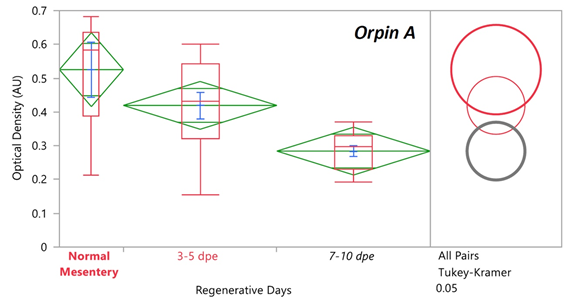
Figure 11: Orpin A expression during intestine regeneration grouped tissues. Semi-quantitative RT-PCR amplification of Orpin A transcripts from mRNA samples from different intestine regenerative days compared to the corresponding expression in samples from normal intestine (NI) and normal mesentery (NM). A statistical high differential expression was found between NM and 7-10 days post evisceration (dpe) (P < .05; P = .002); and between 3-5 dpe and 7-10 dpe (P < .05; P = .03) as indicated in the all pairs Tukey-Kramer HSD test. The large red circle (low number of data points) displays the significant difference between the small grey circles (high number of data points) group means. Red boxes: outlier box plots summarizing the distribution of points at each factor level from the quantiles report. Green diamonds: sample mean and confidence interval ([1 − alpha] x 100). Blue lines: standard mean error. JMP®, Version 12 software was used for the statistical analyses.
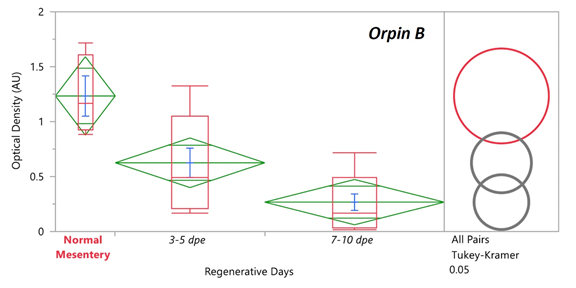
Figure 12: Orpin B expression during intestine regeneration grouped tissues. Semi-quantitative RT-PCR amplification of Orpin B transcripts from mRNA samples from different intestine regenerative days compared to the corresponding expression in samples from normal intestine (NI) and normal mesentery (NM). A statistical high differential expression was found between NM and 3-5 dpe (P < .05; P = .02), and to 7-10 dpe (P < .001) as indicated in the all pairs Tukey-Kramer HSD test. The large red circle (low number of data points) displays the significant difference between the small grey circles (high number of data points) group means. Red boxes: outlier box plots summarizing the distribution of points at each factor level from the quantiles report. Green diamonds: sample mean and confidence interval ([1 − alpha] x 100). Blue lines: standard mean error. JMP®, Version 12 software was used for the statistical analyses.
Interestingly, we found a different expression profile between Orpin A and Orpin B transcript levels. While both Orpin forms show subsequently decreases in their expression to similar levels at 7-dpe to 10-dpe, the decrease of Orpin B seems to occur much faster than that of Orpin A.
3.5 Orpins are found in H. glaberrima draft genome
We were able to identify both Orpin transcripts in the draft genome of H. glaberrima. The complete sequence of Orpin A appeared to be present in our genomic data with a total of three exons (Figure 13). However, we could only identify the first two exons of Orpin B (Figure 13), which could be mainly attributed to the fragmented nature of the draft genome. Nonetheless, the scaffold containing the first two exons of Orpin B only contained an additional 181 nucleotides after the end of the second exon, thus limiting the possibility of finding the third exon. Junctions within each of the identified exons were supported by TopHat, which allowed us to clearly delineate the start/end of each exon. Other than the junctions of these gene structures TopHat identified other multiple junctions within these regions (125 junctions in total).
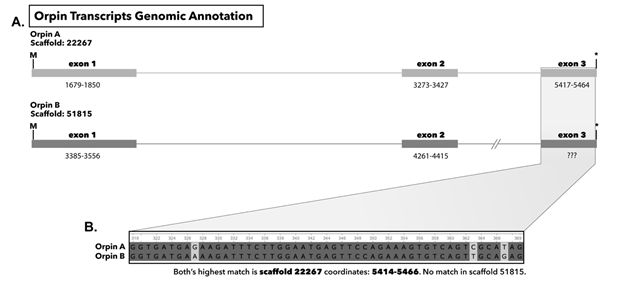
Figure 13 (A): Genomic annotation of Orpin A and Orpin B in the draft genome of H. glaberrima. Numbers under each exon represent coordinates in each scaffold. (B) Alignment of exon 3 of both Orpin transcripts obtained from transcriptomic data; both matched the same sequence on scaffold 22267 coordinates 5414-5466.
4. Discussion
We have now described the presence of two predicted EF-hand domain-containing proteins from the sea cucumber H. glaberrima. These putative proteins apparently belong to a unique group that is present in echinoderms and hemichordates. According to the mRNA distribution in the sea cucumber, the translated proteins are expressed in multiple organs. Moreover, they are highly represented within the mesentery of the normal and regenerating intestine. The possibility that these are Ca2+-binding proteins is discussed in the following sections.
4.1 Orpins are novel genes
When the first Orpin sequence from H. glaberrima was identified, no other sequence that showed significant similarity to it could be found within databases [3, 8]. A few months later, two highly similar sequences (and possible homologs) were identified in the hemichordate Saccoglossus kowalevskii (acorn worm) and were added to the databases. These sequences were annotated with accession numbers XM_006824918.1, XP_006824981 (E-value: 9E-31) and XM_002736690.2, XP_002736736 (E-value: 2E-29). Later on, several homologs from closely related organisms of the Echinodermata phylum were added to the public databases: two from the sea urchin Strongylocentrotus purpuratus, three from other sea cucumber Apostichopus japonicus, and one from the starfish Acanthaster planci. The finding of an additional Orpin sequence in H. glaberrima increased to ten the known sequences and suggested that these sequences belong to a novel family of proteins within a group of metazoans. In all cases, the sequences have been annotated with little or no descriptive information other than their tissue/organism from where they originated. At present, Orpins appear to be restricted to the Ambulacraria clade (the group that encompasses echinoderms and hemichordates), however, it remains to be seen if, with the sequencing of other animal genomes, the specificity of Orpins to the Ambulacraria still stands.
Although the H. glaberrima A and B Orpin variants share a high percentage of similarity at the nucleotide and protein levels, our data suggest that they correspond to distinct genes. First, the nucleotide and putative amino acid differences are distributed throughout the complete sequence of both variants. Second, even though the 5’ UTR sequences of both Orpin sequences share a large similarity, they are not identical, and similar to the coding region, have multiple nucleotide differences distributed along the nucleotide sequences. Third, their 3’ UTR sequences are very different, sharing minimal similarities. Fourth, their exons were found in different scaffolds of our draft genome. Finally, other species also have more than one Orpin-like gene. For example, sequence information from Orpin homologs from S. kowalevskii and S. purpuratus were annotated as located in different loci. These differences are characteristic of different genes rather than allele variants or products from differential splicing. Nonetheless, in spite of these results that suggest two different isoforms originating from two different genes, the genome information is inconclusive since it could be argued that the sequences found in the two scaffolds could be aligned in a single gene that is differentially spliced. Moreover, it should be emphasized that while Orpin A and B mRNAs have been identified and sequenced from various tissues (see below), the S. kowalevskii, A. japonicus, A. planci, and S. purpuratus sequences are hypothetical mRNA/protein-coding sequences that remain to be characterized. Even though it was expected that H. glaberrima Orpin sequences would be more similar to those from the other sea cucumber A. japonicus, the results showed that they shared higher similarity to the hemichordate S. kowalevskii sequences. This can be attributed to longer N-terminal regions from two of the A. japonicus sequences (ARI48335.1 and PIK49419.1) that did not match with any of the other homologs. These additional regions were annotated as part of the corresponding ORFs due to an identified methionine upstream to the one that matched the other homologs. Given the fact that these sequences were not validated, we need to consider the possibility that these first encoding methionine residues could be the result of a PCR artifact, and could be in fact part of their corresponding 5’ UTR regions. Hopefully, the characterization of Orpin isoforms will provide essential insights that eventually would make feasible the characterization of these homolog sequences.
4.2 Are Orpins calcium-binding proteins?
The mainframe of the study of uncharacterized CaBPs is the rigorous analysis of the residue composition of the EF-hand domains coupled to structural and functional experimentation. Thus, understanding the architecture of EF-hand domains provides a hint of the role that a particular EF-hand protein might have. One of the most prominent characteristics of the putative protein Orpin is the presence of EF-hand motifs that are a distinctive signature of calcium-binding proteins. The EF-hand motif has been used as a standard of reference for the description of the calcium-binding loops from the corresponding domain by the residues at key positions for the chelation of each calcium ion. Even though the EF-hand motifs are a characteristic feature of many CaBPs, few of them consist of less than four EF-hands. As mentioned before, Orpin A and Orpin B paralogs comprise a single putative calcium-binding domain composed of two EF-hands. The canonical EF-hand motif topology is a helix-loop-helix conformation, which regularly binds calcium ions [32,33]. Usually, this conformation is composed of a highly conserved 12 residues calcium-binding loop flanked on both sides by alpha-helices [34]. The residues that participate in calcium coordination were labeled as X, Y, Z and −X, −Y, −Z [33,34], and those conserved positions are conventionally used as a reference frame to analyze the calcium-binding potential and dynamics of the EF-hand CaBPs. A typical EF-hand domain is composed of two calcium-binding loops containing motifs flanked by alpha-helices. The adjacent alpha helices are named incoming and exiting helices from the odd (N-terminal) and even (C-terminal) EF-hand motifs [36-39]. H. glaberrima Orpin isoforms odd calcium-binding loops are composed of highly conserved key amino acids of the canonical domain structure. We showed that these domains shared a high level of conservation in the residues that participate in the chelation of calcium ions. However, the “even” calcium-binding loop from sea cucumber Orpins slightly deviates from the canonical pattern. The conserved Gly6 residue that provides for loop flexibility is substituted by a Glu6. This substitution is well conserved throughout all the available Orpin homologs with the exception of the starfish sequence. Also, they share a conserved Trp13 at position −Z+1 of the “odd” EF-hand. Furthermore, this conserved residue seemed to be particular to Orpins after comparison to the other 84 EF-Hand sequences from this study. Interestingly, a Cys11 residue is particular to the holothurian Orpins.
In view of these facts, Orpin homologs can be classified as novel EF-hand proteins. Although the bioinformatics analysis strongly suggests that they might be a new type of CaBPs, in order to assure this, experimental confirmation of the actual binding of calcium ions will be required along with the phylogenetic evidence provided in this study.
4.3 Orpin relationship with other Ca2+ binding proteins
Whether they are indeed CaBPs or not, the sequence comparisons show that Orpins share several characteristics with CaBP subfamilies and that in fact, CaBPs account for the most similar proteins in the database. Several patterns have been developed to accurately classify newly discovered EF-hand proteins. The main pattern is the calmodulin canonical EF motif mostly known as the DXDXDG pattern [40,41], which contrasts with the pseudo-EF-hand binding loops from the most recent vertebrate S100 family of proteins.
One of the main EF-hand protein subfamilies are the S100 proteins. These proteins are small CaBPs containing only two EF-hand motifs. Nevertheless, their N-terminal motifs are considered pseudo-EF-hands, which is the main characteristic of this protein family. At the moment of this study, this subfamily has only been found in vertebrates [42]. Although there was no pseudo-EF-hand predicted from both Orpin isoforms sequences, they share several characteristics with S100 proteins, such as the small size, acidic composition, secretion to extracellular location, and EF-hand motif number. Thus, Orpin sequences share structural features with the main CaBP subfamilies, making it difficult to classify them as any of them. Moreover, we have shown that Orpin and Orpin-like sequences clustered together more closely to osteonectins (BM-40/SPARC) proteins, a group of secreted CaBPs with only two EF-Hand motifs, suggesting that these two groups shared a common ancestral origin. In addition, the best-known CaBPs that grouped closely to Orpin-like sequences were mouse (M. musculus, AAA37432.1) and frog calcineurin A isoforms (X. laevis, AAC23449.1). These two sequences and the human calcineurin A (H. sapiens, AAC37581.1) do not contain EF-hands and were included as outgroups for this analysis. We emphasize the fact that the calcineurin isoform B from zebrafish does contain an EF-hand domain, thus it is separated from the outgroup calcineurin A sequences and is included with the other aligned CaBPs. These results suggest that Orpin sequences comprise a specific EF-hand protein group that is different from the other known subfamilies of calcium-binding proteins. These data suggest that we can be dealing with a new subfamily of EF-hand proteins that is specific to the Ambulacraria clade.
4.4 Orpins as secreted proteins
In addition to their EF-hand motif, an additional feature of Orpins is the presence of a signal peptide. In this respect, Orpin isoforms strongly resemble the groups of EF-hand proteins that are secreted, namely the osteonectins, oncomodulins, and S100s. Similar to Orpin sequences, osteonectins (BM-40/SPARC), oncomodulin, and S100 proteins are small peptides containing two EF-hand motifs each and are secreted to the extracellular milieu. Interestingly, these proteins display calcium-mediated dimerization either as heterodimers as in the case of S100A8/S100A9 [43] or homodimers as in the case of S100P [44], osteonectins [45], and oncomodulin [46]. Usually, EF-hand proteins containing signal peptides are targeted to the outer plasma membrane. There, these CaBPs can act as growth factors recognizing binding targets located on other cell surfaces, thus activating different signaling pathways. Such is the case of osteonectin (BM-40/SPARC), which promotes changes in cell morphology, disrupt cell adhesion, inhibit cell cycle, regulate extracellular matrix, and modulate cell proliferation and migration [47], thus underlying the process of wound repair. Similarly, the secreted (although lacking a signal peptide) oncomodulin and S100 proteins, are involved in a variety of biological processes including: cell proliferation, differentiation, survival, nerve regeneration, interaction with transcription factors, and calcium homeostasis [48-56] among other functions.
4.5 Orpins and Regeneration
Previous results from our laboratory had shown differential expression of the Orpin transcript when mRNAs levels from 3- and 7-day regenerating intestines were compared to normal intestinal tissues. Thus, we had concluded that Orpin was over-expressed during intestinal regeneration. Our new data, where we detect high levels of the transcript in the mesentery region, questions our previous interpretation. Thus, if we consider that the samples containing the 3- and 5-day regenerating intestinal rudiments contain a large proportion of the remaining mesentery (that remains attached to the body wall), then the high expression of Orpin A and B transcript levels that were detected in the 3- or 5-day regenerating tissues can be interpreted as representing the expression in the mesenteric portion and not necessarily in the rudiment itself. As the rudiment itself grows and encompasses a larger proportion of the dissected tissues (in relation to the mesentery) then the Orpin expression would appear to decrease. This is why there is no difference between normal mesentery and early regenerating rudiments. The previously observed difference between regenerating rudiments and “normal” intestine would be merely a reflection of the proportion of mesenterial tissue present in both samples; low in “normal” intestines and high in regenerating ones.
In this respect, the isolated “normal” mesentery is a more appropriate control to compare the relative expression of Orpin transcript sequences between regenerating and normal tissues. This is particularly true in early regenerating stages (3-5 days) when the proportion of tissues corresponding to the mesentery is quite high. While the lack of Orpin differential expression argues against a possible role in the intestinal regeneration process, we cannot completely exclude this possibility. The presence of Musashi binding elements within Orpin isoform sequences suggests that the post-transcriptional regulation of their mRNAs might be controlled by RNA-binding proteins. This element is present in genes that are post-transcriptionally regulated in a spatial and temporal dependent manner [57, 58]. Moreover, this type of regulation has been implicated in the self-renewal of epithelial, neural and hematopoietic stem and progenitor cells [59-65]. Such is the case of the target transcript encoding the transcription factor TTK69 in Drosophila, where translational activation is mediated by the neural Drosophila Musashi. In this way, the Musashi protein induces the differentiation of Drosophila IIb cells as neural precursor cells by repressing the translation of the mRNA of this neural differentiation inhibitory factor [59]. Furthermore, the expression of mammalian Numb protein (m-Numb) induces the expression of regeneration-related genes such as prostate stem cell antigen (PSCA) and metallothionein-2 (Mt2) in gastric mucosal regeneration in mice. Musashi protein (Msi1) enhances the expression of m-Numb during this regenerative process through post-transcriptional regulation. Having stated this, we cannot disregard the possibility that Orpin isoforms play a role during the initial stages of intestine and nerve regeneration. Nonetheless, to explore this possibility, we need to determine if a Musashi protein is present in the H. glaberrima proteome during regenerative processes of the sea cucumber and that it binds to Orpins mRNA.
It is of interest that Orpin isoforms are found to be differentially expressed in a transcriptomic library of regenerating nerve from H. glaberrima [7], also suggesting a possible regeneration-associated function. The two sequences displayed an increase in expression during the regeneration of the radial nerve complex after induced injury. By day 2 and also by day 20 after nerve injury, Orpin A expression was significantly higher than non-regenerating radial nerve (P < .001). In addition, Orpin B was higher in the same samples after day 2, 12, and 20 after nerve injury (P < .001). However, in view of our findings in the intestinal system, it remains to be determined whether this differential expression is also the product of the gene is expressed preferentially in the remaining tissues following injury, and not necessarily of increasing its expression.
In summary, we have identified and characterized a group of Orpin-like proteins from a particular group of invertebrate deuterostomes and shown they all share similarities in size, domain composition, and little significant similarities to other known EF-hand protein sequences. We provide bioinformatics evidence for the presence of signal peptides and cleavage sites in these proteins that suggest secretion of the putative proteins to the extracellular environment. Together, with the identification of predicted EF-hand domains with unique features, we can suggest that these might comprise a novel subfamily of EF-hand containing proteins specific to the Ambulacraria clade. Finally, we studied their expression in normal and regenerating tissues, with the surprise finding that they are highly expressed in the intestinal mesentery.
Acknowledgments
The authors thank Dr. Vladimir S. Mashanov for editorial comments on the manuscript as well as to kindly provide sequence differential expression information from Holothuria glaberrima regenerating nerve.
References
- García-Arrarás JE, Estrada-Rodgers L, Santiago R, Torres II, Díaz-Miranda L, Torres-Avillán I. Cellular mechanisms of intestine regeneration in the sea cucumber, Holothuria glaberrima Selenka (Holothuroidea:Echinodermata). J Exp Zool 281 (1998): 288-304.
- García-Arrarás JE, Greenberg MJ. Visceral Regeneration in Holothurians. Microsc Res Tech. 55(2001): 438-451.
- Rojas-Cartagena C, Ortiz-Pineda P, Ramírez-Gómez F, Suárez-Castillo EC, Matos-Cruz V, Rodríguez C, et al. Distinct profiles of expressed sequence tags during intestinal regeneration in the sea cucumber Holothuria glaberrima. Physiol Genomics 31 (2007): 203-215.
- San Miguel-Ruiz JE, García-Arrarás JE. Common cellular events occur during wound healing and organ regeneration in the sea cucumber Holothuria glaberrima. BMC Dev Biol 7 (2007): 115.
- Mashanov VS, Zueva OR, García-Arrarás JE. Organization of glial cells in the adult sea cucumber central nervous system. Glia 58(2010): 1581-1593.
- Mashanov VS, Zueva OR, García-Arrarás JE. Radial glial cells play a key role in echinoderm neural regeneration. BMC Biol 11 (2013): 49.
- Mashanov VS, Zueva OR, García-Arrarás JE. Transcriptomic changes during regeneration of the central nervous system in an echinoderm. BMC Genomics 15 (2014): 1-21.
- Ortiz-Pineda PA, Ramírez-Gómez F, Pérez-Ortiz J, González-Díaz S, Santiago-De Jesús F, Hernández-Pasos J, et al. Gene expression profiling of intestinal regeneration in the sea cucumber. BMC Genomics. 10 (2009): 262.
- Suárez-Castillo EC, García-Arrarás JE. Molecular evolution of the ependymin protein family: a necessary update. BMC Evol Biol 7 (2007): 23.
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9 (2012): 671-675.
- Sayers EW, Agarwala R, Bolton EE, Brister JR, Canese K, Clark K, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 47 (2019): D23-D28.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 215 (1990): 403-410.
- Bias C, Gish W. Combined Use of Sequence Similarity and Codon Bias for Coding Region Identification. J Comput Biol 1 (1994): 39-50.
- Marchler-bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, et al. CDD / SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acid Res 45 (2017): 200-203.
- Huang H-Y, Chien C-H, Jen K-H, Huang H-D. RegRNA: an integrated web server for identifying regulatory RNA motifs and elements. Nucleic Acids Res 34 (2006): W429-434.
- Bengert P, Dandekar T. A software tool-box for analysis of regulatory RNA elements. Nucleic Acids Res 31 (2003): 3441-3445.
- Buchan DWA, Jones DT. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res 47 (2019): 402-407.
- Jones DT. Protein Secondary Structure Prediction Based on Position-specific Scoring Matrices. J Mol Biol 292 (1999): 195-202.
- Castro E De, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-genevaux PS, Gasteiger E, et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34 (2006): 362-365.
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, et al. InterProScan: protein domains identifier. Nucleic Acids Res 33 (2005): 116-120.
- Jones P, Binns D, Chang H, Fraser M, Li W, Mcanulla C, et al. Sequence analysis InterProScan 5: genome-scale protein function classification. Bioinformatics 30 (2014): 1236-1240.
- Yao X, Liu Y, Tan Y, Song Y, Corlett RT. The complete chloroplast genome sequence of Helwingia himalaica (Helwingiaceae, Aquifoliales) and a chloroplast phylogenomic analysis of the Campanulidae. PeerJ 4 (2016): e2734.
- Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37 (2019): 420-423.
- Madeira F, Park Y mi, Lee J, Buso N, Gur T, Madhusoodanan N, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47 (2019): W636-W641.
- Edgar RC, Drive RM, Valley M. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32 (2004): 1792-1797.
- Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability Article Fast Track. Mol Biol Evol 30 (2013): 772-780.
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0 New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst Biol 59 (2010): 307-321.
- Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47 (2019): W256-W259.
- Medina-Feliciano JG, Pirro S, García-Arrarás JE, Mashanov V, Ryan JF. Draft Genome of the Sea Cucumber Holothuria glaberrima, a Model for the Study of Regeneration. Front. Mar. Sci 8 (2021): 603410.
- Ortiz-Pineda PA. Analysis and Characterization of Genes Associated with Intestinal Regeneration in the Sea Cucumber (Echinodermata: Holothuroidea). PhD Thesis. University of Puerto Rico, Rio Piedras Campus (2010). 364 p.
- Petersen TN, Brunak S, Heijne G Von, Nielsen H. Correspondence SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Publ Gr 8 (2011): 785-786.
- Persechini A, Moncrief ND, Kretsinger RH. The EF-hand family of calcium-modulated proteins. Trends Neurosci 12 (1989): 462-467.
- Kretsinger R.H., Tolbert D., Nakayama S., Pearson W. (1991) The EF-Hand, Homologs and Analogs. In: Heizmann C.W. (eds) Novel Calcium-Binding Proteins. Springer, Berlin, Heidelberg.
- Nelson MR, Chazin WJ. Structures of EF-hand Ca2+-binding proteins: Diversity in the organization, packing and response to Ca2+ binding. BioMetals 11 (1998): 297-318.
- Malmendal A, Carlström G, Hambraeus C, Drakenberg T, Forsén S, Akke M. Sequence and Context Dependence of EF-Hand Loop Dynamics. an 15 N Relaxation Study of a Calcium-Binding Site Mutant of Calbindin D9k. Biochemistry 37 (1998): 2586-2595.
- Gifford JL, Walsh MP, Vogel HJ. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem J 405 (2007): 199-221.
- Moncrief ND, Kretsinger RH, Goodman M. Evolution of EF-hand Calcium-modulated Proteins. I. Relationships Based on Amino Acid Sequences. J Mol Evol 30 (1990): 522-562.
- Grabarek Z. Structural Basis for Diversity of the EF-hand Calcium-binding Proteins. J Mol Biol 359 (2006): 509-525.
- Kawasaki H, Nakayama S, Kretsinger RH. Classification and evolution of EF-hand proteins. BioMetals 11 (1998): 277-295.
- Rigden DJ, Galperin MY. The DxDxDG motif for calcium binding: Multiple structural contexts and implications for evolution. J Mol Biol 343 (2004): 971-984.
- Denessiouk K, Permyakov S, Denesyuk A, Permyakov E, Johnson MS. Two Structural Motifs within Canonical EF-Hand Calcium- Binding Domains Identify Five Different Classes of Calcium Buffers and Sensors. PLoS One 9 (2014): 1-14.
- Morgan RO, Martin-Almedina S, García M, Jhoncon-Kooyip J, Fernandez MP. Deciphering function and mechanism of calcium-binding proteins from their evolutionary imprints. Biochim Biophys Acta - Mol Cell Res 1763 (2006): 1238-1249.
- Edgeworth J, Gorman M, Bennett R, Freemont P, Hogg N. Identification of p8,14 as a highly abundant heterodimeric calcium binding protein complex of myeloid cells. J Biol Chem 266 (1991): 7706-7713.
- Koltzscher M, Gerke V. Identification of Hydrophobic Amino Acid Residues Involved in the Formation of S100P Homodimers in Vivo. Biochemistry 39 (2000): 9533-9539.
- Maurer P, Hohenadl C, Hohenester E, Göhring W, Timpl R, Engel J. The C-terminal Portion of BM-40 (SPARC/Osteonectin) is an Autonomously Folding and Crystallisable Domain that Binds Calcium and Collagen IV. J Mol Biol 253 (1995): 347-357.
- Mutus B, Palmer EJ, MacManus JP. Disulfide-Linked Dimer of Oncomodulin: Comparison to Calmodulin. Biochemistry 27 (1988): 5615-5622.
- Phan E, Ahluwalia A, Tarnawski AS. Role of SPARC-matricellular protein in pathophysiology and tissue injury healing: Implications for gastritis and gastric ulcers. Med Sci Monit 13 (2007): RA25-30.
- Yin Y, Cui Q, Li Y, Irwin N, Fischer D, Harvey AR, et al. Macrophage-Derived Factors Stimulate Optic Nerve Regeneration. J Neurosci 23 (2003): 2284-2293.
- Yin Y, Cui Q, Gilbert H-Y, Yang Y, Yang Z, Berlinicke C, et al. Oncomodulin links inflammation to optic nerve regeneration. Proc Natl Acad Sci USA 106 (2006): 19587-19592.
- Kurimoto T, Yin Y, Habboub G, Gilbert H-Y, Li Y, Nakao S, et al. Neutrophils express oncomodulin and promote optic nerve regeneration. J Neurosci 33 (2013): 14816-14824.
- Yin Y, Cui Q, Li Y, Irwin N, Fischer D, Harvey AR, et al. Macrophage-derived factors stimulate optic nerve regeneration. J Neurosci. 23(2003): 2284-2293.
- Donato R. S100: A multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int J Biochem Cell Biol 33 (2001): 637-668.
- Donato R. Functional roles of S100 proteins, calcium-binding proteins of the EF-hand type. Biochim Biophys Acta - Mol Cell Res 1450 (1999): 191-231.
- Donato R, Cannon BR, Sorci G, Riuzzi F, Hsu K, Weber DJ, et al. Functions of S100 proteins. Curr Mol Med 13 (2013): 24-57.
- Donato R, Sorci G, Riuzzi F, Arcuri C, Bianchi R, Brozzi F, et al. S100B’s double life: Intracellular regulator and extracellular signal. Biochim Biophys Acta - Mol Cell Res 1793 (2009): 1008-1022.
- Schaub MC, Heizmann CW. Calcium, troponin, calmodulin, S100 proteins: From myocardial basics to new therapeutic strategies. Biochem Biophys Res Commun 369 (2008): 247-264.
- MacNicol MC, Cragle CE, MacNicol AM. Context-dependent regulation of Musashi-mediated mRNA translation and cell cycle regulation. Cell Cycle 10 (2011): 39-44.
- Takahashi T, Suzuki H, Imai T, Shibata S, Tabuchi Y, Tsuchimoto K, et al. Musashi-1 post-transcriptionally enhances phosphotyrosine-binding domain-containing m-Numb protein expression in regenerating gastric mucosa. PLoS One 8 (2013): e53540.
- Okano H, Kawahara H, Toriya M, Nakao K, Shibata S, Imai T. Function of RNA-binding protein Musashi-1 in stem cells. Exp Cell Res 306 (2005): 349-356.
- Nakamura M, Okano H, Blendy JA, Montell C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron 13 (1994): 67-81.
- Sakakibara S, Nakamura Y, Yoshida T, Shibata S, Koike M, Takano H, et al. RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc Natl Acad Sci USA 99 (2002): 15194-15199.
- Asai R, Okano H, Yasugi S. Correlation between Musashi-1 and c-hairy-1 expression and cell proliferation activity in the developing intestine and stomach of both chicken and mouse. Dev Growth Differ 47 (2005): 501-510.
- Wang X-Y, Yin Y, Yuan H, Sakamaki T, Okano H, Glazer RI. Musashi1 modulates mammary progenitor cell expansion through proliferin-mediated activation of the Wnt and Notch pathways. Mol Cell Biol 28 (2008): 3589-3599.
- Kharas MG, Lengner CJ, Al-Shahrour F, Bullinger L, Ball B, Zaidi S, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med 16 (2010): 903-908.
- Hope KJ, Cellot S, Ting SB, MacRae T, Mayotte N, Iscove NN, et al. An RNAi screen identifies msi2 and prox1 as having opposite roles in the regulation of hematopoietic stem cell activity. Cell Stem Cell 7 (2010): 101-113.


 Impact Factor: * 4.2
Impact Factor: * 4.2 Acceptance Rate: 77.66%
Acceptance Rate: 77.66%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 