Computer-Aided Molecular Design of CCR2 - CCR5 Dual Antagonists for the Treatment of NASH
Sonia Kumari, Elizabeth Sobhia M*
Department of Pharmacoinformatics, National Institute of Pharmaceutical Education and Research, Punjab, India
*Corresponding author: Dr. M Elizabeth Sobhia, Department of Pharmacoinformatics, National Institute of Pharmaceutical Education and Research (NIPER), Sector - 67, S. A. S. Nagar (Mohali), Punjab - 160062, India
Received: 13 January 2022; Accepted: 25 January 2022; Published: 31 March 2022
Article Information
Citation: Sonia Kumari, Elizabeth Sobhia M. Computer-Aided Molecular Design of CCR2 - CCR5 Dual Antagonists for the Treatment of NASH. Journal of Bioinformatics and Systems Biology 5 (2022): 63-77.
View / Download Pdf Share at FacebookAbstract
Non-alcoholic fatty liver disease (NAFLD), one of the most common liver diseases, is caused by the disruption of hepatic lipid homeostasis by various metabolic disorders. The progression of NAFLD into Non-alcoholic steatohepatitis (NASH) is mediated by inflammatory chemokines, cytokines, mitochondrial dysfunction, and oxidative stress resulting in hepatocyte inflammation, ballooning, apoptosis, and activation of hepatic stellate cells (HSC). NASH can further lead to cirrhosis, hepatic carcinoma, and also it is predicted to be a major cause of liver transplantation over the next 10 years. Chemokine receptors are majorly involved in recruiting the monocytes in the liver where they are converted into pro-inflammatory macrophages, which further activate the hepatic stellate cell (HSCs) to promote their survival while activating collagen production and fibrogenesis. Thus, chemokines and their receptor play a vital role in the pathogenesis of NASH and can be a potential target for the treatment of NASH. Herein, in this study, we have carried out a structure-based design of CCR2 and CCR5 dual antagonists. We performed pharmacophore mapping studies followed by virtual screening of commercial database to obtain novel molecules which can potentially act as CCR2 and CCR5 dual antagonists. We also performed molecular docking studies of newly obtained hits molecules to see their interactions with both CCR2 and CCR5 receptors.
Keywords
<p>NASH; Structure-Based Drug Design; Pharmacophore Mapping; Molecular Docking</p>
Article Details
Abbreviations:
NASH: Non-alcoholic steatohepatitis; NAFLD: Non- alcoholic fatty liver disease; BMI: Body Mass Index; HRQoL: Health-Related Quality of Life; HSC: Hepatic Stellate Cells; CCR-2: Chemokine Receptor- 2; CCR-5: Chemokine Receptor- 5; CVC: Cenicriviroc; EF: Enrichment Factor; SP: Standard Precision; EP: Extra Precision; EMBOSS: European Molecular Biology Open Software Suite; RCSB: Research Collaboratory for Structural Bioinformatics; NCI: National Cancer Institute
1. Introduction
Non-alcoholic steatohepatitis (NASH) is a liver disease in the absence of alcohol. Accumulation of excess fat in the liver is known as fatty liver [1]. Non- alcoholic fatty liver disease (NAFLD) is a common liver diseases which mainly caused by the disruption of hepatic lipid homeostasis. NAFLD is associated with obesity, insulin resistance or type2 diabetes, and other metabolic abnormalities, such as dyslipidemia and hypertension [2]. Cirrhosis, hepatocellular carcinoma, and liver-related mortality are all consequences of NASH, a progressive subtype of NAFLD. Hepatic fibrosis is the only histologic feature of NASH that has been found to be independently linked to long-term overall mortality, liver transplantation, and liver-related deaths [3]. The occurrence of NAFLD and NASH is increasing in western countries due to high dietary fat intake and low physical exercise which leads to insulin resistance [4]. When there is only steatosis in the liver, it is known as a less severe form of Non-alcoholic fatty liver disease (NAFLD) [5]. The progression of NAFLD into Non-alcoholic steatohepatitis (NASH) is mediated by inflammatory cytokines, mitochondrial dysfunction secondary to nutrient excess, and oxidative stress resulting in hepatocyte inflammation, ballooning, apoptosis, and activation of hepatic stellate cells (HSC) [6]. The probability of developing hepatic fibrosis is significantly high in individuals with steatohepatitis (NASH) than in those with simple steatosis. The fibrosis stage is the strongest predictor for cause and disease specific mortality in NASH patients [7].
Currently, the leading cause for liver transplantation includes NASH [8]. Although NASH is largely unrecognized and underdiagnosed, it is believed to affect at least three to five percent of the global population [9]. In its early stages, NASH is frequently thought to be asymptomatic and "silent", but symptoms such as persistent fatigue, malaise, apparent hepatomegaly, and upper-right quadrant abdominal fullness and discomfort have been recorded [10, 11]. Because NASH is usually asymptomatic, there is little published research on how patients feel it. However, emerging evidence suggests that individuals with NASH have a higher symptom burden and a lower health-related quality of life (HRQoL) [12]. Patients who are overweight, obese, have metabolic syndrome, insulin resistance, Type 2 diabetes, high levels of triglycerides or blood cholesterol, are more likely to be at risk for NASH [13, 14].
The approximation says that the global prevalence of NAFLD is as high as one billion [15]. According to a meta-analysis, the global prevalence of NAFLD is 25.24 percent, with the highest prevalence observed in the Middle East (31.78 percent) and South America (30.45 percent), and the lowest in Africa (13.48 percent) [16,17]. A smaller subset of these people may develop non-alcoholic steatohepatitis (NASH), which is one of the most common causes of hepatocellular carcinoma (HCC) in the United States and a reason for liver transplantation [18]. As obesity became more prevalent earlier in life, NAFLD/NASH prevalence began to rise in younger groups. NAFLD is predicted to affect 3–10 percent of youngsters in Western countries [19]. In America, the rate among those aged 18–35 years is at 24%, which has also been underestimated [20].
In India, the prevalence of NAFLD in the general population varies between 9% and 53%. The majority of studies conducted in urban areas have found a higher frequency than those conducted in rural areas. Rural West Bengal was the site of one of the first population-based studies in India, which revealed an obesity prevalence rate of 8.7% in predominantly nonobese populations [21]. After adjusting for sex, body mass index (BMI), diabetes mellitus (DM), and metabolic syndrome, population-based research from coastal south India indicated an overall NAFLD prevalence rate of 49.8%; the urban residence was shown to be linked with a greater risk for NAFLD [22]. The prevalence of NAFLD was found to be greater in urban populations (53.7%) than rural groups in the ongoing community-based Prospective Urban Rural Epidemiology (PURE) cohort research in north India (30.2 percent) [23, 24].
NAFLD is defined as fat accumulation in the hepatocytes when fat import or synthesis surpasses fat export or degradation. In these lipotoxic hepatocytes, a series of events occur, including immune mediator activation and inflammation, matrix remodeling via fibrogenesis and fibrinolysis, angiogenesis, and mobilization of liver progenitor cells [25].
The Pathophysiology of NAFLD is the accumulation of hepatic free fatty acids and triglycerides. The pathogenesis of progression of simple fatty liver to NASH is not fully understood, yet a "two-hit hypothesis" was initially proposed.
"Two hit hypothesis" suggests a "First hit" involves the lipid accumulation in the liver cells, which increases the risk for a "Second hit", which leads to hepatic injury, inflammation, and fibrosis. However, this theory has been criticized for being too simplistic in describing the pathogenesis of NAFLD. As a result, it has been replaced by the 'multiple-parallel hits' model, which appears to be a more exact portrayal of the process of NAFLD development and progression in subjects with genetic predisposition, in which various factors act in parallel and coherent ways [26, 27]. This multiple-hit hypothesis is based on the idea that obesity, insulin resistance, and changes in the intestinal microbiome are caused by genetic and environmental factors linked to dietary habits [28]. Insulin resistance promotes hepatic de novo lipogenesis and adipose tissue lipolysis which resulting into an increased flux of fatty acids to the liver. Insulin resistance will also lead to adipose tissue dysfunction inducing secretion of inflammatory cytokines [29].
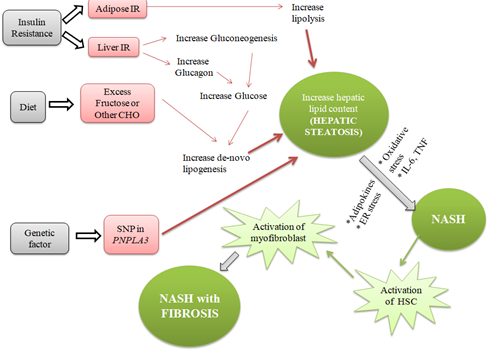
Figure1: Detailed pathophysiology of Non-alcoholic Steatohepatitis.
2. Methodology
2.1 Sequence analysis of CCR2 and CCR5 receptors
To find out the identity and similarity between the CCR2 and CCR5 receptor the Pairwise Sequence Alignment using EMBOSS Needle [30] (Online Sever) was done. The primary sequences for CCR2 and CCR5 receptors were retrieved from the UniprotKB database [31]. The accession numbers for CCR2 and CCR5 sequences were P41597 and P51681respectively.
2.2 Molecular docking
Docking studies were carried out using the Glide Module [32] of Maestro11.2. Prior to undergoing Protein-Ligand docking, protein structures for CCR2 (PDB ID: 5T1A) having resolution2.81 Å and CCR5 (PDB ID: 4MBS) having resolution 2.71 Å, obtained from the RCSB Protein Data Bank (www.rcsb.org) [33] was prepared using the Protein Preparation Wizard (Schrödinger Suite 2016 Protein Preparation Wizard) accessed via Maestro. Ligands including dual antagonist (CVC) and hits from virtual screening were subjected to the LigPrep module of Maestro. For geometry optimization, the OPLS3 force field was used. The grid box was generated at the ligand-binding site by selecting the co-crystallized ligand molecule and extended up to 15 Å as the inner box and 20 Å as the outer box for covering the binding site cavity entirely.
2.2.1 Validation of Docking Protocol: The validation of the docking protocol for both the receptors was done. To perform molecular docking validation protocol, the ligand in native co-crystallized structure BMS-681(in CCR2 receptor) [34] and Maraviroc (in CCR5 receptor) [35], were extracted and re-docked with the active cavity of CCR2 and CCR5 respectively.
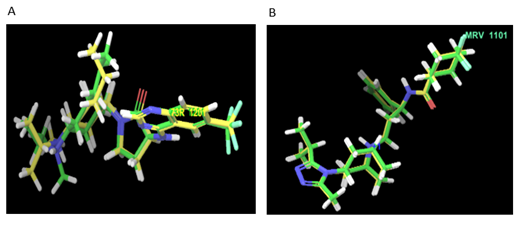
Figure 2: Superimposition of native and re-docked conformer (CCR2) having RMSD 1.64Å and (CCR5) having RMSD 2.1 Å.
2.3 Pharmacophore mapping
2.3.1 E- Pharmacophore generation (Structure-based pharmacophore): Phase [36] module of Schrodinger was used to generate the pharmacophore hypotheses from the docked poses of dual antagonist Cenicriviroc using the default set of six chemical features in Phase: hydrogen bond acceptor (HBA), hydrogen bond donor (HBD), negative ionizable group, positive ionizable group, aromatic ring, and hydrophobic group. The e- Pharmacophores [37] method utilizes the Glide XP scoring function to accurately characterize protein-ligand interactions and to generate the potential pharmacophoric feature, resulting in improved database screening enrichments.
2.3.2 Validation of hypothesis: Pharmacophore validation was done to determine whether the generated hypothesis is able to identify the actives from the database of ligands. The docked ligand (CVC) was considered as active which was then combined to decoys set having 1000 drug-like compounds (Drug-Like Ligand Decoys Set) [38] retrieved from Schrodinger to form an internal library of 1001 compounds. For validating the screening protocol, the ability of the e- pharmacophore to differentiate the actives from the internal library was evaluated. Enrichment factor (EF) is employed for recovering a fraction of the known actives after a fraction of the database has been screened and it was calculated using the following equation, EF=(a/n)/(A/N), where a is the number of actives retrieved in a sample size of n, A is the total number of actives, and N is the number of ligands in decoy dataset.
3. Results and Discussion
3.1 Sequence analysis of CCR2 and CCR5 receptors
The result of pairwise alignment shows that both receptors (CCR2 and CCR5) have the identity of 61.6% and similarity of 70.1%.

Figure 3: Sequence alignment of CCR5 (UniProtKB: P51681) with CCR2 (UniProtKB: P41597).
3.2 Binding site alignment
The binding site alignment showed that the most of amino acid residues of CCR2 at orthosteric site are identical with the binding site of the CCR5 receptor which has Maraviroc as a crystallized ligand. The residues which are not identical include the Ser101/Tyr89, His121/Phe109, Arg206/Ile198, PHE116/LEU104, THR287/MET279, and GLY191/SER179 in CCR2/CCR5 respectively.
3.3 Docking studies
Docking of dual antagonist Cenicriviroc [39] was done into the active pockets of both the receptors to find the important residues required for antagonism. The precision mode used was XP (Extra precision).
3.3.1 Molecular docking of dual antagonist cenicriviroc (CVC):
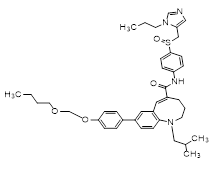
(5E)-8-[4-(2-butoxyethoxy)phenyl]-1-(2-methylpropyl)-N-[4-[(S)-(3-propylimidazol-4-yl)methylsulfinyl]phenyl]-3,4-dihydro-2H-1-benzazocine-5-carboxamide
|
Compound |
Receptor |
Docking score |
XP Score |
Glide Emodel |
|
Cenicriviroc |
CCR2 |
-9.931 |
-10.736 |
-116.605 |
|
CCR5 |
-7.259 |
-8.064 |
-102.550 |
Table 1: Docking results of dual antagonist (Cenicriviroc).
From the Docking of CCR2 known antagonist and CCR5 known antagonist, we got the important residues which are vital for binding of antagonist. The results showed that various residues involved in salt bridge interaction, π-π stacking interaction, π-cation interactions, and hydrogen bonding interactions. In the docking of dual antagonist (CVC), it was found that CVC was forming salt bridge interaction and hydrogen bond interactions with acidic residues Glu283 (in CCR5) and Glu291 (in CCR2) respectively. Tyr89 (in CCR5) and Trp98 (in CCR2) are involved in π-π stacking interaction. In the CCR5 receptor, it was found that CVC formed the π-cation interaction with Trp86. Apart from these interactions, hydrophobic interactions were also found. In the CCR2 receptor, CVC forms hydrophobic interaction with Trp98, Pro192, Val189, Leu 45, Tyr120, Ile263, Val289, and Phe194. Similarly, in CCR5 receptor Ile198, Phe112, Tyr108, Phe109, Trp248, Phe113, Leu255, and Tyr 251 were involved in hydrophobic interactions with CVC. From these docking studies, the important residues for antagonist binding have been identified.
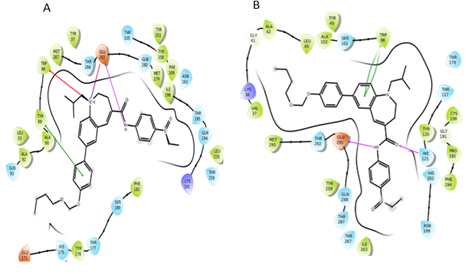
Figure 4: Interactions of CVC with CCR5 receptor and CCR2 receptor.
3.4 Pharmacophore mapping
3.4.1 E- Pharmacophore generation (Structure-based pharmacophore): The E-pharmacophore was generated for both receptors using receptor-ligand complex (CCR2 with CVC and CCR5 with CVC). The seven maximum features were selected to generate with 2Å as the minimum distance between two features. To account for the shape of the active site, a van der Waals scaling of 0.5 was used for receptor-based excluded volumes in each hypothesis.
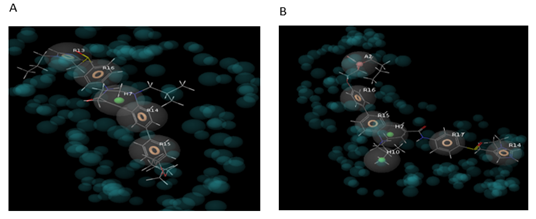
Figure 5: Pharmacophore map of CCR2 and CVC complex (HRRRR) and CCR5 and CVC complex (AHHRRRR).
Firstly, the docked complex of CVC with CCR2 receptor was subjected to the E– Pharmacophore and generated five features pharmacophore map (HRRRR) with four aromatic ring features and one hydrophobic feature. Similarly, the docked complex of CVC with CCR5 receptor was subjected to the E- pharmacophore and generated seven features pharmacophore map (AHHRRRR) with four aromatic ring features, one hydrophobic feature, and one hydrogen bond acceptor feature.
3.4.2 Merging of E- Pharmacophores: Two e- pharmacophores were prepared one for CCR2 (HRRRR) and one for CCR5 (AHHRRRR). These two pharmacophores were first aligned and then merged to obtain the dual antagonist. The merged hypothesis was prepared by selecting three aromatic ring features, one hydrophobic and one acceptor feature (AHRRR) using the Merged Hypothesis tool of the maestro.
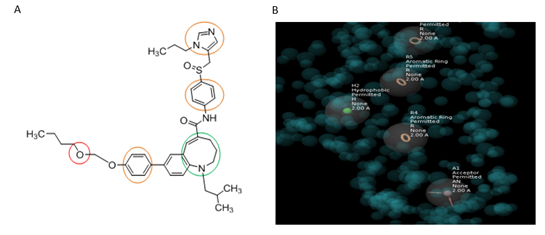
Figure 6: Cenicriviroc merged pharmacophore (AHRRR).
3.5 Pharmacophore based virtual screening
The NCI database was screened using the generated pharmacophore model (AHRRR) in order to search for potent dual antagonists. Phase Ligand screening module was used to screen the databases requiring a minimum of four feature matches with the hypothesis. The pharmacophore model (AHRRR) was able to screen 59 hits from the NCI database. The screened molecules showing the negative fitness score were rejected and the remaining 50 screened molecules were docked into active pockets of both receptors (CCR2 and CCR5) using glide standard precision (SP) scoring function. Then 20 ligands were selected from them according to their docking scores comparable in both receptors and docked again using glide extra precision (XP) scoring function. Then from XP docking, 5 hits have binding scores comparable to that of reference ligand cenicriviroc in both receptors (glide score -8.064 (in CCR5) and -10.736 (in CCR2) using glide XP scoring function) were selected for detailed evaluation.
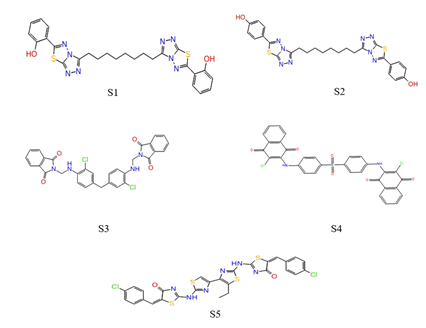
Figure 7: Top five screened molecules by XP docking.
Figure 8: Binding poses of top five screened molecules in pockets of CCR5 and CCR2 receptor.
The interactions of five top-scored hits were analyzed from the ligand interactions diagram. The pink-colored lines represent hydrogen bonds, green-colored lines represent π-π stacking interactions, and orange-colored lines represent halogen bond interactions. The protein-ligand interaction of the top five molecules is shown as:
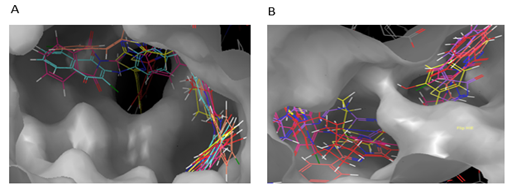
Figure 9: Interactions of S1 molecule with CCR5 and CCR2 receptor respectively.
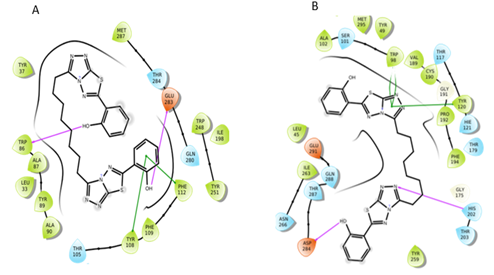
Figure 10: Interactions of S2 molecule with CCR5 and CCR2 receptor respectively
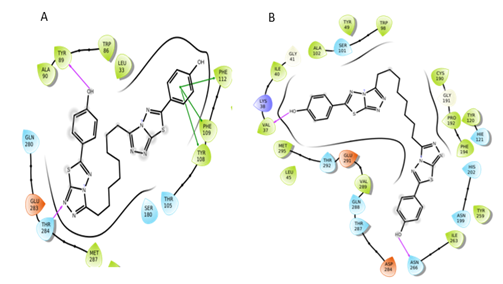
Figure 11: Interactions of S3 molecule with CCR5 and CCR2 receptor respectively.
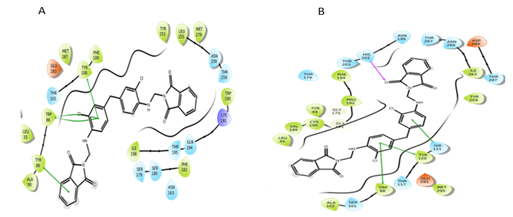
Figure 12: Interactions of S4 molecule with CCR5 and CCR2 receptor respectively.
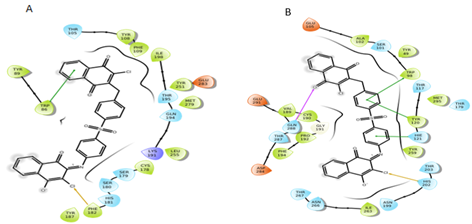
Figure 13: Interactions of S5 molecule with CCR5 and CCR2 receptor respectively.
By evaluating the chemical structures of the top five molecules, it was observed that all five molecules possess C2 symmetry. The docking results of the top five molecules showed that Thr284, Trp86, Tyr89, and Glu283 (in CCR5) and Asp283, Val37, Asn286, His202, and Gln288 (in CCR2) residues were involved in hydrogen bond interactions. The molecules also showed π-π stacking interaction with key residues Phe112, Tyr108, Phe109, Trp86, and Tyr89 (in CCR5) and HIE121, Trp98, Tyr120 (in CCR2). Additionally, in some molecule’s halogen bond was also observed. The residues which formed the halogen bond include Phe182, Thr195 (in CCR5), and Lys38 (in CCR2). The screened molecules showed the interactions with some key residues i.e., Phe112, Tyr108, Phe109 (in CCR5) and Trp98, Tyr120 (in CCR2) as same that of CVC interactions with CCR5 and CCR2 receptor.
|
Compound |
Receptor |
Docking score |
XP Score |
Glide Emodel |
|
S1 |
CCR5 |
-9.59 |
-9.629 |
-107.074 |
|
CCR2 |
-7.826 |
-9.629 |
-95.048 |
|
|
S2 |
CCR5 |
-9.507 |
-9.515 |
-101.872 |
|
CCR2 |
-7.233 |
-9.515 |
-91.588 |
|
|
S3 |
CCR5 |
-7.551 |
-7.551 |
-101.281 |
|
CCR2 |
-9.022 |
-7.551 |
-102.392 |
|
|
S4 |
CCR5 |
-5.723 |
-6.744 |
-93.791 |
|
CCR2 |
-6.859 |
-6.744 |
-92.53 |
|
|
S5 |
CCR5 |
-5.894 |
-6.714 |
-94.786 |
|
CCR2 |
-5.929 |
-6.714 |
-91.691 |
Table 2: Docking results for selected five hits molecules.
4. Conclusion
The objective of this study was to discover and design new dual antagonists which can bind to both CCR2 and CCR5 receptors. With a systematic integration of pharmacophore generation and virtual screening tools with multiple filters, an attempt was made to find molecules that can effectively bind to both receptors. For this study, we took cenicriviroc (CVC) as a reference molecule, which is clinically approved as a dual antagonist for CCR2 and CCR5 receptors. In this quest, we obtained five hits having comparable binding affinity as that of cenicriviroc (CVC) in both CCR2 and CCR5 receptors. These hits had diverse scaffolds related to structural features of cenicriviroc (CVC) and may act as dual antagonists.
Acknowledgments
We thank the National Institute of Pharmaceutical Education and Research (NIPER) SAS Nagar, Department of Pharmaceuticals, Ministry of Chemicals and Fertilizers, New Delhi, Government of India for providing the provision to work. We also thank CSIR, DST and DBT, New Delhi for the financial support offered to establish the infrastructure of the Department.
Declaration of Interests
All authors declare no competing interests.
References
- Lee J, Hong SW, Rhee EJ, Lee WY. GLP-1 receptor agonist and non-alcoholic fatty liver disease. Diabetes & metabolism journal 36 (2012): 262-267.
- Esler WP, Bence KK. Metabolic targets in nonalcoholic fatty liver disease. Cellular and molecular gastroenterology and hepatology 8 (2019): 247-267.
- Angulo P, Kleiner DE, Dam-Larsen S, Adams LA, Bjornsson ES, Charatcharoenwitthaya P, et al. Liver fibrosis, but no other histologic features, is associated with long-term outcomes of patients with nonalcoholic fatty liver disease. Gastroenterology 149 (2015): 389-397.
- Svegliati-Baroni G, Saccomanno S, Rychlicki C, Agostinelli L, De Minicis S, Candelaresi C, et al. Glucagon-like peptide-1 receptor activation stimulates hepatic lipid oxidation and restores hepatic signalling alteration induced by a high-fat diet in nonalcoholic steatohepatitis. Liver International 31 (2011): 1285-1297.
- Caligiuri A, Gentilini A, Marra F. Molecular pathogenesis of NASH. International journal of molecular sciences 17 (2016): 1575.
- Townsend S, Newsome P. new treatments in non-alcoholic fatty liver disease. Alimentary pharmacology & therapeutics 46 (2017): 494-507.
- Boubia B, Poupardin O, Barth M, Binet J, Peralba P, Mounier L, et al. Design, synthesis, and evaluation of a novel series of indole sulfonamide peroxisome proliferator activated receptor (PPAR) α/γ/δ triple activators: discovery of lanifibranor, a new antifibrotic clinical candidate. Journal of medicinal chemistry 61 (2018): 2246-2265.
- Noureddin M, Vipani A, Bresee C, Todo T, Kim IK, Alkhouri N, et al. NASH leading cause of liver transplant in women: updated analysis of indications for liver transplant and ethnic and gender variances. Official journal of the American College of Gastroenterology ACG 113 (2018): 1649-1659.
- Chalasani N, Younossi Z, Lavine J E, Diehl A M, Brunt EM, Cusi K, et al. The diagnosis and management of non-alcoholic fatty liver disease: Practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology 55 (2012): 2005-2023.
- Heeringa M, Hastings A, Yamazaki S, De Koning P. Serum biomarkers in nonalcoholic steatohepatitis: value for assessing drug effects?. Biomarkers in medicine 6 (2012): 743-757.
- Nagaratnam N, Nagaratnam K, Cheuk G. Geriatric diseases: evaluation and management. Springer (2018).
- Balp MM, Krieger N, Przybysz R, Way N, Cai J, Zappe D, et al. The burden of non-alcoholic steatohepatitis (NASH) among patients from Europe: A real-world patient-reported outcomes study. JHEP Reports 1 (2019): 154-161.
- Gastaldelli A, Cusi K. From NASH to diabetes and from diabetes to NASH: Mechanisms and treatment options. JHEP Reports 1 (2019): 312-328.
- Godoy-Matos AF, Silva Júnior WS, Valerio CM. NAFLD as a continuum: from obesity to metabolic syndrome and diabetes. Diabetology & Metabolic Syndrome 12 (2020): 1-20.
- Perumpail BJ, Khan MA, Yoo ER, Cholankeril G, Kim D, Ahmed A. Clinical epidemiology and disease burden of nonalcoholic fatty liver disease. World journal of gastroenterology 23 (2017): 8263.
- Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease—meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 64 (2016): 73-84.
- Pappachan JM, Babu S, Krishnan B, Ravindran NC. Non-alcoholic fatty liver disease: a clinical update. Journal of Clinical and Translational Hepatology 5 (2017): 384.
- Anstee QM, Targher G, Day CP. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nature reviews Gastroenterology & hepatology 10 (2013): 330-344.
- Alisi A, Feldstein AE, Villani A, Raponi M, Nobili V. Pediatric nonalcoholic fatty liver disease: a multidisciplinary approach. Nature reviews Gastroenterology & hepatology 9 (2012): 152-161.
- Doycheva I, Watt KD, Alkhouri N. Nonalcoholic fatty liver disease in adolescents and young adults: the next frontier in the epidemic. Hepatology 65 (2017): 2100-2109.
- Duseja A, Singh SP, Saraswat VA, Acharya SK, Chawla YK, Chowdhury S, et al. Non-alcoholic fatty liver disease and metabolic syndrome—position paper of the Indian National Association for the Study of the Liver, Endocrine Society of India, Indian College of Cardiology and Indian Society of Gastroenterology. Journal of clinical and experimental hepatology 5 (2015): 51-68.
- Chalmers J, Ban L, Leena KB, Edwards KL, Grove JL, Aithal GP, et al. Cohort profile: the Trivandrum non-alcoholic fatty liver disease (NAFLD) cohort. BMJ open 9 (2019): e027244.
- Mahajan R, Duseja A, Kumar R, Chakraborti A, Lakshmi PA. Community based study to determine the prevalence of nonalcoholic fatty liver disease (NAFLD) and its metabolic risk factors in urban and rural communities of north India. In Journal of Gastroenterology and Hepatology 34 (2019): 310-310.
- De A, Duseja A. nonalcoholic Fatty liver Disease: indian Perspective. Clinical Liver Disease 18 (2021): 158.
- Machado MV, Diehl AM. Pathogenesis of nonalcoholic steatohepatitis. Gastroenterology 150 (2016): 1769-1777.
- Parthasarathy G, Revelo X, Malhi H. Pathogenesis of nonalcoholic steatohepatitis: an overview. Hepatology communications 4 (2020): 478-492.
- Li Q, Dhyani M, Grajo JR, Sirlin C, Samir AE. Current status of imaging in nonalcoholic fatty liver disease. World journal of hepatology 10 (2018): 530.
- Buzzetti E, Pinzani M, Tsochatzis EA. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD). Metabolism 65 (2016): 1038-1048.
- Bugianesi E, Moscatiello S, Ciaravella M, Marchesini G. Insulin resistance in nonalcoholic fatty liver disease. Current pharmaceutical design 16 (2010): 1941-1951.
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. Journal of molecular biology 48 (1970): 443-453.
- Morgat A, Lombardot T, Coudert E, Axelsen K, Neto TB, Gehant S, et al. Enzyme annotation in UniProtKB using Rhea. Bioinformatics 36 (2020): 1896-1901.
- Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, et al. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein− ligand complexes. Journal of medicinal chemistry 49 (2006): 6177-6196.
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The protein data bank. Nucleic acids research 28 (2000): 235-242.
- Zheng Y, Qin L, Zacarías NVO, de Vries H, Han GW, Gustavsson M, et al. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature 540 (2016): 458-461.
- Tan Q, Zhu Y, Li J, Chen Z, Han GW, Kufareva I, et al. Structure of the CCR5 chemokine receptor–HIV entry inhibitor maraviroc complex. Science 341 (2013): 1387-1390.
- Dixon SL, Smondyrev AM, Rao SN. PHASE: a novel approach to pharmacophore modeling and 3D database searching. Chemical biology & drug design 67 (2006): 370-372.
- Salam NK, Nuti R, Sherman W. Novel method for generating structure-based pharmacophores using energetic analysis. Journal of chemical information and modeling 49 (2009): 2356-2368.
- Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. Journal of medicinal chemistry 47 (2004): 1739-1749.
- Anstee QM, Neuschwander-Tetri BA, Wong VWS, Abdelmalek MF, Younossi ZM, Yuan J, et al. Cenicriviroc for the treatment of liver fibrosis in adults with nonalcoholic steatohepatitis: AURORA Phase 3 study design. Contemporary clinical trials 89 (2020): 105922.


 Impact Factor: * 4.2
Impact Factor: * 4.2 Acceptance Rate: 77.66%
Acceptance Rate: 77.66%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 