Transcriptional and Translational Regulation of Differentially Expressed Genes in Yucatan Miniswine Brain Tissues following Traumatic Brain Injury
Vikrant Rai, Yssel Mendoza-Mari, Mohamed M. Radwan, James Brazdzionis, David A. Connett, Dan E. Miulli, and Devendra K. Agrawal*
Department of Translational Research, College of Osteopathic Medicine of the Pacific, Western University of Health Sciences, Pomona CA 91766
*Corresponding author: Devendra K Agrawal, Department of Translational Research Western University of Health Sciences, Pomona CA.
Received: 17 January 2024; Accepted: 29 January 2024; Published: 05 March 2024
Article Information
Citation: Vikrant Rai, Yssel Mendoza-Mari, Mohamed M. Radwan, James Brazdzionis, David A. Connett, Dan E. Miulli, and Devendra K. Agrawal. Transcriptional and Translational Regulation of Differentially Expressed Genes in Yucatan Miniswine Brain Tissues following Traumatic Brain Injury. Journal of Bioinformatics and Systems Biology. 7 (2024): 81-91.
View / Download Pdf Share at FacebookAbstract
Traumatic brain injury (TBI) is a leading cause of morbidity, disability, and mortality worldwide. Motor and cognitive deficits and emotional disturbances are long-term consequences of TBI. A lack of effective treatment for TBI-induced neural damage, functional impairments, and cognitive deficits makes it challenging in the recovery following TBI. One of the reasons may be the lack of knowledge underlying the complex pathophysiology of TBI and the regulatory factors involved in the cellular and molecular mechanisms of inflammation, neural regeneration, and injury repair. These mechanisms involve a change in the expression of various proteins encoded by genes whose expression is regulated by transcription factors (TFs) at the transcriptional level and microRNA (miRs) at the mRNA level. In this pilot study, we performed the RNA sequencing of injured tissues and non-injured tissues from the brain of Yucatan miniswine and analyzed the sequencing data for differentially expressed genes (DEGs) and the TFs and miRs regulating the expression of DEGs using in-silico analysis. We also compared the effect of the electromagnetic field (EMF) applied to the injured miniswine on the expression profile of various DEGs. The results of this pilot study revealed a few DEGs that were significantly upregulated in the injured brain tissue and the EMF stimulation showed effect on their expression profile.
Keywords
<p>Differentially expressed genes, Electromagnetic field, MicroRNA, Transcription factors,Traumatic brain injury.</p>
Article Details
1. Introduction
Traumatic brain injury (TBI), a leading cause of morbidity, disability, and mortality in the military and civilian population, affects approximately 1.5 million Americans each year, and nearly 3.2–5.3 million individuals in the United States are living with a TBI-related disability as per the Center for Disease Control and Prevention (CDC). Worldwide, nearly 50 million people suffer from TBI each year. The post-traumatic complications of TBI include neurological and psychological issues contributing to long-term disability. Despite the increasing understanding of the pathophysiology underlying primary and secondary TBI and the development of advanced therapies, there is a need to develop better therapeutics to improve clinical outcomes [1, 2]. Injuries to cerebral brain tissues, diffuse axonal injury, loss of neuronal and glial cells, brain edema, and ischemic brain damage after primary brain injury contribute to secondary brain injuries resulting in cognitive deficits, behavioral changes, and hemiparesis. Increased oxidative stress, excitotoxicity, mitochondrial dysfunction, lipid peroxidation, neuroinflammation, axonal degeneration, inhibition of myelination and axonal growth, increased apoptosis of neurons and oligodendrocytes, and impairment of autophagy and lysosomal pathways contribute to secondary brain injuries [1, 2]. These processes are regulated by several proteins including enzymes, cytokines, chemokines, and growth factors generated by activated microglial cells. The expression of genes coding these proteins is regulated by transcription factors (TF) at the transcriptional level and microRNAs (miR) at the post-transcriptional level [3].
Liu et. al. [4] reported the association of time-dependent increased expression of Forkhead box transcription factors, class O (FOXOs), and FOXO proteins, important in regulating neurite outgrowth, axonal degeneration, and learning and memory, within 24 hours after TBI. Pennypacker et al. [5] reported prolonged activation (days to months) of TFs Fos-related antigen-2 (FRA2 from AP-1 family TF and involved in cell death and repair) and p65 and p50 subunits of nuclear factor kappa B (NF-κB) (involved in neuronal survival and/or repair) after neurotoxic, excitotoxic or ischemic injuries. Liu et al. [6] reported increased expression of transcription initiation factor IIB (TFIIB) in association with proliferating cell nuclear antigen (PCNA) after brain injury with its implications in astrocytes and microglia proliferation and neurological recovery. Activating transcription factor 3 (ATF3) is another TF involved in protecting neuronal damage after an injury by suppressing neuronal apoptosis and microglia activation via targeting CCL2 involving TLR4/NF-κB signaling [7]. Further, the role of NF-κB, SMAD1, ATF3, ATF4, c-JUN, Olig1, and Arpc1b in spinal cord injuries [8] and of CEBPD, Pax6, Spi1, NF-κB, NRF2, and PPAR-γ in TBI [8, 9] as well as their potential to be used as a therapeutic target has been discussed. A genome-wide methyl binding domain-sequencing and RNA-sequencing at 3 months after TBI in adult male Sprague-Dawley rats reported upregulation of Cebpd, Pax6, Spi1, and Tp73 and the potential of targeting these TFs to improve post-TBI recovery [10]. These findings suggest that the regulation of TFs and their signaling could be therapeutic targets to protect brain damage and improve clinical outcomes [11].
Additionally, microRNAs (miRNAs), long noncoding RNAs (lncRNAs), circular RNAs (circRNAs), and other non-coding RNAs (ncRNAs) play a diagnostic and therapeutic role in TBI [12]. Mehta et al. [13] reported that lncRNA FosDT interacts with REST-associated chromatin-modifying proteins and enhances ischemic brain injury. LncRNA TUG1 plays a detrimental role in aggravating ischemic reperfusion brain injury [14]. Further, the role of lncRNAs including metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), maternally expressed gene 3 (MEG3), homeobox (HOX) transcript antisense RNA (HOTAIR), Gm4419, H19, N1LR, and antisense non-coding RNA in the INK4 locus (ANRIL) in stroke and of ENSRNOG00000021987, BC088414, growth arrest-specific 5 (GAS5), H19, nuclear-enriched abundant transcript 1 (NEAT1), and others in hypoxia-related brain tissue damage and neuronal cell fate has been documented [15]. Zhang et al. reported differential expression profile of miRNAs and mRNAs in human TBI and that lncRNA tubulin beta 6 class V (TUBB6)/nuclear factor E2-related factor 2 (NRF2) are closely related to the pathogenesis of TBI [16]. Overexpression of lncRNA H19-mediated inhibition of miR-107 results in increased expression of vascular endothelial growth factor (VEGF) and protects brain tissue from hypoxic-ischemic injury [17]. These results suggest that targeting ncRNAs, lncRNAs, miRNAs, and circRNA could be a potential therapeutic strategy to improve the clinical outcome post-TBI or injury after stroke [18]. This suggests that TFs and miRNAs may play a critical role in TBI in improving clinical outcomes.
Electromagnetic field (EMF) plays a critical role in TBI and has a beneficial effect on improving clinical outcomes [19, 20], however, further research is warranted to standardize the frequency and time of application. We recently developed a swine model of TBI and reported the findings on the effect of simulated TBI and the effect of EMF on TBI [21]. In this study, we performed the RNA sequencing of the brain tissues collected from the injured areas and non-injured areas and investigated the differential gene expression profile and associated TFs and non-coding RNAs.
2. Materials and Methods
2.1 Animal model and tissue harvesting:
Male Yucatan minipigs obtained from Premier BioSource (Romona, CA) were housed in the animal care facility of Western University of Health Sciences, Pomona CA. The swine were kept with 12 hours of light and dark cycle at 72°-74° F temperature and were fed with the Mini-Pig Grower Diet (Test Diet # 5801) and water ad libitum. The experimental protocol was approved by the Institutional Animal Care and Use Committee of the Western University of Health Sciences (# R23IACUC003). Induction of traumatic brain injury (TBI), EMF application, and signal detection were done as previously described [21-23]. After completion of the experiment, the swine were euthanized and brain cortex tissues from the site of injury and contralateral hemisphere were collected in 10% formalin for histological analysis, in RNAlater for RNA isolation, and dry freezing for protein isolation. The tissues in RNAlater were kept at 40C followed by -200C and then at -800C.
2.2 RNA sequencing and data analysis:
The tissues from injured (n=3) and control non injured (n=3) sites were sent for RNA sequencing to the University of California at Los Angeles (UCLA). The RNA was extracted and RNAs with RIN >6 were used for sequencing. Libraries for RNA-Seq were prepared with KAPA Stranded mRNA-Seq Kit. The workflow consisted of mRNA enrichment and fragmentation, first-strand cDNA synthesis using random priming followed by second-strand synthesis converting cDNA: RNA hybrid to double-stranded cDNA (dscDNA) and incorporated dUTP into the second cDNA strand. cDNA generation was followed by end repair to generate blunt ends, A-tailing, adaptor ligation, and PCR amplification. Different adaptors were used for multiplexing samples in a half-lane. Sequencing was performed on Illumina Novaseq 6000 for PE 2x50 run. A data quality check was done on Illumina SAV. Demultiplexing was performed with Illumina Bcl2fastq v2.19.1.403 software. The alignment was performed using STAR [24] with pig reference genome Sscrofa11.1. The Ensembl Transcripts release Sus_scrofa.Sscrofa11.1.110 GTF was used for gene feature annotation. For normalization of transcripts counts, CPM normalized counts were generated by adding 1.0E-4 followed by counts per million (CPM) normalization. Differential Gene expression was determined by Partek Flow GSA algorithm (Partek® Flow® software, version 7.0 Copyright ©; 2019 Partek Inc., St. Louis, MO, USA.). A p-value of 0.05 and log2 Fold-change (FC) were used to filter for differentially expressed gene lists. Network analysis was done using Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) for protein-protein interaction, SIGnaling Network (SIGNOR), and Networkanalyst.ca for investigating the association between DEGs, transcription factors (TFs), and microRNA.
3. Results and Discussion
3.1 RNA sequencing analysis revealed differentially expressed genes (DEGs):
The analysis of RNA seq data comparing control and experimental gene expression revealed 175 DEGs (p<0.05) and 13 significantly upregulated (p<0.05, log2FC>1.5) DEGs (CYP19A1, ADGRG3, DNAAF6 (PIH1D3), ADGRG5, LRRC2, CCDC194, C1orf56, OR10A4, PRSS53, LIPG, ATP8B3, CFAP157, and PDCL2) in the experimental tissues compared to control out of a total of 24309 genes (Sheet 2, Supplementary file).
3.2 STRING network reveals protein-protein interaction between various proteins:
STRING network showed an interaction between various proteins encoded by significantly DEGs (CYP19A1, ADGRG3, DNAAF6 (PIH1D3), ADGRG5, LRRC2, CCDC194, C1orf56, OR10A4, PRSS53, LIPG, ATP8B3, CFAP157, and PDCL2) identified after RNAseq analysis (Figure 1, Supplementary File Sheet Folds Change Comparison). CYP19A1 gene encodes the aromatase cytochrome P450 enzyme extensively expressed in astrocytes and a subpopulation of excitatory and inhibitory neurons in the human brain mainly in the cortex, thalamus, hypothalamus, amygdala, and hippocampus. This enzyme plays a role in the regulation of sensory integration, body homeostasis, social behavior, cognition, and language [25]. RNAseq analysis revealed increased expression in injured tissues suggesting its role to regulate sensory integration as a response to injury.
Adhesion G Protein-Coupled Receptors are expressed in the plasma membrane and is involved in various signaling pathway and regulation of cell migration [26, 27]. Adhesion GPCR GPR125 upregulates after brain injury in the choroid plexus and its expression decreases after 2 days while its expression in the hippocampus increases 1 day after brain injury [28]. An increased expression of ADGRG3 (Adhesion G Protein-Coupled Receptor G3) and ADGRG5 in the cortex after injury suggests a response to the injury to promote healing by increasing cell migration. Further, an increased expression in experimental animals 2 and 3, who started receiving EMF stimulation after 2 days and the same day, respectively, suggests the beneficial role of EMF. This notion is supported by the fact that the role of ADGRG5 has been implicated in the pathogenesis of glioblastoma and is a therapeutic target in neurological diseases [29]. Leucine Rich Repeat Containing 2 (LRRC2) localized in the mitochondria is transcriptionally regulated by the mitochondrial master regulator Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1-Alpha (PGC-1α) [30] which in turn is a marker of mitochondrial biogenesis. The protein levels of nuclear PGC-1α increased from 6 to 24 hours after TBI and decreased at 48 hours after TBI [31]. We found an increased expression of LRRC2 in swine after injury with no EMF while the expression was decreased with EMF. This suggests that mitochondrial biogenesis increased after injury and application of EMF helped in repair which leads to decreased need of mitochondria and thus decreased LRRC2. Increased expression of lipoprotein lipase (LPL) and endothelial lipase (EL) has been documented in association with brain injury mainly in the brain cortex as a pathophysiological response to brain injury [32]. We found an increased expression of Lipase G, endothelial type (LIPG) after TBI, however, the results of the effects of EMF on LIPG expression were equivocal. Mutations in ATP8B3 (Phospholipid-transporting ATPase 8B3) is associated with axonal degeneration [33]. We found an increased expression of ATP8B3 after injury and a beneficial effect of EMF (Figure 1, Supplementary File Sheet Folds Change Comparison).
C1orf56 (chromosome 1 open reading frame 56) protein is involved in the pathogenesis of multiple system atrophy (MSA), a rare oligodendroglial synucleinopathy of unknown etiopathogenesis including two major clinical variants with predominant parkinsonism (MSA-P) or cerebellar dysfunction (MSA-C) [34]. Mutations in dynein (axonemal) assembly factors (DNAAFs) are involved in impaired chordotonal neuron function [35]. Olfactory Receptor, Family 10, Subfamily A, Member 4 (OR10A4) is a specific epigenomic target of postprandial hyperglycemia in peripheral blood leukocytes [36]. As per GeneCards, OR10A4 plays a role in smell sensation. The expression of serine protease 53 (PRSS53) is associated with Alzheimer’s disease [37] and is a mitochondrial protein in islet beta cells [38]. Increased expression of CFAP157 is associated with human prodromal Huntington’s disease where dramatic and unique transcriptional changes occur in the caudate nucleus [39]. Phosducin-like protein 2 (PDCL2) protein is associated with germ cell aplasia [40] and is essential for spermiogenesis and male fertility in mice [41]. We found an increased expression of C1orf56, DNAAF6, OR10A4, CFAP157, and PDCL2 after brain injury but how they play a role in the pathogenesis of TBI warrant further investigation.
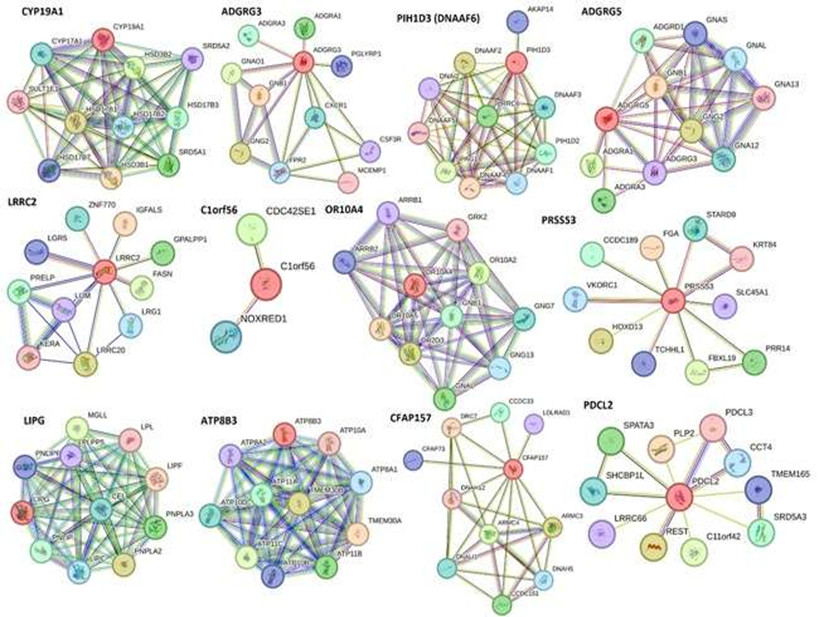
Figure 1: Significantly upregulated differentially expressed genes (DEGs) in experimental brain cortex tissues after traumatic brain injury (TBI).
Further network analysis of 175 significantly expressed DEGs using STRING networking showed interaction of various proteins encoded by these genes, however, the interaction between upregulated 13 genes was minimal, and we only found interaction between LRRC2 with FBXO8, ADGRG3, KISS1R; GPR26 and OR10A4 with GRM2; and CYP19A1, AREG, IL5, and CRP (Figure 2). The association between LRRC2 with FBXO8, ADGRG3, and KISS1R indicates that FBXO8 (negatively regulating Arf6 GTPase) [42] and KISS1R (associated with glucose metabolism) [43] may play a role in the pathogenesis of TBI because of their co-expression with LRRC2 and ADGRG3 whose expression is increased after TBI in our study. GPR26 plays a role in CNS disorders [44] and activation of GRM2 (Glutamate Metabotropic Receptor 2) protects rat brain from oxidative stress injury [45], reduces apoptosis, and regulates BDNF and GDNF levels after hypoxia-ischemia injury [46]. The interaction between GPR26 and OR10A4 with GRM2 suggests that OR10A4 whose expression is increased after TBI may play a protective role after TBI and warrants investigation. Increased expression of amphiregulin (AREG) [47], interleukin (IL)-5 [48], and C-reactive protein [49] is associated with brain injury and inflammation. It should be noted that IL-5 and CRP are biomarkers and prognostic markers of TBI. The interaction of CYP19A1, whose expression is increased after TBI in this study, with IL-5, CRP, and AREG suggests that CYP19A1 might play a protective role after TBI.
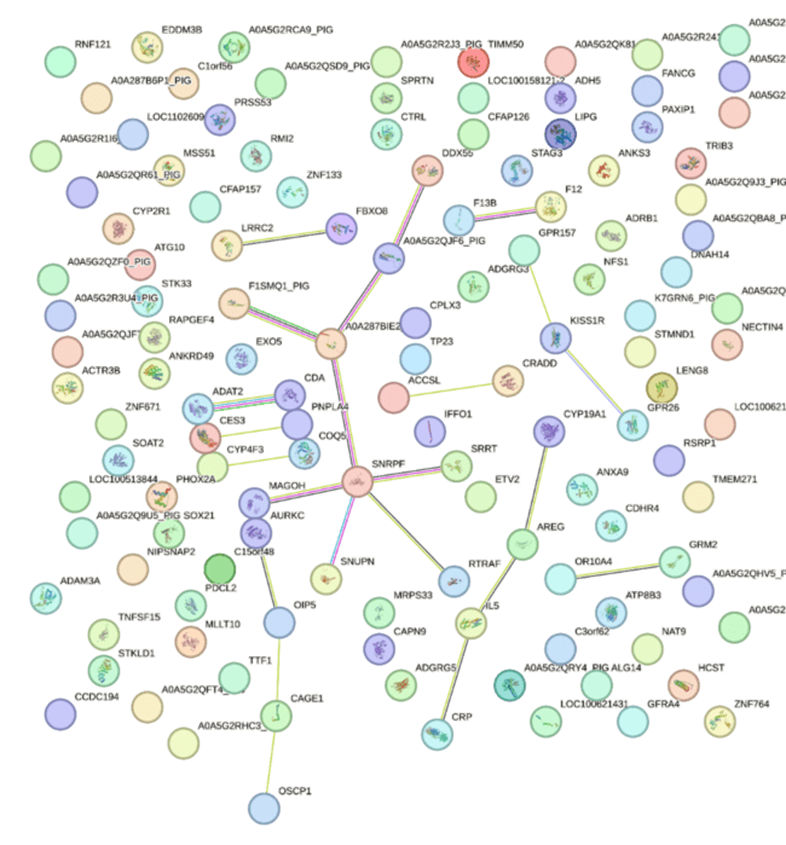
Figure 2: STRING network analysis of significant differentially expressed genes (DEGs) after traumatic brain injury (TBI) in swine.
Analysis of these genes using Networkanalyst.ca with the STRING dataset revealed more interactions between these proteins (gene-protein interaction) along with various drugs associated with their expression (Figure 3, Supplementary File Fold change comparison). Src family kinases play a significant role in brain edema after acute brain injury [50] and its association with CYP19A1 and AREG (Figure 3 panel A) suggests the synergistic role of these proteins in brain injury. GNA14, GNA11, and GNAQ mutations are associated with benign but not malignant cutaneous vascular tumors [51] while blockade of β1 receptor (ADRB1) attenuates atherosclerosis progression after TBI [52]. The association between KISS1R with GNAQ and GNA14 along with other molecules (Figure 3 panel B) may play a significant role in the pathogenesis of TBI.
As discussed above, IL-5 and CRP are associated with brain injury, their association with Akt, a cytoplasmic kinase, and degranulation (Figure 3 panel C) suggests their role in inflammation after TBI. Similarly, the association of CRP with CXCL8 and IL-10 (Figure 3 panel E) suggests a response to injury with an increased IL-10, an anti-inflammatory cytokine, and CXCL8, a pro-inflammatory cytokine but also involved in angiogenesis after an injury suggest their role in the pathogenesis of TBI [53]. However, this association is not known in TBI and must be investigated. Increased expression of transcription factor achaete-scute family bHLH transcription factor 1 (ASCL1) after an injury mediates a transitory cell state needed for repair of the neonatal cerebellum [54]. Cakir et al. [55] reported that reprogramming of ETV2-induced endothelial cells is a potent way to generate vascularized human cortical organoids, and this suggests that ETV2 may induce angiogenesis and tissue repair. Further, its association with differentiation (Figure 3 panel F) supports the notion that targeting the ETV2 transcription factor may have a therapeutic effect on TBI.
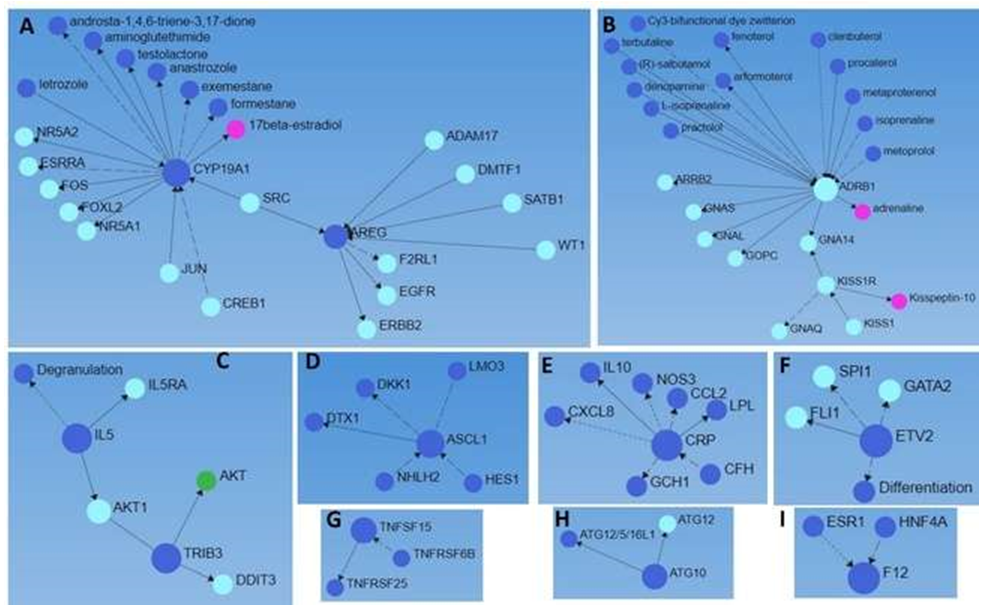
Figure 3: A network analysis of 175 differentially expressed genes (p<0.05, DEGs) expressed in injured brain tissues after traumatic brain injury with Networkanalyst.ca using the STRING dataset. Larger blue circles (input DEGs encoding proteins), smaller blue circles (output genes encoding proteins, drugs, and cellular functions), green circles (cytoplasmic kinases), light blue circles (transcription factors), and pink circles (associated hormones).
TNFSF15 and TNFSF25 (Figure 3 panel G) are associated with co-stimulatory signals for activating, proliferative activity, and cytokine production by lymphocytes along with influencing the development and suppressive function of Tregs [56]. TNFSF6B is a negative regulator of TNFSF15 and TNFSF25 [57]. The expression of TNFSF15, TNFSF25, and TNFSF6B in the brain tissues suggests that lymphocyte activation and their regulation are involved in the pathogenesis of TBI though their functions warrant investigation. ATG12-ATG5-ATG16L1 conjugate facilitated by ATG7 and ATG10 plays a critical role in autophagosome formation and autophagy [58] and upregulation of ATG10 in the injured brain tissues in this study suggest the involvement of autophagy after TBI (Figure 3 panel H). Increased secretion of bradykinin by activated factor XII (F12) plays an important role in secondary brain damage by enhancing brain edema and inflammation [59] and targeting activated F12 has been proposed as a therapeutic target to reduce edema and inflammation. An increased F12 expression in injured cortex tissues after TBI in this study suggests F12 be an attractive target to prevent secondary damage and enhance recovery (Figure 3 panel I).
3.3 Network analysis revealed the association of DEGs with transcription factors:
We performed the gene-transcription factor network analysis using the Encyclopedia of DNA Elements (ENCODE) dataset (Networkanalyst.ca). We find the association of various TFs associated with the DEGs (Figure 4, Network Analysis Output Supplementary File Columns W-Z). Since adhesion molecules ADGRG3 and ADGRG5, ATP8B3, LIPG, and LRRC2 play a crucial role in the pathogenesis of TBI, we looked for the betweenness of genes and TFs and found that ATP8B3 has a betweenness (influence of TF on gene) of 239.4165, LRRC2 of 675.1977, and LIPG of 4.479487. Forty-five genes were found to have betweenness higher than LRRC2 (Supplementary File) and among them, the probable role of KISS1R, ETV2, and F12 (an attractive therapeutic target) in association with upregulated DEGs in TBI has been discussed above. Increased betweenness of GPR114 and GPR97 was of interest because of the upregulation of adhesion GPCRs in the injured tissues of the brain after TBI. However, these correlations should be investigated using in-vitro studies. Other associated TFs of importance were the ones playing a role in regeneration, cell reprogramming, and mRNA processing (TRIB3, MLLT10, NFS1, NAT9, KLF9 (suppress axon regeneration), LENG8, KDM5B, KLF16, etc.) and since neurons regenerate after TBI, investigating these associations will be of importance.

Figure 4: Network analysis for the association of DEGs with transcription factors using the Encyclopedia of DNA Elements (ENCODE) dataset (Networkanalyst.ca). Red circles (input genes) and blue squares (output transcription factors).
3.4 Network analysis revealed the association of DEGs with microRNAs:
The network analysis using Networkanalyst.ca revealed an association between DEGs and miRNAs. We found an association of various miRs associated with the DEGs (Figure 5, Network Analysis Output Supplementary File Columns R-U).
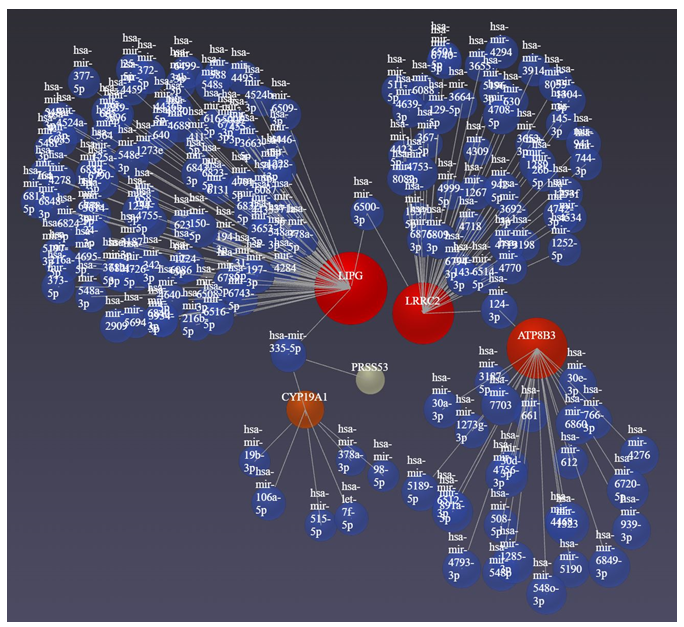
Figure 5: Network analysis for the association of DEGs with microRNAs using Networkanalyst.ca. Red circles (input genes) and blue circles (output miRs).
The network analysis for DEGs and miRNAs revealed LRRC2, LIPG, ATP8B3, and CYP19A1 in association with several miRs with 207 having betweenness (Supplementary File Network analysis). Of these, the regulatory role of miR-130a, miR-223, miR-433, miR-451, miR-541, miR-16, miR-21, miR-9, miR-27, etc. [60-62] has been reported in the context of brain injury. LRRC2 gene expression is regulated by various miRs (MiRTarBase microRNA Targets dataset. https://maayanlab.cloud/Harmonizome/gene/LRRC2) and is also associated with atrial fibrillation being regulated by LOC101928304/miR-490-3p/LRRC2 [63]. The presence of miR-490-3p in our network analysis supports the notion that miR-490-3p regulates LRR2, regulating mitochondrial biogenesis, in TBI. miR-27a and miR-29a regulate adipogenesis and miR-655-3p and mR-497-5p inhibit cell proliferation [64] by regulating the expression of LIPG. The presence of these miRs in association with LIPG in our study indicate that these miRs may play a role in the pathogenesis of TBI by regulating cell proliferation and brain activation pattern [65]. Src family kinases play a significant role in brain edema after acute brain injury [50], an association of Src kinase with CYP19A1 which is negatively regulated by miR-326 [66] appearing in our analysis indicates the probable role of miR-326 in TBI. Since miRs play a critical role in the pathophysiology of TBI, hypoxic-ischemic brain injuries, secondary brain injuries, and even post-traumatic stress disorder and may serve as diagnostic and prognostic biomarkers and therapeutic targets [60, 67-69]; the role, diagnostic and prognostic utility, and therapeutic targetability of additional miRs identified in this study should be investigated.
3.5 Network analysis revealed the association of TFs with microRNAs:
We did the network analysis to investigate the association between transcription factors and miRs using Networkanalyst.ca. We found an association of various TFs associated with the miRs (Figure 6, Network Analysis Output Supplementary File Columns H-K). Since TFs and miRs, both regulate the expression of various genes encoding proteins, we analyzed the data for the association/co-expression of TFs and miRs. With the input of DEGs, Networkanalyst.ca generated a co-expression of TFs (n=260, Network Analysis Output Supplementary File Columns M-P) and miRs (n=364, Network Analysis Output Supplementary File Columns R-U). LRRC2 has a betweenness of 5847.164, LIPG of 1442.449, and ATP8B3 of 1528.56. The betweenness of co-expression of TF and miR higher than LIPG was for MLLT10, STK33, CYP19A, PAXIP1, AREG, ADRB1, RAPGEF4, SOAT2, SOX21, FBXO8, C1orf63, ANKRD49, TRIB3, LENG8, IL5, CRADD, ETV2, and DEM1 (Supplementary File Network analysis results). Of these genes/TFs, we have discussed the role of CYP19A1, AREG, FBXO8, IL-5, and ETV2 above in the context of the central nervous system and brain injury. Thus, the role of these genes/TFs and others and their regulation by microRNAs in the pathogenesis of TBI warrant investigations. Furthermore, epigenetics, mutation variants, gene transcription, miRs-mRNA interaction, and competing endogenous RNA (ceRNA) or miRNA sponges [70] regulate the expression of miRs, the role of epigenetics and other regulatory mechanisms warrant investigation in the pathogenesis of TBI. Phosphorylation and ligand binding are the main mechanisms regulating the levels and expression of TFs [71], investigating the receptors and their ligands involved in TBI pathogenesis will enhance our understanding of the expression of genes involved in TBI pathogenesis.
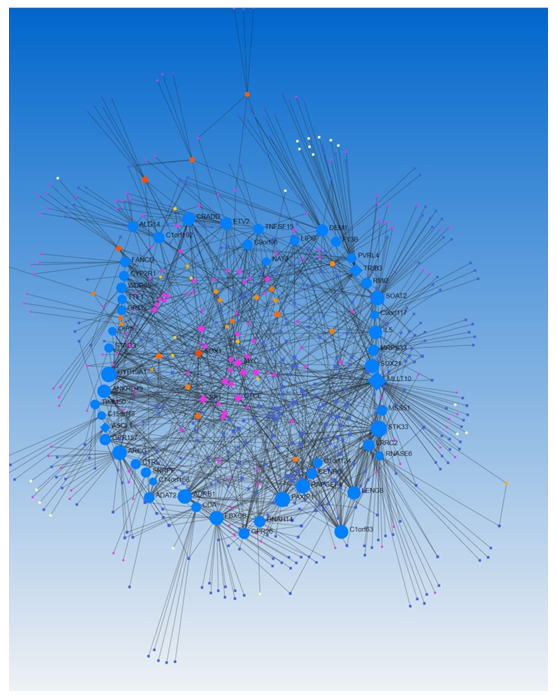
Figure 6: A network analysis of transcription factors co-expressed with microRNAs regulating differentially expressed genes in traumatic brain injury using Networkanalyst.ca.
3.6 Effect of EMF on gene expression:
The fold change (experiment/control) in ADGRG5 was 2.5 in an animal without EMF (hereafter swine 1) compared to 5.3 in swine received EMF after 2 days (hereafter swine 2) and 0 in swine receiving EMF the same day just after injury (hereafter swine 3). Looking at the gene raw count, it was obvious that read counts increased after injury and were more in swine 2 while decreased in swine 3. The fold change of LRRC2 was 7 in swine 1, 2.3 in swine 2, and 0 in swine 3 and this suggests an effect of EMF on gene expression. The read count of LRRC2 was also decreased in swine 3 compared to swine 1 and swine 2 (4 vs 7 and 7, respectively) in the experimental tissues. A similar trend was observed for C1orf56 whose fold change was 7.15 in swine 1, 5.27 in swine 2, and 1.5 in swine 3 suggesting the effect of EMF. A similar trend was with the raw gene count (3 in swine 3 vs 7.15 in swine 1, 5.27 in swine 2) in the experimental tissues. The read count for ATP8B3 was decreased with EMF (4 in swine 3 vs 5 in swine 2, and 6 in swine 1). The fold change in other DEGs showed no trend and this was due to the limited number of tissues we used for RNA sequencing.
4. Conclusion
The pathogenesis of TBI is multifactorial and the mechanisms involved are regulated by various differentially expressed genes. The results of this study suggest that DEGs ADGRG3, ADGRG5, ATP8B3, LIPG, CYP19A1, and LRRC2 significantly upregulated in injured tissues (logFC >2 and p<0.05) and CALB1, CFAP126, INSC, TTR, CDH19, and SERPINE1 upregulated in the control tissues (FC >2 and p<0.05). Since the expression of genes is regulated by transcription factors and microRNAs, the association between these genes and the TFs and miRs regulating them should be investigated. Additionally, there was an effect of EMF on the expression of these genes, the effect of EMF and the most effective frequency should be investigated.
Limitation of the study and future directions:
One of the limitations of this study is the limited number of swine (n=1 in each experimental category). This makes it difficult to delineate the generalized effect of EMF on gene expression. Another limitation is analyzing the data by combining the controls and experimental gene expressions with different treatment strategies to look for fold change and log2 fold change. Future studies must be conducted to increase the number of swine and the data should be pooled to validate the findings of this study. The results of this study pave the way for targeting a single DEG with a small molecule or drug and investigating the effect of various frequencies on the gene expression profile to choose the most effective therapeutic frequency with improved clinical outcomes.
Funding:
This research work was partially supported by the funds to DAC from the Office of Research & Biotechnology, Western University of Health Sciences, Pomona, CA. The research work of DKA is supported by the R01 HL144125 and R01 HL147662 grants from the National Institutes of Health, USA. The contents of this original research article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Competing interest:
All the authors have read the manuscript and declare no conflict of interest. No writing assistance was utilized in the production of this manuscript.
Consent for publication:
All the authors have read the manuscript and consented for publication.
Supplementary Data
https://www.fortunejournals.com/suppli/suppliJBSB_10090.pdf
References
- Ng SY, Lee AYW. Traumatic Brain Injuries: Pathophysiology and Potential Therapeutic Targets. Front Cell Neurosci 13 (2019): 528.
- Algattas H, Huang JH. Traumatic Brain Injury pathophysiology and treatments: early, intermediate, and late phases post-injury. Int J Mol Sci 15 (2013): 309-341.
- Zhao M, Kim P, Mitra R, et al. TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res 44 (2016): D1023-1031.
- Liu XL, Gao CC, Qi M, et al. Expression of FOXO transcription factors in the brain following traumatic brain injury. Neurosci Lett 753 (2021): 135882.
- Pennypacker KR, Kassed CA, Eidizadeh S, et al. Brain injury: prolonged induction of transcription factors. Acta Neurobiol Exp (Wars) 60 (2000): 515-30.
- Liu Z, Wang D, Shao B, et al. Increased expression of transcription initiation factor IIB after rat traumatic brain injury. J Mol Histol 42 (2011): 265-71.
- Ma N, Li G, Fu X. Protective role of activating transcription factor 3 against neuronal damage in rats with cerebral ischemia. Brain Behav 12 (2022): e2522.
- Kumar S, Fritz Z, Sulakhiya K, et al. Transcriptional Factors and Protein Biomarkers as Target Therapeutics in Traumatic Spinal Cord and Brain Injury. Curr Neuropharmacol 18 (2020): 1092-105.
- Ratliff WA, Qubty D, Delic V, et al. Repetitive Mild Traumatic Brain Injury and Transcription Factor Modulation. J Neurotrauma 37 (2020): 1910-7.
- Lipponen A, El-Osta A, Kaspi A, et al. Transcription factors Tp73, Cebpd, Pax6, and Spi1 rather than DNA methylation regulate chronic transcriptomics changes after experimental traumatic brain injury. Acta Neuropathol Commun 6 (2018): 17.
- Wei J, Zhang Y, Jia Q, et al. Systematic investigation of transcription factors critical in the protection against cerebral ischemia by Danhong injection. Scientific reports 6 (2016): 29823.
- Li S, Qiu N, Ni A, et al. Role of regulatory non-coding RNAs in traumatic brain injury. Neurochem Int (2023): 105643.
- Mehta SL, Kim T, Vemuganti R. Long Noncoding RNA FosDT Promotes Ischemic Brain Injury by Interacting with REST-Associated Chromatin-Modifying Proteins. J Neurosci 35 (2015): 16443-9.
- Chen M, Wang F, Fan L, et al. Long Noncoding RNA TUG1 Aggravates Cerebral Ischemia/Reperfusion Injury by Acting as a ceRNA for miR-3072-3p to Target St8sia2. Oxid Med Cell Longev. 2022;2022:9381203.
- Lim KH, Yang S, Kim SH, Chun S, Joo JY. Discoveries for Long Non-Coding RNA Dynamics in Traumatic Brain Injury. Biology (Basel) 9 (2020).
- Zhang L, Tian M, Wang H, et al. Expression characteristics of long noncoding RNA and messenger RNA in human traumatic brain injury. Neuroreport 33 (2022): 90-100.
- Fang H, Li HF, Pan Q, et al. Long Noncoding RNA H19 Overexpression Protects against Hypoxic-Ischemic Brain Damage by Inhibiting miR-107 and Up-Regulating Vascular Endothelial Growth Factor. Am J Pathol 191 (2021): 503-14.
- Wang Y, Pan WY, Ge JS, et al. A review of the relationship between long noncoding RNA and post-stroke injury repair. J Int Med Res 47 (2019): 4619-24.
- Patchana T, Agrawal DK, Connett D, et al. Immunomodulatory Effect of Electromagnetic Field in the Treatment of Traumatic Brain Injury. J Biotechnol Biomed 6 (2023): 32-46.
- Siddiqi I, Marino M, Agrawal DK, et al. Cellular mechanisms of electromagnetic field in traumatic brain injury. J Biotechnol Biomed 6 (2023): 95-104.
- Brazdzionis J, Radwan MM, Thankam F, et al. A Swine Model of Traumatic Brain Injury: Effects of Neuronally Generated Electromagnetic Fields and Electromagnetic Field Stimulation on Traumatic Brain Injury-Related Changes. Cureus 15 (2023): e42544.
- Brazdzionis J, Radwan MM, Thankam F, et al. A Swine Model of Neural Circuit Electromagnetic Fields: Effects of Immediate Electromagnetic Field Stimulation on Cortical Injury. Cureus 15 (2023): e43774.
- Brazdzionis J, Radwan MM, Thankam FG, et al. A Swine Model of Changes in the Neuronal Electromagnetic Field After Traumatic Brain Injury: A Pilot Study. Cureus 15 (2023): e41763.
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat methods 9 (2012): 357-9.
- Azcoitia I, Mendez P, Garcia-Segura LM. Aromatase in the Human Brain. Androg Clin Res Ther 2 (2021): 189-202.
- Syrovatkina V, Alegre KO, Dey R, et al. Regulation, Signaling, and Physiological Functions of G-Proteins. J Mol Biol 428 (2016): 3850-3868.
- Cotton M, Claing A. G protein-coupled receptors stimulation and the control of cell migration. Cell Signal 21 (2009): 1045-1053.
- Pickering C, Hagglund M, Szmydynger-Chodobska J, et al. The Adhesion GPCR GPR125 is specifically expressed in the choroid plexus and is upregulated following brain injury. BMC Neurosci 9 (2008): 97.
- Stephan G, Ravn-Boess N, Placantonakis DG. Adhesion G protein-coupled receptors in glioblastoma. Neurooncol Adv 3 (2021): vdab046.
- McDermott-Roe C, Leleu M, Rowe GC, et al. Transcriptome-wide co-expression analysis identifies LRRC2 as a novel mediator of mitochondrial and cardiac function. PLoS One 12 (2017): e0170458.
- Fan YW, Sun KJ, Mao L, et al. The expression of PGC-1alpha in the mice brain after traumatic brain injury. Brain Inj 26 (2012): 1267-1272.
- Paradis E, Clavel S, Julien P, et al. Lipoprotein lipase and endothelial lipase expression in mouse brain: regional distribution and selective induction following kainic acid-induced lesion and focal cerebral ischemia. Neurobiol Dis 15 (2004): 312-325.
- Zhu X, Libby RT, de Vries WN, et al. Mutations in a P-type ATPase gene cause axonal degeneration. PLoS Genet 8 (2012): e1002853.
- Perez-Soriano A, Arnal Segura M, Botta-Orfila T, et al. Transcriptomic differences in MSA clinical variants. Sci Rep 10 (2020): 10310.
- Lennon J, Zur Lage P, von Kriegsheim A, et al. Strongly Truncated Dnaaf4 Plays a Conserved Role in Drosophila Ciliary Dynein Assembly as Part of an R2TP-Like Co-Chaperone Complex With Dnaaf6. Front Genet 13 (2022): 943197.
- Shim SM, Cho YK, Hong EJ, et al. An epigenomic signature of postprandial hyperglycemia in peripheral blood leukocytes. J Hum Genet 61 (2016): 241-6.
- Sun Y, Zhu J, Zhou D, et al. A transcriptome-wide association study of Alzheimer's disease using prediction models of relevant tissues identifies novel candidate susceptibility genes. Genome Med 13 (2021): 141.
- Mizusawa N, Harada N, Iwata T, et al. Identification of protease serine S1 family member 53 as a mitochondrial protein in murine islet beta cells. Islets 14 (2022): 1-13.
- Agus F, Crespo D, Myers RH, et al. The caudate nucleus undergoes dramatic and unique transcriptional changes in human prodromal Huntington's disease brain. BMC Med Genomics 12 (2019): 137.
- Yang Y, Qin S, Wu H, et al. Identification of PDCL2 as a candidate marker in Sertoli cell-only syndrome by chromatin immunoprecipitation-sequencing and bioinformatics analysis. Transl Androl Urol 12 (2023): 1127-1136.
- Li M, Chen Y, Ou J, et al. PDCL2 is essential for spermiogenesis and male fertility in mice. Cell Death Discov 8 (2022): 419.
- Randle SJ, Laman H. F-box protein interactions with the hallmark pathways in cancer. Semin Cancer Biol 36 (2016): 3-17.
- Dudek M, Ziarniak K, Sliwowska JH. Kisspeptin and Metabolism: The Brain and Beyond. Front Endocrinol (Lausanne) 9 (2018): 145.
- Alavi MS, Shamsizadeh A, Azhdari-Zarmehri H, et al. Orphan G protein-coupled receptors: The role in CNS disorders. Biomed Pharmacother 98 (2018): 222-32.
- Bratek E, Ziembowicz A, Bronisz A, et al. The activation of group II metabotropic glutamate receptors protects neonatal rat brains from oxidative stress injury after hypoxia-ischemia. PLoS One 13 (2018): e0200933.
- Bratek-Gerej E, Ziembowicz A, Salinska E. Group II Metabotropic Glutamate Receptors Reduce Apoptosis and Regulate BDNF and GDNF Levels in Hypoxic-Ischemic Injury in Neonatal Rats. Int J Mol Sci 23 (2022).
- Yu Y, Zhang X, Han Z, et al. Expression and regulation of miR-449a and AREG in cerebral ischemic injury. Metab Brain Dis 34 (2019): 821-832.
- Siva Sai Sujith Sajja B, Tenn C, McLaws LJ, et al. IL-5; a diffuse biomarker associated with brain inflammation after blast exposure. Biomed Sci Instrum 50 (2014): 375-82.
- Xu LB, Yue JK, Korley F, et al. High-Sensitivity C-Reactive Protein is a Prognostic Biomarker of Six-Month Disability after Traumatic Brain Injury: Results from the TRACK-TBI Study. J Neurotrauma 38 (2021): 918-27.
- Liu D, Zhang X, Hu B, et al. Src Family Kinases in Brain Edema After Acute Brain Injury. Acta Neurochir Suppl 121 (2016): 185-90.
- Jansen P, Muller H, Lodde GC, et al. GNA14, GNA11, and GNAQ Mutations Are Frequent in Benign but Not Malignant Cutaneous Vascular Tumors. Front Genet 12 (2021): 663272.
- Wang J, Venugopal J, Silaghi P, et al. Beta1-receptor blockade attenuates atherosclerosis progression following traumatic brain injury in apolipoprotein E deficient mice. PLoS One 18 (2023): e0285499.
- Rizzo M, Varnier L, Pezzicoli G, et al. IL-8 and its role as a potential biomarker of resistance to anti-angiogenic agents and immune checkpoint inhibitors in metastatic renal cell carcinoma. Front Oncol 12 (2022): 990568.
- Bayin NS, Mizrak D, Stephen DN, et al. Injury-induced ASCL1 expression orchestrates a transitory cell state required for repair of the neonatal cerebellum. Sci Adv 7 (2021): eabj1598.
- Cakir B, Xiang Y, Tanaka Y, et al. Engineering of human brain organoids with a functional vascular-like system. Nat Methods 16 (2019): 1169-1175.
- Valatas V, Kolios G, Bamias G. TL1A (TNFSF15) and DR3 (TNFRSF25): A Co-stimulatory System of Cytokines With Diverse Functions in Gut Mucosal Immunity. Front Immunol 10 (2019): 583.
- Richard AC, Ferdinand JR, Meylan F, et al. The TNF-family cytokine TL1A: from lymphocyte costimulator to disease co-conspirator. J Leukoc Biol 98 (2015): 333-345.
- Fraiberg M, Elazar Z. Genetic defects of autophagy linked to disease. Prog Mol Biol Trans Sci 172 (2020): 293-323.
- Hopp S, Nolte MW, Stetter C, et al. Alleviation of secondary brain injury, posttraumatic inflammation, and brain edema formation by inhibition of factor XIIa. J Neuroinflammation 14 (2017): 39.
- Atif H, Hicks SD. A Review of MicroRNA Biomarkers in Traumatic Brain Injury. J Exp Neurosci 13 (2019): 1179069519832286.
- Pinchi E, Frati P, Arcangeli M, et al. MicroRNAs: The New Challenge for Traumatic Brain Injury Diagnosis. Curr Neuropharmacol 18 (2020): 319-331.
- Zhu Z, Huang X, Du M, et al. Recent advances in the role of miRNAs in post-traumatic stress disorder and traumatic brain injury. Mol Psychiatry 28 (2023): 2630-44.
- Ke X, Zhang J, Huang X, et al. Construction and Analysis of the lncRNA-miRNA-mRNA Network Based on Competing Endogenous RNA in Atrial Fibrillation. Front Cardiovasc Med 9 (2022): 791156.
- Gajera M, Desai N, Suzuki A, et al. MicroRNA-655-3p and microRNA-497-5p inhibit cell proliferation in cultured human lip cells through the regulation of genes related to human cleft lip. BMC Med Genomics 12 (2019): 70.
- Kasahara M, Menon DK, Salmond CH, et al. Traumatic brain injury alters the functional brain network mediating working memory. Brain Inj 25 (2011): 1170-87.
- Chaurasiya V, Kumari S, Onteru SK, et al. miR-326 down-regulate CYP19A1 expression and estradiol-17b production in buffalo granulosa cells through CREB and C/EBP-beta. J Steroid Biochem Mol Biol 199 (2020): 105608.
- Martinez B, Peplow PV. MicroRNAs as diagnostic markers and therapeutic targets for traumatic brain injury. Neural Regen Res 12 (2017): 1749-61.
- Li S, Qiu N, Ni A, et al. Role of regulatory non-coding RNAs in traumatic brain injury. Neurochem Int 172 (2023): 105643.
- Peeples ES. MicroRNA therapeutic targets in neonatal hypoxic-ischemic brain injury: a narrative review. Pediatr Res 93 (2023): 780-8.
- Misiewicz-Krzeminska I, Krzeminski P, Corchete LA, et al. Factors Regulating microRNA Expression and Function in Multiple Myeloma. Noncoding RNA 5 (2019).
- Calkhoven CF, Ab G. Multiple steps in the regulation of transcription-factor level and activity. Biochem J 317 (1996): 329-342.


 Impact Factor: * 4.2
Impact Factor: * 4.2 Acceptance Rate: 77.66%
Acceptance Rate: 77.66%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 