Conserved Structural Motifs across Diverse Vitamin B12 Binding Proteins
Pushya Pradeep1, Deepesh Nagarajan1,2*
1Department of Biotechnology, M.S. Ramaiah University of Applied Sciences, Bangalore - 560054
2Department of Microbiology, St. Xaviers College, Mumbai - 400001
*Corresponding author: Deepesh Nagarajan Department of Biotechnology, M.S. Ramaiah University of Applied Sciences, Bangalore - 560054.
Received: 12 October 2023; Accepted: 21 October 2023; Published: 06 November 2023
Article Information
Citation: Pushya Pradeep, Deepesh Nagarajan. Conserved Structural Motifs across Diverse Vitamin B12 Binding Proteins. Journal of Bioinformatics and Systems Biology. 6 (2023): 312-326.
View / Download Pdf Share at FacebookAbstract
Vitamin B12, a significant organometallic porphyrin derivative, is an essential growth factor in most organisms. Its primary function is as a cofactor in a diverse range of enzymes, belonging to diverse protein families. Understanding the conserved binding-site characteristics that enable different proteins to recognise the same ligand is therefore of significant importance. In-depth binding-site comparisons, ligand-based site alignments, clustering, and tree computing were performed employing a non-redundant dataset of known vitaminB12 binding proteins to derive the principles for vitamin B12 recognition. The 53 protein structures that bind to vitamin B12 can be clustered into 14 categories, and contain 8 unique binding motifs. Knowledge of these binding-site determinants could be used to detect the function of unknown proteins. An example analysis on the Swiss-Prot database revealed 15 proteins from pathogenic species with identified sequence motifs, indicating that they may have potential vitamin B12 binding activity and have potential as therapeutic targets.
Keywords
<p>Structural Bioinformatics, Structural Motifs, Ligand Binding, Binding Site Signature, Vitamin B12, Cobalamin, Phylogenetic Database, Sequence Motif.</p>
Article Details
1. Introduction
Vitamin B12 (cobalamin) is a large ligand with a complex structure[1]. It consists of a corrin ring co-ordinately bound to cobalt. This imparts an intense red colouration, making cobalamin, and cobalamin-binding proteins easily recognizable in solution. The corrin ring is covalently linked to an S-adenosyl methionine (AdoMet) moiety. This moiety is further composed of amide, phosphatidyl, ribosyl, and dimethyl-benzimidazole moieties. The AdoMet chain may interact with the corrin ring via a benzimidazole to cobalt co-ordinate bond.
It is well established that vitamin B12 is essential for fungal, plant and animal life[2]. Vitamin B12 acts as a cofactor for several classes of enzymes, including but not limited to, dehydratases, mutases, transferases, lyases, synthases, and reductases. Excluding enzymes, vitamin B12 is also found bound to specific transporters[3][4] that absorb the molecule in the ileum for distribution in tissues. A diverse range of folds host vitamin B12, including alpha-beta proteins like TIM barrels, rossmann folds, flavodoxin-like folds, up-down bundles, triple helical bundles, and several folds only associated with vitamin B12 binding.
Vitamin B12 binds to proteins with diverse sequences, structures, and enzymatic activities. The detection of common motifs that allow vitamin B12 binding is therefore a significant question.
Although several vitamin B12 dependent enzymes have been identified and characterised, little work has been done in identifying common sequence or structural motifs. An early study based on two crystallised vitamin B12 binding structures, namely a methionine synthase and methylmalonyl CoA mutase, detected a catalytic triad composed of asp-his-ser residues[5]. Two distinct conformations of the vitamin B12 ligand were noted. The catalytic triad occurred in the open conformation, but was replaced by corrin's dimethyl benzimidazole moiety in the closed conformation. Further, a conserved sequence motif asp-X-his-X(2)-gly-(41)-ser-X-leu-(26,28)-gly-gly was detected based on a subset of four known vitamin B12 binding enzymes. An AdoMet binding motif arg-X(3)-gly-tyr was also identified surrounding the benzimidazole moiety. Later work identified the role of vitamin B12 in three more enzyme classes: isomerases, methyltransferases, and reductive dehalogenases[6].
This study aims to identify further characteristics of vitamin B12 binding sites, in order to derive useful sequence and structural motifs. Such motifs would be of great use in the functional annotation of uncharacterised genes and proteins. With the exponential increase in both the number of solved protein structures[7], and annotated protein sequences, it has become possible to further the analysis of vitamin B12-binding motifs at both a sequence and structural level. Motifs identified in this study may help annotate function to uncharacterised sequences, especially those from clinically significant organisms. As an example survey, we have used sequence and structural motifs detected to identify 15 sequences from pathogenic organisms that may have the ability to bind vitamin B12.
2. Methods:
2.1 Creation of a non-redundant dataset:
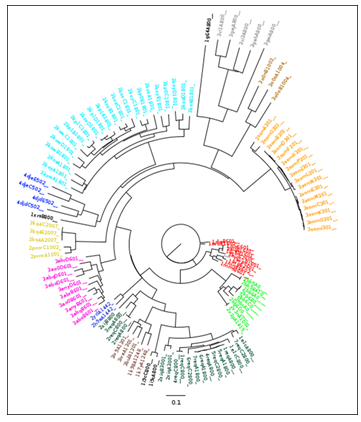
Figure 1: A neighbour joining tree produced from PocketMatch scores for all known vitamin B12 binding pockets.Colour coding is explained in Table 1. It should be noted that this data was previously described for the validation of the Pocketmatch (version 2.0) algorithm [10].
|
Colour (figure 1) |
PDB ID |
Protein function |
Sequence similarity |
E.C. Number |
||
|
95.00% |
70.00% |
50.00% |
||||
|
red |
1DIO |
Diol dehydratase |
4.2.1.28 |
|||
|
dark cyan |
1E1C |
Methylmalonyl-CoA mutase |
5.4.99.2 |
|||
|
1G64 |
ATP:corrinoid adenosyltransferase |
1G64 |
1G64 |
1G64 |
2.5.1.17 |
|
|
119C |
Glutamate mutase |
119C |
119C |
119C |
5.4.99.1 |
|
|
red |
1IWB |
Diol dehydratase |
1IWB |
1IWB |
1IWB |
4.2.1.28 |
|
red |
1IWP |
Glycerol dehydratase |
1IWP |
4.2.1.30 |
||
|
purple |
1K7Y |
MetH C-terminal fragment |
2.1.1.13 |
|||
|
purple |
1K98 |
MetH C-terminal fragment |
2.1.1.13 |
|||
|
red |
1MMF |
Glycerol dehydratase |
4.2.1.30 |
|||
|
dark cyan |
1REQ |
Methylmalonyl-CoA mutase |
1REQ |
1REQ |
5.4.99.2 |
|
|
1XRS |
Lysine 5,6-aminomutase |
1XRS |
1XRS |
1XRS |
5.4.3.4 |
|
|
green |
2BB5 |
Transcobalamin |
2BB5 |
NA |
||
|
green |
2BB6 |
Transcobalamin |
2BB6 |
2BB6 |
2BB6 |
NA |
|
green |
2BBC |
Transcobalamin |
NA |
|||
|
blue |
2H9A |
Corrinoid iron-sulfur protein |
2H9A |
2H9A |
2H9A |
NA |
|
yellow |
2PMV |
Human intrinsic factor |
2PMV |
2PMV |
2PMV |
NA |
|
dark cyan |
2REQ |
Methylmalonyl-CoA mutase |
5.4.99.2 |
|||
|
green |
2V3N |
Transcobalamin |
NA |
|||
|
green |
2V3P |
Transcobalamin |
NA |
|||
|
dark cyan |
2XIJ |
Methylmalonyl-CoA mutase |
2XIJ |
2XIJ |
2XIJ |
5.4.99.2 |
|
dark cyan |
2XIQ |
Methylmalonyl-CoA mutase |
5.4.99.2 |
|||
|
blue |
2YCL |
Corrinoid iron-sulfur protein |
NA |
|||
|
magenta |
3ABO |
Ethanolamine ammonia-lyase |
4.3.1.7 |
|||
|
magenta |
3ABQ |
Ethanolamine ammonia-lyase |
3ABQ |
3ABQ |
3ABQ |
4.3.1.7 |
|
magenta |
3ABR |
Ethanolamine ammonia-lyase |
4.3.1.7 |
|||
|
magenta |
3ANY |
Ethanolamine ammonia-lyase |
4.3.1.7 |
|||
|
magenta |
3AO0 |
Ethanolamine ammonia-lyase |
4.3.1.7 |
|||
|
red |
3AUJ |
Diol dehydratase |
4.2.1.28 |
|||
|
purple |
3BUL |
MetH C-terminal fragment |
3BUL |
3BUL |
3BUL |
2.1.1.13 |
|
gray |
3CI1 |
PduO-type ATP:co(I)rrinoid adenosyltransferase |
2.5.1.17 |
|||
|
gray |
3CI3 |
PduO-type ATP:co(I)rrinoid adenosyltransferase |
3CI3 |
3C13 |
3CI3 |
2.5.1.17 |
|
gray |
3GAH |
PduO-type ATP:co(1)rrinoid adenosyltransferase |
2.5.1.17 |
|||
|
gray |
3GAI |
PduO-type ATP:co(I)rrinoid adenosyltransferase |
2.5.1.17 |
|||
|
gray |
3GAJ |
PduO-type ATP:co(1)rrinoid adenosyltransferase |
2.5.1.17 |
|||
|
purple |
3IV9 |
B12-dependent Methionine Synthase (MetH) |
2.1.1.13 |
|||
|
purple |
3IVA |
B12-dependent methionine synthase (MetH) |
2.1.1.13 |
|||
|
cyan |
3KOW |
Omithine 4,5 aminomutase |
5.4.3.5 |
|||
|
cyan |
3KOX |
Omithine 4,5 aminomutase |
5.4.3.5 |
|||
|
cyan |
3KOY |
Omithine 4,5 aminomutase |
5.4.3.5 |
|||
|
cyan |
3KOZ |
Omithine 4,5 aminomutase |
5.4.3.5 |
|||
|
cyan |
3KP0 |
Omithine 4,5 aminomutase |
5.4.3.5 |
|||
|
cyan |
3KP1 |
Omithine 4,5 aminomutase |
3KP1 |
3KP1 |
3KP1 |
5.4.3.5 |
|
yellow |
3KQ4 |
Intrinsic Factor |
NA |
|||
|
dark yellow |
3O0N |
Ribonucleotide reductase |
1.17.4.1 |
|||
|
dark yellow |
3O0O |
Ribonucleotide reductase |
3O0O |
3O0O |
3O0O |
1.17.4.1 |
|
dark cyan |
3REQ |
Methylmalonyl-CoA mutase |
5.4.99.2 |
|||
|
3SOM |
Methylmalonic aciduria and homocystinuria type C (mmachc) |
3SOM |
3SOM |
3SOM |
NA |
|
|
blue |
4DJD |
Folate-free corrinoid iron-sulfur protein (cfesp) |
4DJD |
4DJD |
NA |
|
|
blue |
4DJE |
Folate-free corrinoid iron-sulfur protein (cfesp) |
NA |
|||
|
dark cyan |
4REQ |
Methylmalonyl-CoA mutase |
5.4.99.2 |
|||
|
dark cyan |
5REQ |
Methylmalonyl-CoA mutase |
5.4.99.2 |
|||
|
dark cyan |
6REQ |
Methylmalonyl-CoA mutase |
5.4.99.2 |
|||
|
dark cyan |
7REQ |
Methylmalonyl-CoA mutase |
5.4.99.2 |
|||
Table 1: Clustering of vitamin B12 binding sites into 14 categories (colour coded) based on sequence similarity and Enzyme classification data. Protein structures of the same function, but with a low sequence similarity are highlighted using dashed borders. It should be noted that this data was previously used to validate the Pocketmatch (version 2.0) algorithm [10].
An advanced search through the PDB revealed 53 structures containing the B12 ligand. The structures obtained were classified on the basis of percentage sequence similarity, at a 70% threshold, and E.C. Number[8]. A literature survey was used to functionally classify those structures lacking annotated E.C. Numbers. The E.C based and percentage sequence similarity based classification schemes yielded identical results (Table 1).
This classification scheme, reflecting sequence and enzymatic diversity, reflected the structural diversity of the B12 ligand binding sites. All amino acid residues within 5Å of B12 ligands were extracted. All such binding sites were compared using the PocketMatch algorithm[9][10] for the detection of binding site similarities, and clustered using the Neighbour Joining algorithm[11] implemented in the Phylip software package[12] (Figure 1). It should be noted that nodes are labelled by the following convention: (PDB ID)(chain)(residue number). Clustering of binding sites within the neighbour joining tree correlates well with the E.C based and percentage sequence similarity based classification schemes, despite dual clustering for methylmalonyl-CoA mutases and corrinoid iron sulphur proteins.
It was observed that all structures could be clustered into 14 different categories (Table 1). In order to create a non-redundant dataset, each category was represented by a single structure of the highest resolution. However, in the case of methylmalonyl-CoA-mutase, two structures (PDB IDs: 1req and 2xij) show relatively low sequence similarity (<70%). Similarly, for corrinoid iron-sulphur proteins, two structures (PDB IDs: 2h9a and 4djd) show relatively low sequence similarity (<70%).
In both cases, both structures have been included in the final dataset. Their low sequence similarities would help identify conserved binding-site residues, presumably with important ligand-binding or catalytic functions.
2.2 Binding site alignment:
From the 16 proteins identified, all residues located within 5Å of the B12 ligand were extracted. In cases where multiple binding sites occur per structure, the first binding site was extracted and analysed. A ligand based structural superimposition was carried out on these 16 binding sites. The B12 ligand contains two distinct moieties: a large corrin and a relatively smaller adomet. The adomet moiety is further divided into three distinct moieties: a phosphatidyl, ribosyl, and dimethyl benzimidazole moiety. The B12 ligand adopts two main conformations: open and closed. In the open conformation, the dimethyl benzimidazole moiety is displaced far from the corrin moiety. The corrin moieties' cobalt may be coordinately bonded with the NE2 atom of histidine. In the closed conformation, the benzimidazole moiety is located proximal to the corrin moiety, and stabilizes the cobalt atom via a coordinate bond with it's N3B atom. In order to fully account for conserved residues in both these conformations, two distinct ligand-based binding-site alignments were carried out: a corrin based and benzimidazole based alignment. An Octave implementation of the Kabsch algorithm[13] was used to align atoms C1-19, N20-23, and CO1 of the corrin ring to an ideal template. Similarly, atoms N1B, N3B, C2B, and C4-9B of the benzimidazole ring were aligned to an ideal template. The crystal structure of an ATP:co(I)rrinoid adenosyltransferase (PDB ID: 3ci3) lacked a benzimodazole moiety and was not aligned to the benzimidazole template.
2.3 Interpretation of aligned binding sites:
A 99% confidence interval (P-value of 0.01) has been chosen for this study.
Conventionally, the clustering of aligned binding sites is performed by visual inspection and subjective interpretation of residue proximities. Here, we have developed a method to unambiguously cluster binding site residues into cliques based on the graph-theoretic principles.
Table 2: In red: residue-cliques identified as belonging primarily to the upper catalytic triad. In blue: residue-cliques identified as belonging primarily to the lower catalytic triad. In green: residue-cliques identified as forming the hydrophobic shroud. In yellow: residue-cliques without any functional description
A MATLAB implementation of the Bron-Kerbosch algorithm[14] was adapted to Octave, for use in deriving conserved structural motifs from ligand-based binding site alignments. The Bron-Kerbosch algorithm is used for finding maximal cliques in an undirected graph. Here, a clique is defined as a subset of vertices that are connected to each and every other vertice in the subset by an edge.
In order to adapt the algorithm for use on aligned binding sites, all amino acid residues within an alignment were considered to be nodes. The 20 amino acids were treated as 20 separate, mutually exclusive, graphs. Every graph would contain a single type of amino acid aligned over 15/16 binding sites. Within any given graph, if the all-atom to all-atom RMSD between any two residues was measured at 3Å or less, an edge was drawn between them. The Bron-Kerbosch algorithm was then used on these graphs to detect any cliques of residues within the same spatial location, bounded within a 3Å limit. These cliques are listed in Table 2.
In order to determine the statistical significance of the cliques detected, random rotations of the aligned binding sites were carried out. In one iteration of random rotation, aligned binding sites are rotated around the centroid of the corrin or benzimidazole moiety, using randomised euclidean rotation angles. Bron-Kerbosch clique detection is then carried out, as previously described. This entire process is repeated for 1000 iterations.
In randomly rotated pockets, all cliques are formed by chance. The total number of cliques formed by chance, and the number of residues within these cliques is tallied to establish the null hypothesis at the selected confidence interval. P-value cutoffs for different residues are listed in Table 3. All cliques containing more residues than accounted for by the null hypothesis are hereafter reported.
|
Percentage probability of locating a clique by chance with: |
|||||
|
1 residue |
2 residues |
3 residues |
4 residues |
5 residues |
|
|
GLY |
91.00813 |
8.68842 |
0.30021 |
0.00323 |
0 |
|
ALA |
91.76163 |
7.98996 |
0.24584 |
0.00256 |
0 |
|
VAL |
96.91933 |
3.04712 |
0.03356 |
0 |
0 |
|
LEU |
97.84656 |
2.14777 |
0.00567 |
0 |
0 |
|
ILE |
98.81775 |
1.17647 |
0.00578 |
0 |
0 |
|
MET |
99.36754 |
0.63246 |
0 |
0 |
0 |
|
PHE |
99.11822 |
0.87805 |
0.00374 |
0 |
0 |
|
TYR |
98.41241 |
1.58452 |
0.00308 |
0 |
0 |
|
TRP |
99.66891 |
0.33109 |
0 |
0 |
0 |
|
PRO |
97.64766 |
2.34869 |
0.00365 |
0 |
0 |
|
SER |
95.26298 |
4.68476 |
0.05226 |
0 |
0 |
|
CYS |
98.93913 |
1.06087 |
0 |
0 |
0 |
|
THR |
96.68963 |
3.28527 |
0.0251 |
0 |
0 |
|
ASN |
97.89683 |
2.09751 |
0.00567 |
0 |
0 |
|
GLN |
99.29087 |
0.70913 |
0 |
0 |
0 |
|
ASP |
96.01296 |
3.90813 |
0.0706 |
0.00831 |
0 |
|
GLU |
99.04394 |
0.95012 |
0.00594 |
0 |
0 |
|
LYS |
99.82828 |
0.17172 |
0 |
0 |
0 |
|
ARG |
99.47453 |
0.52547 |
0 |
0 |
0 |
|
HIS |
96.17198 |
3.71924 |
0.10878 |
0 |
0 |
Table 3: P-value cut-offs for different residues with random rotations. All clique sizes falling within the level of statistical significance have their corresponding P-values are coloured orange.
The Ligand Protein Contacts (LPC)[15] tool has been used to describe the nature of any protein-ligand interactions identified within cliques. Hydrogen bonding and Van der Waals interactions have been quantified using LPC. In this study, we have described all hydrogen bonds as such: (Donor [D]: [residue/ligand] [atom], Acceptor [A]: [residue/ligand] [atom]). Protein-protein interactions within a binding-site have been analysed using the Rosetta 3.5 software suite[16]. Aromatic interactions have been described based on available literature and chemical intuition.
2.4 Detection of structural motifs and homology modelling:
The PATTINPROT search webserver[17] was used to detect two identified, conserved vitamin-B12 binding sequence motifs from amongst the Swiss-Prot database sequences[18]. The ModBase webserver [19] was used to obtain homology-based structural models of select sequences. Mustang (v3.2.1) [20] was used for structural alignments of homology-based structural models with known vitamin B12 binding proteins.
3. Results:
16 non-redundant vitamin B12 binding sites have been aligned using Kabsch's algorithm. Conserved patterns have been interpreted using various tools including Ligand Protein Contacts, Bron-Kerbosch's algorithms, and Rosetta3.5. Conserved structural patterns have been interpreted as cliques within graphs. Here, multiple cliques have been grouped into a few structural motifs that best describe their collective nature and function in relation to their parent proteins. A full account of structural motifs is given below.
An analysis of amino-acid composition of these sites (Figure 2) was carried out to give insight into the assignment of cliques within structural motifs. The ratios of residues found within the binding sites was compared to the ratios of residues within the parent proteins. It was observed that glycine, tyrosine, and histidine were found at statistically significantly higher frequencies inside binding sites. Conversely, methionine, glutamine and lysine were found at statistically significantly lower frequencies inside binding sites. The catalytic triad can account for the abundance of glycine and histidine. A 'hydrophobic shroud' surrounding the B12 ligand can account for the abundance of tyrosine and the scarcity of glutamine and lysine. The scarcity of methionine, however, is unexplained.
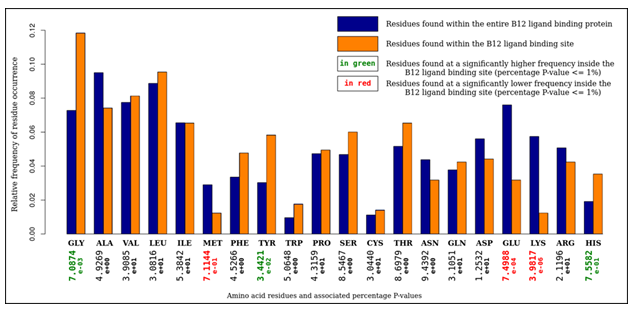
Figure 2: An analysis of amino acid composition of selected vitamin B12 binding sites. It is observed that glycine, tyrosine, and histidine residues were overrepresented, whereas methionine, glutamine, and lysine residues were underrepresented.
3.1 The catalytic triad:
Five structures were identified with an asp-his-ser catalytic triad, and with the B12 ligand in the open conformation. These structures include glutamate mutase (PDB ID: 1i9c), methylmalonyl CoA mutase (PDB ID: 1req, 2xij), Lysine 5,6-aminomutase (PDB ID: 1xrs), and ornithine 4,5 aminomutase (PDB ID: 3kp1). Bron Kerbosch clustering identified several residues that were spatially conserved within the B12 binding sites of these enzymes. For ease of description, these residues have been described separately, based on their location around the B12 ligand.
3.2 Conserved residue interactions with the corrin ring:
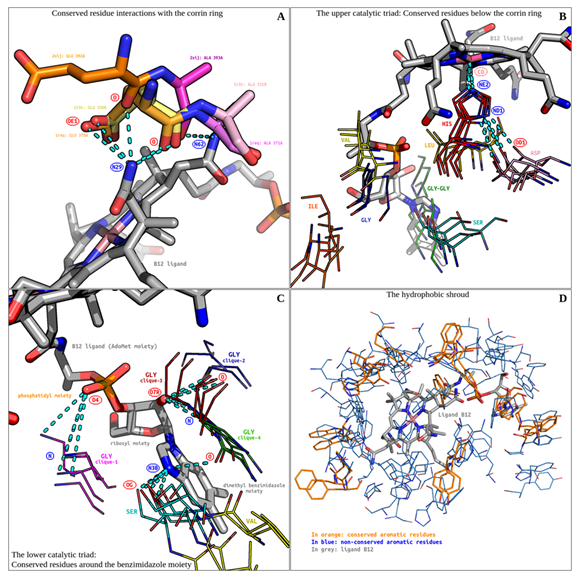
Figure 3: The vitamin B12 binding site. (A) Conserved residue interactions with the corrin ring are shown. (B) The upper catalytic triad's conserved residues are shown. (C) The lower Catalytic triad's conserved residues are shown. (D) The hydrophobic shroud is shown. All hydrogen bonds are displayed as cyan-coloured broken lines.
Bron-Kerbosch clustering revealed two distinct alanine and glutamate residue cliques, neighbouring the N29 and N62 atoms of the corrin ring (Figure 3). All residues belong to three catalytic triad containing enzymes (PDB ID: 1i9c, 1req, 2xij). Since the residues are contiguous and share common hydrogen bonding donors, they have been discussed together. A bonding cluster is formed involving (D: ligand N29, A: glutamine OE1), (D: ligand N29, A: glutamine/alanine O) hydrogen bonds, and a single (D: ligand N62, A: alanine O) hydrogen bond. Of particular interest is the ligand N29 atom. It acts as a hydrogen bonding donor to atoms O and OE1 of a conserved glutamine residue (PDB ID: 1i9c, 1req). The third residue pair (PDB ID: 2xij) shows an unusual spatial translocation. The alanine backbone is superposed upon the previous two glutamate backbones. This results in a the backbone O hydrogen bonding with both the N29 and N62 atoms of the ligand. Although the translocated glutamate is incapable of forming hydrogen bonds with its OE1 and OE2 atoms, its backbone O hydrogen bonds with N29. Ligand atom N29 thus acts as a lynchpin in this local conserved hydrogen bonding network. Unlike glutamine, the side chain of alanine does not directly interact with the ligand, and therefore the reason for their conservation is not apparent. Alanine could be conserved solely for structural reasons.
3.2 The upper catalytic triad: Conserved residues below the corrin ring:
This cluster comprises all five enzymes identified with catalytic triads (Figure 3). Bron Kerbosch clustering confirmed the presence of spatially conserved asp-his-ser residues. This triad was first described for methionine synthase and methylmalonyl CoA-mutase[5], which were described to fall within a conserved Asp-X-His-X(2)-Gly-X(41)-Ser-X-Leu-X(26,28)-Gly-Gly sequence pattern. It was reported that the triad may regulate conformational changes, allowing switching between enzyme-catalytic and enzyme-activation cycles. This was supported by mutational studies[6] confirming the essentiality of the residues in the catalytic triad. The glycine residues were further described as playing a role in accommodating the S-Adenosyl methionine (AdoMet) side chain of the B12 ligand.
Amongst all the residues present in the catalytic triad and the conserved sequence, only histidine makes direct contact with the corrin moiety via an NE1 to CO coordination bond. A conserved inter-residue (D: histidine ND1, A: aspartate OD1) hydrogen bond further stabilises this complex.
Hydrophobic residues also appear to play an important role in B12 binding-site structure. Previous studies have reported a conserved Leucine and a functionally important valine. The valine residue was reported in methionine synthase, being modified when the cob(I)alamin form of the enzyme reacts with nitrous oxide[21]. However, the conservation of valine across different enzymes was not reported..
Here, we confirm the conservation of leucine across enzymes glutamate mutase (PDB ID: 1i9c) and methylmalonyl CoA-mutase (PDB ID: 1req, 2xij). We detected the conservation of valine across enzymes glutamate mutase (PDB ID: 1i9c), lysine 5,6-aminomutase (PDB ID: 1xrs), and ornithine 4,5 aminomutase (PDB ID: 3kp1). Valine was conserved at the same location reported, approximately 4Å distant from the C3P atom on the AdoMet side chain of the B12 ligand. This confirms valine’s essential functional role at this position. Further, we detected a previously unreported conservation on isoleucine. Isoleucine from four distinct enzymes (PDB ID: 1i9c, 1req, 1xrs, 2xij) remain conserved at a position approximately 4Å distant from the C5R atom of the S-Adenosyl AdoMet side chain of the B12 ligand. Although the functions of the conserved leucine and isoleucine are unreported, we believe that they may simply play a role in shape-complementarity of the B12 binding-pocket, to help accommodate largely hydrophobic portions of the B12 ligand.
The protein-ligand interactions involving residues serine, glycine, and the dimethyl benzimidazole moiety are discussed later. These residues are shown for perspective only.
3.3 The lower catalytic triad: Conserved residues around the benzimidazole moiety:
All residue clusters discussed so far were derived from a corrin moiety based structural alignment. However, ligand B12 adopts multiple conformations, and may show distinct rotamers even within the open conformation. A corrin-centric alignment may fail to detect residues interacting with the AdoMet segment of the B12 ligand, specifically the dimethyl benzimidazole ring. An alternate, dimethyl benzimidazole based alignment was used prior to Bron-Kerbosch clustering. Cliques from both alignment schemes were then combined into the description of the lower catalytic triad.
Glycine and serine residues were found to be clustered around the benzimidazole ring (Figure 3).
Serine residues are part of the catalytic triad previously discussed. Glycine residues are ubiquitous and fall into 4 cliques. Glycines of the clique-1 are conserved near the phosphatidyl moiety of the AdoMet chain. They form a (D: glycine O, A: ligand O4) hydrogen bond. Glycines from clique-2 sparsely interacted with the ligand. A sole residue interacts with the phosphatidyl moiety via a (D: glycine O, A: ligand O5) hydrogen bond. These bonds may help stabilize the polar, charged moiety inside a relatively apolar B12-binding pocket.
Clique-3 and clique-4 are composed of contiguous, neighbouring glycine residues. The glycines of clique-3 extensively interact with the ribosyl moiety of the AdoMet chain, through (D: glycine O, A: ligand O7R) and (D: glycine N, A: ligand O7R) conserved hydrogen bonds. The glycines of clique-4 interact sparingly with the ligand. A single (D: glycine O, A: ligand N3B) hydrogen bond links this clique with the dimethyl benzimidazole moiety.
Dimethyl benzimidazole-centric alignments revealed that another methionine synthase (PDB ID: 3bul) possesses spatially conserved glycine and serine residues, although lacking a catalytic triad. In this case, the catalytic histidine dissociates from the corrin moiety and forms intermodular contacts with the AdoMet moiety[22], which is described as helping generate a distribution of conformers required for the enzyme's catalytic and reactivation cycles.
Dimethyl benzimidazole-centric alignments revealed more glycine cliques than a corrin-based alignment. Glycine is especially abundant around the benzimidazole ring. It has previously been reported that glycine residues may help accommodate the neighbouring benzimidazole ring, due to their absence of CA atoms[5]. We believe that glycine residues play additional roles in the B12 ligand binding site. As previously described, numerous, conserved glycine backbone to ligand hydrogen bonds have been observed. Amino acids with CA atoms and long side-chains may interfere with hydrogen bonding by destabilizing the interaction-space.
Furthermore, multiple backbone interactions per glycine residue would be impossible without the glycine backbone's ability to adopt extreme Ramachandaran angles. The Ramachandaran angles for all glycine residues have been superimposed onto a standard (non-glycine) Ramachandaran map[23] (Figure 4). This was done to compare the backbone conformations of glycine with standard, non-glycine residues. Glycine residues of clique-1 and clique-4 adopt most stable conformations. Glycine residues of clique-2, while not adopting highly stable conformations, nevertheless are located in the allowed region. Glycine residues of clique-3 adopt conformations that place them in the completely disallowed region, with some conformations overlapping the generously allowed region. Interestingly, glycine residues of clique-3 form hydrogen bonds with the B12 ligand using both N and O backbone atoms. This double-backbone hydrogen bonding may only be possible under extreme Ramachandaran angles, adoptable only by glycine.
With the exception of methionine synthase (PDB ID: 3bul), all serine residues interact with the benzimidazole ring through a (D: serine OG, A: ligand N3B) hydrogen bond. Serine is part of the catalytic triad, and a highly conserved interaction of this nature is expected.
All serine residues are followed in sequence by valine. Valine residues are located approximately 5Å away from the closest atom on the dimethyl benzimidazole ring, and are pointed away from it. This makes conventional valine-ligand interactions unlikely. Valine may be conserved at this position owing to inter-residue interactions. Methyltransferase (PDB ID: 4djd) is the only protein containing a conserved valine, in the absence of other residues associated with the catalytic triad.
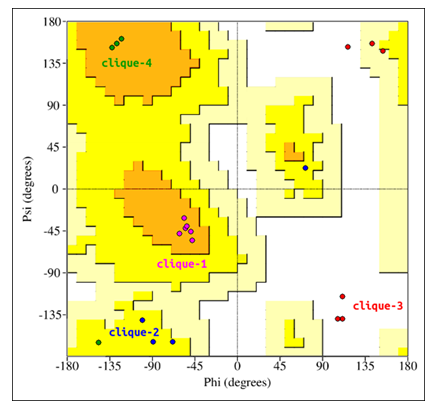
Figure 4: Glycine residues of the lower catalytic triad. Glycine residues belonging to cliques 1 to 4 have been superimposed onto a standard (non-glycine) Ramachandaran map.
Taking into consideration these findings, we would like to expand the conserved sequence associated with the catalytic triad to the following: Asp-X-His-X-Val-Gly-X(2,3)-Ile-X(37,38)-Ser-Val-Leu-X(26,28)-Gly-Gly.
3.4 The hydrophobic shroud:
With the exception of motifs associated with the catalytic triad, composed entirely of mutase enzymes, there is little in common between all classes of B12 ligand binding proteins. Vitamin B12 is large, with several hydrophobic and aromatic groups included in its structure. This would suggest that B12 binding sites would include several large hydrophobic, aromatic residues involved in nonspecific hydrophobic and aromatic interactions.
It was observed that in all binding sites included in this study, the aromatic residues namely phenylalanine, tyrosine, tryptophan, and histidine, form an envelope around the entire B12 ligand. This envelope shows some conservation in residue location, however the majority appear to be involved in non-specific, non catalytic hydrophobic interactions (Figure 3).
Aromatic interactions between residues, ligands, and small molecules have been extensively studied. One study on closest contact distances analysed inter-residue aromatic interactions in the hydrophobic cores of proteins[24]. It was concluded that such interactions occur at distances of less than 4.5Å. The porphyrin ring itself is known to be aromatic. It forms pi-stacked aggregates with the individual molecules spaced 3.4-3.6Å apart[25]. Other studies[26][27][28] involving various aromatic interactions place the distances at which pi-stacking occurs at 3-6Å.
Interactions between the B12 ligand and aromatic residues were found to be predominantly non-aromatic. The corrin ring of the B12 ligand is a non-aromatic porphyrin derivative. Unlike porphyrin, corrin contains a C19 to C1 covalent linkage, eliminating the C20 atom. This eliminates C20-related pi-bonding, breaking conjugation over the entire ring. LPC predicted 112 non-polar interactions between aromatic residues and corrin, excluding histidines interacting with cobalt. Since corrin is non-aromatic itself, these interactions must be interpreted as solely hydrophobic in nature.
The benzimidazole ring, however, is known to be aromatic. LPC predicted 15 non-polar interactions between ligand aromatic rings and the aromatic benzimidazole ring. In order to interpret the nature of these interactions, the distances of all benzimidazole non-polar interactions were compared to the distances of all corrin non-polar interactions. Corrin, being non-aromatic, acted as an ideal control. It should be noted that all non-polar interactions occur within 3-6Å. The Welch two sample T-test was applied to both datasets, in order to determine if they form two distinct distributions. The Welch test calculated a p-value of 0.127, larger than the p-value of 0.01 adopted fot this study. A full description of the statistical analysis, with source code, can be found in the supplementary data.
Therefore pi-stacking is not a statistically significant mode of protein-B12 ligand interaction. Most aromatic residues, excluding histidine, around the B12 ligand are only involved in non-specific, hydrophobic interactions that enhance shape-complementarity, and as such would vary considerably from protein to protein.
3.5 Pairwise comparison of vitamin B12 binding sites
The PocketAlign algorithm was used for the pairwise comparison of binding sites, in order to detect similarities shared between small groups of binding sites. 120 binding site comparisons were performed, capturing all combinations of binding site pairs. For every pairwise alignment, all
residue-correspondences scoring 2 or above on the BLOSUM62 matrix were tallied to determine the similarity score for the binding site pair (BLOSUM-score). A pairwise alignment matrix was constructed based on the BLOSUM-scores. Differing BLOSUM-scores for the same pairwise comparison was resolved by accepting the higher score, creating a 120-unit BLOSUM-score matrix (figure 5A). It was observed that the scores followed a Poisson distribution with a mode of 4 (figure 5B). Analysis of the distribution revealed that all binding site pairs with a BLOSUM-score of 8 and above passed the P-value threshold of 0.05. An undirected graph (figure 5C) was constructed from all such binding site pairs.
Of the 16 binding sites (nodes), 11 formed the largest connected component (major component), 2 were isolated, sharing a common edge (minor component), and 3 were unconnected. Based on PocketAlign superpositions, these nodes were further classified based on shared aligned residues.
4PocketAlign-based subgraphs (PA-subgraph I-IV) were annotated. All 6 binding sites of the catalytic triad are represented in PA-subgraph I, forming a clique. A mutase, lyase, and dehydratase (PDB ID: 1i9c, 3abq, 1iwb) form PA-subgraph II. Despite their divergent substrates and enzymatic function, they share 5 conserved residues (figure 5D). A mutase and two methyltransferases (PDB ID: 1iwb, 4djd, 2h9a) form PA-subgraph III (figure 5E). 5 residues are conserved, all of which are hydrophobic in nature. The minor component contains two vitamin B12 transporters (PDB ID: 2pmv, 2bb6), which show considerable structural conservation. For all connected nodes, the SCOP classification for all nodes was noted or assigned (Gough et al., 2001). It was observed that despite sequence, structural and functional diversity, 9 binding sites belonged to 3 unique SCOP folds. The remaining binding sites were unclassifiable. Therefore, 16 binding sites were classified into 8 unique motifs: the catalytic triad/PA-subgraph I (6), PA-subgraph I (3), PA-subgraph II(3), PA- subgraph III(2), and 4 singletons.
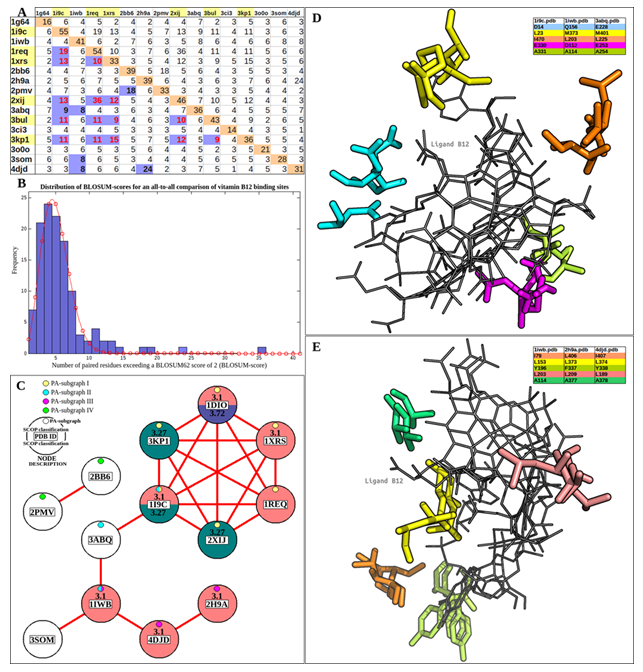
Figure 5: PocketAlign analysis of vitamin B12 binding sites. (A) A 120-unit BLOSUM-score matrix created from PocketAlign residue pairings. Binding sites belonging to the catalytic triad are highlighted in yellow. Scores for catalytic triad residues are highlighted in red. (B) The distribution of BLOSUM-scores has been fitted to a poisson distribution (red line) with a mode of 4. (C) An undirected graph representing all statistically significant binding-site relations, as determined from BLOSUM-scores. (D) PA-subgraph II, sharing 5 conserved residues. 5E: PA-subgraph III, sharing 5 conserved residues.
3.6 Database-wide detection of structural motifs:
The previously mentioned, the vitamin-B12 related catalytic triad: Asp-X-His-X(44)-Ser, was expanded to include other functionally relevant residues, resulting in an Asp-X-His-X-X-Gly-X(41)-Ser-X-Leu-X(26,28)-Gly-Gly sequence motif. This study expanded the sequence motif to the following: Asp-X-His-X-Val-Gly-X(2,3)-Ile-X(37,38)-Ser-Val-Leu-X(26,28)-Gly-Gly. This expansion was based on observations after ligand superposition and Bron-Kerbosch clique detection. It should be noted that a sequence alignment alone would not be able to detect such a motif. A sequence alignment cannot help identify any protein ligand interactions, nor the functional relevance of these residues.
The expanded sequence motif was scanned against the Swiss-Prot database using the PATTINPROT search webserver. Protein sequences with a motif similarity of at least 60%, corresponding to a maximum of 4 residue substitutions within the query motif, were selected (Table 4). Selected protein sequences accepted only if they contained the residues of the catalytic triad. However, asp to asn mutations were accepted.
The motif search revealed 874 sequences that passed the search criteria. The vast majority (665) of these sequences were found to be confirmed enzymes. Ligases (90), dehydratases (68), synthases (52), mutases (43), and transferases (35) were found to be the most abundant confirmed enzyme classes. Non-enzymatic proteins including transporters (25), Zinc fingers (8), and regulators (4) were also observed. Many proteins belonged to clinically relevant, pathogenic organisms, including, but not limited to, HIV (21), Mycobacterial species (34), Plasmodium species (4), Chlamydia species (11), Shigella species (2), Salmonella species (2) and Legionella pneumophila (3).
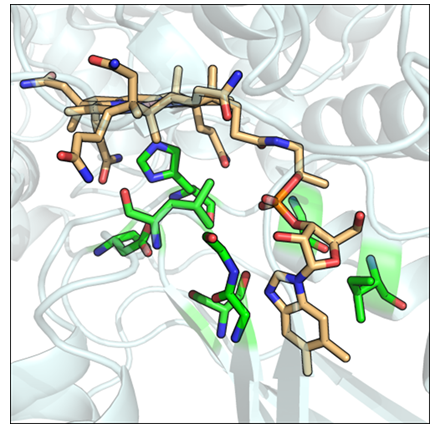
Figure 6: Modbase generated structural model for a probable methylmalonyl-CoA mutase (Swiss-Prot ID: MUTB_MYCBO). Conserved structural motif residues are in green. The vitamin B12 ligand is colored orange.
Of interest to this study are proteins whose functions are classified as probable (79), putative (20), and uncharacterised (19). Nearly all such proteins have been inferred from homology. The classification and functional annotation of these proteins may be aided by the discovery of possible vitamin B12 binding functions. Especially important are insufficiently characterised proteins from pathogenic organisms, which may serve as potential drug targets. The motif search revealed such proteins, from organisms including Plasmodium falciparum, Mycobacterium tuberculosis, Mycobacterium leprae, Chlamydia trachomatis, Legionella pneumophila, and Mycoplasma genitalium (Table 2). Model organisms and closely related organisms are also included. The protein sequences were then queried for homologous structures using ModBase. For all but three sequences, homologous structures were found. Modbase predictions are poor. Most assigned template structures do not match the putative functions of their sequence counterparts. For example, a probable cytosol aminopeptidase sequence (Swiss-Prot ID: AMPA_LEGPA) was assigned the homologous structure of a beta amyloid peptide fragment (PDB ID: 1qyt-A). However, for two enzyme types, namely a probable fructose-bisphosphatase aldolase class I and a probable methylmalonyl-CoA mutase, the template structures selected by ModBase match the functions of their homologous sequence counterparts. For two probable methylmalonyl-CoA mutase sequences originating from Mycobacterial species (Swiss-Prot ID: MUTB_MYCBO, MUTB_MYCTU), ModBase models contained a correctly arranged vitamin B12 binding structural motif. The B12 molecule was fitted into the modelled binding sites using the Mustang(v3.2.1) structural alignment program (Figure 6). Further refinements were made within Pymol. A single figure is shown, as both methylmalonyl-CoA mutases share a 100% sequence identity. Both sequences, however, share only a 68.93% sequence identity with their template. The close structural compatibility is considered to be further evidence of the vitamin B12 binding abilities of these proteins.
|
Swiss Prot ID |
Putative Protein function |
Organism |
Motif similarity |
Homologous structure(s) PDB ID |
|
ALTH1_PLAF7 |
E3 ubiquitin ligase |
Plasmodium falciparum |
61% |
None |
|
Y089_MYCSS |
methyltransferase |
Mycobacterium sp. |
62% |
None |
|
Y098_MYCSK |
methyltransferase |
Mycobacterium sp. |
62% |
None |
|
Y1496_MYCTU |
GTPase |
Mycobacterium tuberculosis |
69% |
1qzx(A), 2ffh(A), 1rij(A) |
|
Y1533_MYCBO |
GTPase |
Mycobacterium bovis |
69% |
1qzx(A), 2ffh(A), 1rij(A) |
|
ALF1_CHLMU |
fructose-bisphosphate aldolase class 1 |
Chlamydia muridarum |
61% |
1ojx(A) |
|
ALF1_CHLPN |
fructose-bisphosphate aldolase class 1 |
Chlamydia pneumoniae |
61% |
1ojx(A) |
|
ALF1_CHLTR |
fructose-bisphosphate aldolase class 1 |
Chlamydia trachomatis |
61% |
1ojx(A) |
|
AMPA_LEGPA |
cytosol aminopeptidase |
Legionella pneumophila |
70% |
1qyt(A) |
|
AMPA_LEGPC |
cytosol aminopeptidase |
Legionella pneumophila |
70% |
1qyt(A) |
|
AMPA_LEGPL |
cytosol aminopeptidase |
Legionella pneumophila |
70% |
1qyt(A) |
|
MUTB_MYCBO |
methylmalonyl-CoA mutase |
Mycobacterium bovis |
61% |
1req(A) |
|
MUTB_MYCTU |
methylmalonyl-CoA mutase |
Mycobacterium tuberculosis |
61% |
1req(A) |
|
Y246_MYCGE |
Uncharacterized |
Mycoplasma genitalium |
62% |
1hp1(A) |
|
Y392_MYCLE |
glycosyl hydrolase |
Mycobacterium leprae |
62% |
1h54(A) |
Table 4: Protein sequences extracted from the Swiss-Prot database using the PATTINPROT webserver. Only sequences from clinically significant organisms are shown.
4. Discussion
This study has analysed the structural motifs required to constitute a vitamin B12 binding site, improving upon previous work[5]. A structural motif based around a core catalytic triad was detected and expanded. Its sequence counterpart was queried across the Swiss Prot database, revealing multiple enzymes with similar motifs. Uncharacterised proteins from clinically significant organisms were identified, and structural models were constructed. In the case of two methylmalonyl-CoA mutases, the modelled structures contained vitamin B12 binding structural motifs that closely matched natural counterparts, providing further evidence of vitamin B12 binding abilities.
Future work will involve the synthesis, cloning, and expression of the genes of these insufficiently characterised proteins, especially those inferred from homology, in order to assay for vitamin B12 binding and enzymatic action. Several insufficiently characterised proteins from pathogens have the potential to act as drug targets, especially if they serve a critical enzymatic function.
Author contribution statement:
The authors confirm contribution to the paper as follows: study conception and design: DN; data collection: PP, DN; analysis and interpretation of results: PP, DN; draft manuscript preparation: PP, DN. All authors reviewed the results and approved the final version of the manuscript.
Competing interests:
The authors declare no competing interests.
References
- Hodgkin DC, Pickwirth J, et al. Structure of Vitamin B12: The Crystal Structure of the Hexacarboxylic Acid derived from B12 and the Molecular Structure of the Vitamin. Nat 176 (1955): 325-328.
- Martens JH, Barg H, et al. Microbial production of vitamin B12. Appl Microbiol. Biotechnol 58 (2002): 275-285.
- Mathews FS, Gordon MM, et al. Crystal structure of human intrinsic factor: cobalamin complex at 2.6Å resolution. PNAS 104 (2007): 17311-17316.
- Wuerges J, Garau G, et al. Structural basis for mammalian vitamin B12 transport by transcobalamin. PNAS 103 (2006): 4386-4391.
- Ludwig ML, Matthew RG. Structure-based perspectives on B12-dependent enzymes. Annu Rev Biochem 66 (1997): 269-313.
- Banerjee R, Ragsdale SW. The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu Rev Biochem 72 (2003): 709-247.
- Berman HM, Westbrook J, et al. The Protein Data Bank. Nucleic Acids Res 28 (2000): 235-242.
- Webb Edwin C, et al. Enzyme nomenclature 1992. Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes. No. Ed. 6. Academic Press (1992).
- Yeturu K, Chandra N. PocketMatch: a new algorithm to compare binding sites in protein structures. BMC bioinformatics 9 (2008): 543.
- Nagarajan D, Chandra N. PocketMatch (version 2.0): A parallel algorithm for the detection of structural similarities between protein ligand binding-sites. National Conference on Parallel Computing Technologies (2013): 177-187.
- Naruya S, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4 (1987): 406-425.
- Plotree D, Plotgram D. PHYLIP-phylogeny inference package (version 3.2) (1989).
- Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Crystallographica Section A: Crystal Physics, Diffraction, Theoretical and General Crystallography 32 (1976): 922-923.
- Bron C, Kerbosch J. Algorithm 457: finding all cliques of an undirected graph. Communications of the ACM 16 (1973): 575–577.
- Sobolev V, Sorokine A, et al. Automated analysis of interatomic contacts in proteins. Bioinformatics 15 (1999): 327-332.
- Kaufmann KW, Lemmon GH, et al. Practically useful: what the Rosetta protein modeling suite can do for you. Biochemist 49 (2010): 2987-2998.
- Combet C, Blanchet C, et al. NPS@: Network Protein Sequence Analysis. Trends Biochem Sci 25 (2000): 147-149.
- Boeckmann B, Bairoch A, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 31 (2003): 365-370.
- Pieper U, Eswar N, et al. MODBASE: a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res 34 (2006): 291-295.
- Konagurthu AS, Whisstock J C. MUSTANG: a multiple structural alignment algorithm. Proteins: Structure, Function, and Bioinformatics 64 (2006): 559-574.
- Drummond JT, Matthews RG. Nitrous Oxide Inactivation of Cobalamin-Dependent Methionine Synthase from Escherichia coli: Characterization of the Damage to the Enzyme and Prosthetic Group. Biochem 33 (1994): 3742-3750.
- Datta S, Koutmos M, et al. A disulfide-stabilized conformer of methionine synthase reveals an unexpected role for the histidine ligand of the cobalamin cofactor. PNAS 105 (2008): 4115-4120.
- Laskowski R A, MacArthur MW, et al. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography 26 (1992): 283-291.
- McGaughey GB, Gagné M, et al. pi-Stacking interactions alive and well in proteins. J Biol Chemist 273 (1998): 15458-15463.
- Hunter C A, Sanders JK. The nature of pi-pi interactions. JACS 112 (1990): 5525-5534.
- Cauët E, Rooman M, et al. Histidine-aromatic interactions in proteins and protein-ligand complexes: quantum chemical study of x-ray and model structures. J Chemical Theory Computation 1 (2005): 472-483.
- Hunter CA, Singh J, et al. pi-pi interactions: The geometry and energetics of phenylalanine-phenylalanine interactions in proteins. J Mol Biol 218 (1991): 837-846.
- Sinnokrot M O, Valeev EF, et al. Estimates of the ab initio limit for π-π interactions: The benzene dimer. J Am Chem Soci 124 (2002): 10887-10893.


 Impact Factor: * 4.2
Impact Factor: * 4.2 Acceptance Rate: 77.66%
Acceptance Rate: 77.66%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 