Fungal Contaminants Associated with Groundnut (Arachis Hypogaea) Seeds
Isalar OF.1,3*, Ogbuji NG.2, Okungbowa FI.3, Ataga AE2.4
1Department of Biological Sciences, Edwin Clark University, Kiagbodo, Delta State, Nigeria
2Department of Plant Science and Biotechnology, University of Port Harcourt, Rivers State, Nigeria
3Department of Plant Biology and Biotechnology, University of Benin, Edo State, Nigeria
4Regional Center for Biotechnology and Bio-resources Research Laboratory, University of Port Harcourt, Rivers State
*Corresponding author: Isalar Oluwatoyin Funmilayo, Department of Biological Sciences, Edwin Clark University, Kiagbodo, Delta State, Nigeria.
Received: 18 November 2021; Accepted: 30 November 2021; Published: 29 December 2021
Article Information
Citation: Isalar OF, Ogbuji NG, Okungbowa FI, Ataga AE. Fungal Contaminants Associated with Groundnut (Arachis hypogaea) Seeds. Journal of Bioinformatics and Systems Biology 4 (2021): 182-193.
View / Download Pdf Share at FacebookAbstract
Groundnut (Arachis hypogaea L.) commonly known as ‘peanut’ is one of the staple foods in Nigeria. This study is aimed at isolating and identifying the fungi associated with groundnut (Arachis hypogaea) using cultural techniques and molecular method. Fungal organisms were isolated from the seeds using both Standard Blotter and Potato Dextrose Agar methods. Fungi were identified using molecular tools. Fungal DNA extraction was carried out using Quick-DNA Fungal/Bacterial MiniPrepTM Kit (Zymo Research Group, California, USA). Polymerase chain reaction amplification of the Internal transcribed spacer genes was done using universal primer pair; ITS4 and ITS5. Sequencing of the PCR products allowed for species identification on National Centre for Biotechnology Information (NCBI) database. The findings from this study revealed six fungal organisms viz Aspergillus tamarii, Lasiodiplodia iranensis, Macrophomina phaseolina, Penicillium citrinum, Aspergillus oryzae, and Aspergillus pennicillioides. Phylogenetic analysis of the obtained sequences showed the relationship that exists among the fungal isolates. The results obtained from this study shows that groundnut harbors several fungal organisms some of which are pathogenic and may diminish its productivity or possibly cause harm to man through consumption.
Keywords
<p>Chain reaction; Fungi; Groundnut; Internal transcribed spacer; Molecular identification; Polymerase</p>
Article Details
1. Introduction
Groundnut (Arachis hypogaea L.), is an important food and fodder crop in the farming systems of developing countries [1]. Groundnut is an annual legume which is also known as peanut, earthnut, monkeynut and goobers. It is the 13th most important food crop and 4th most important oilseed crop of the world. Groundnut seeds are a nutritional source of vitamin E, niacin, calcium, phosphorus, magnesium, zinc, iron, riboflavin, thiamine and potassium. Groundnut kernels are consumed directly as raw, roasted or boiled kernels or oil extracted from the kernel is used as culinary oil. It is also used as animal feed (oil pressings, seeds, green material and straw) and industrial raw material (oil cakes and fertilizer). These multiple uses of groundnut plant make it an excellent cash crop for domestic markets as well as for foreign trade in several developing and developed countries [2]. One of the major constraints facing the productivity and availability of healthy groundnut produce worldwide are the losses and spoilage caused by fungi, bacteria, viruses, insects, nematodes and parasitic weeds. Seed-borne disease have been found to affect the growth and productively of crop plants. A seed-borne pathogen present externally or internally or associated with the seed as contaminant may cause seed abortion, seed rot, seed necrosis, reduction or elimination of germination capacity as well as seedling damage resulting in development of disease at later stages of growth by systemic or local infection [3]. Fungi like Aspergillus niger, Aspergillus flavus, Alternari adianthicola, Curvularia lunata, Curvularia pellescens, Fusarium oxysporum, Fusarium equiseti, Macrophomina phaseolina, Rhizopus stolonifer, Penicillum digitatum and Penicillum chrysogenum cause discoloration, rotting, shrinking, seed necrosis, loss in germination capacity and toxification to oilseeds [4]. Arachis hypogaea is a geocarpic crop grown by farmers in various parts of the world especially in West Africa. Highly susceptible to infection by pathogenic fungi, groundnut is widely consumed by a lot of people without knowledge of the fungal pathogens associated with them which can be detrimental to their health. The use of spores or pictures in identifying these fungi organisms can prove abortive in most cases, therefore molecular characterization is absolutely necessary for proper identification of the fungi associated with groundnut seeds. This study was carried out to isolate and identify the fungal organisms associated with groundnut Arachis hypogaea L. using cultural techniques and molecular method.
2. Materials and Methods
2.1. Study Area
The study was carried out at the Regional Center for Biotechnology and Bio-resources Research Laboratory, University of Port Harcourt, Choba, Rivers State. Sequencing of the PCR products was done at the International Institute of Tropical Agriculture (IITA), Ibadan.
2.2. Collection of Materials
Raw groundnut seeds were purchased at Ubogo market, Delta State. Other materials such as forceps, conical flask, Petri dishes, filter paper, measuring cylinder, distilled water, inoculating loop, Bunsen burner, aluminum foil paper, cotton wool, masking tape, Quick DNA Fungal/Bacterial Mini-prep Kit (Zymo Research Laboratories, California, USA), refrigerated centrifuge, micro-pipettes, micro-tips, micro-centrifuge tubes, weighing balance, surgical blade, agarose powder, measuring flask, electrophoresis tank, gel documentation system, viewing dye (EZ vision gel- blue light), lactic acid and ethanol (70%) were used in the study.
2.3. Isolation of Fungi from Arachis hypogaea Seeds
Standard Blotter Method as Recommended by International Seed Health Testing Association [5] was used to isolate fungi from groundnut seeds. The groundnut seeds were surface sterilized in a measuring cylinder with 70% ethanol for 2-3 minutes and rinsed three times with sterile distilled water. The Petri-dishes were lined with 3 layers of sterilized Whatman’s filter paper (9cm). Five groundnut seeds were plated per Petri-dish equidistantly. The plated groundnut seeds were incubated at room temperature (25±2oC) in the laboratory for 7 days. Frequency of occurrence of fungi was determined based on the score method recommended by [6]; where numeral ‘1’ represents absence of fungi and ‘2’ represents presence of fungi, therefore any value above ‘1’ indicates presence of fungi. After incubation, the isolated fungi were sub-cultured on Potato Dextrose Agar to obtain pure cultures of fungi.
2.4. Extraction of Fungal DNA
Genomic DNA was extracted following the protocol of Quick-DNATM Fungal/Bacterial MiniPrep Kit (Zymo Research Group, California, USA) with modifications. The fungal mycelium was scrapped off from the surface of the culture using a sterilized surgical blade and transferred into a sterilized mortar. Seven hundred and fifty (750μl) of lysis solution was added to the sample before homogenizing with liquid nitrogen. The homogenized sample was added to microcentrifuge tubes (1.5ml) and placed in a centrifuge at 10,000 x g for 1 minute. 400μl of the supernatant was transferred to a Zymo-Spin IV Filter (orange top) in a collection tube and centrifuged at 7,000 x g for 1 minute. 1,200μl of genomic lysis buffer was added to the filtrate in the collection tube obtained from the last step. 800μl of the mixture was transferred to a Zymo-Spin IIC Column in a collection tube and centrifuged at 10,000 x g for 1 minute. The flow through was discarded and the process repeated. 200ul of DNA pre-wash buffer was added to the Zymo-Spin IIC Column in a new collection tube and centrifuged at 10,000 x g for 1 minute. 500ul of g-DNA wash buffer was added to the Zymo-Spin IIC Column and centrifuged at 10,000 x g for 1 minute. The Zymo-Spin IIC Column was transferred to a clean micro-centrifuge tube and 100μl of DNA elution buffer was added directly to the column matrix and centrifuged at 10,000 x g for 30 seconds to elute the DNA. DNA was stored at 4oC till when needed. DNA quantity (concentration) and quality was determined using Nanodrop 2000c spectrophotometer (Thermo Fisher Scientific, USA) and gel electrophoresis (1% agarose gel) respectively.
2.5. PCR Amplification and Sequencing
Amplification of the ITS genes was carried out using the primer pair; ITS4 (5’-CCTCCGCTTATTGATATGS-3’) - forward and ITS5 (5’-GAAGTAAAAGTCGTAACAAGG-3’) - reverse. The PCR reaction mix contained 1 µl of DMSO, 2.5 µl of 10× PCR buffer, 2 µl of 2.5 mM NTPs, 1 µl of 25 mM MgCl2, 1 µl each of forward primer and reverse primer, 0.1 µl of 5 µ µl- 1 of Taq DNA polymerase, 13.4 µl of Nuclease-free water and 3 µl of 10 ng µl-1 DNA. The cycling parameters includes initial denaturation at 94oC for 5 min., 36 cycles of denaturation, annealing and elongation at 94oC for 30 s, 54oC for 30 s and 72oC for 45 s respectively. These were followed by final elongation at 72oC for 7 min. and the samples were held in the thermal cycler (Eppendorf) at 10oC. Amplified products were viewed on 1.5% agarose gel under Ultraviolet (UV) light. Sequencing of PCR products was carried out on an ABI 3500 genetic analyzer (Thermo Fisher Scientific, Massachusetts, U.S.A.). Sanger sequencing was performed using single-stranded DNA template, a DNA primer, deoxynucleotide triphosphates (dNTPs), di-deoxynucleotide triphosphates (ddNTPs) and a DNA polymerase.
2.6. Data Analysis
Raw sequences of the ITS1-2 genes were edited on MEGA X software [7] to remove sequencing errors. Sequences were blasted on National Centre for Biotechnology information (NCBI) database for species identification. The sequences of the fungal ITS genes obtained from the study were compared with sequences in GenBank. Sequences were aligned on MEGA X using Clustal W. BLAST hits with the highest identity threshold and querry cover were used for the construction of phylogenetic tree.
3. Results
3.1. Fungi Isolated from Arachis hypogaea Seeds
Five fungi were isolated from groundnut seeds (Table 1 and Plate 1). After one week of inoculation and incubation, sample 1 grew rapidly, had a surface color of greenish-yellow to olive with a woolly texture, sample 2 had a fluffy dark mouse grey texture with a sky grey color, sample 3 had a surface colour of light brown or dark brown, sample 4 had a grey green inconspicuous mycelium and sample 5 had a dark yellow-green color with a wrinkled and floccose texture and a slight odor. Sample 5 (1.8) had the highest frequency of occurrence while sample 3 (1.3) had the least.
|
Sample ID |
Frequency of Occurrence |
|
1 |
1.7 |
|
2 |
1.6 |
|
3 |
1.3 |
|
4 |
1.5 |
|
5 |
1.8 |
Table 1: Fungi Isolated from Arachis hypogaea seeds at room temperature (25+2OC).
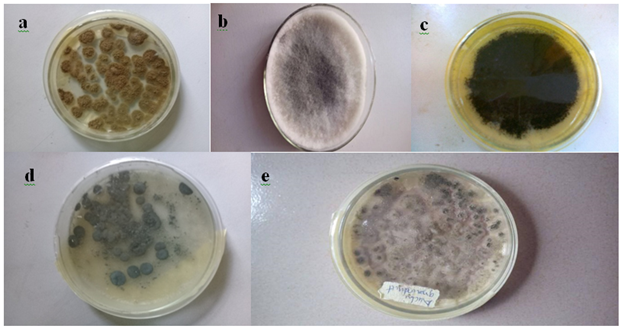
Plate 1: Pure cultures of fungi obtained from Arachis hypogaea seeds
Alphabets a to e represent the isolates viz sample 1 (a) Aspergillus tamarii; sample 2 (b) Lasidioplodia iranensis; sample 3 (c) Macrophomina phaseolina; sample 4 (d) Penicillium citrinum and sample 5 (e) Aspergillus oryzae
3.2. DNA Concentration and Purity
DNA was extracted from the five fungal isolates and also from groundnut seed to ascertain whether fungal DNA can be directly extracted from the seeds without going through isolating the organism. The concentration and purity (absorbance at 260/280nm) of genomic DNA extracted from fungal isolates and groundnut seed are presented in Table 2.
|
Sample ID |
Nucleic acid conc. (ng/µl) |
Absorbance at 260/280nm |
|
1 |
31.7 |
1.63 |
|
2 |
126.1 |
1.53 |
|
3 |
32.7 |
1.62 |
|
4 |
101.2 |
1.82 |
|
5 |
5.3 |
1.47 |
|
6 |
122.6 |
1.37 |
Table 2: Concentration of DNA extracted from fungal isolates and groundnut seed.
3.3. Amplified Products
The result of the amplified ITS1-2 gene sequences of the fungal isolates is presented in Plate 2. The amplified DNA showed clear bands on gel when observed under Ultraviolet (UV) light.
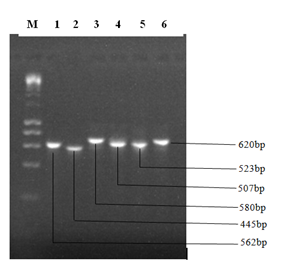
Plate 2: Gel electrophoresis showing bands generated from PCR amplification.
DNA Marker (1Kb Ladder). The numbers 1 to 6 represent the sample ID
3.4. Species Identification using BLAST
BLAST results revealed the species identity of the DNA samples to be:
Sample 1: Aspergillus tamarii; sample 2: Lasiodiplodia iranensis; sample 3: Macrophomina phaseolina; sample 4: Penicillium citrinum; sample 5: Aspergillus oryzae, and sample 6: Aspergillus pennicillioides.
|
Sample ID |
Putative taxonomic affinity(Gene bank no) |
Similarity % |
|
1 |
Aspergillus tamarii (KX010797.1) |
94 |
|
2 |
Lasiodiplodia iranensis (MH464274.1) |
99 |
|
3 |
Macrophomina phaseolina (KF951750.1) |
99 |
|
4 |
Penicillium citrinum (LT558895.1) |
99 |
|
5 |
Aspergillus oryzae (JF899327.1) |
99 |
|
6 |
Aspergillus pennicilliodes (MH864329.1) |
97 |
Table 3: Species obtained from BLAST searches of ITS sequences.
The nucleotide lengths of the DNA samples were determined to be 562 base pairs for Aspergillus tamarii (sample 1), 445 base pairs for Lasidioplodia iranensis (sample 2), 580 base pairs for Macrophomina phaseolina (sample 3), 507 base pairs for Penicillium citrinum (sample 4), 523 for Aspergillus oryzae (sample 5) and 620 base pairs for Aspergillus penicilliodes (sample 6).
Fungal sequences were submitted to GenBank and accession numbers were obtained. A strain number was also assigned to each isolate.
Aspergillus tamarii (MK454907) Strain RCBBR_AEATY1
Lasiodiplodia iranensis (MK454908) Strain RCBBR_AEATY2
Macrophomina phaseolina (MK454909) Strain RCBBR_AEATY3
Penicillium citrinum (MK454910) Strain RCBBR_AEATY4
Aspergillus oryzae (MK454911) Strain RCBBR_AEATY5
Aspergillus penicilliodes (MK454912) Strain RCBBR_AEATY6
3.5. Phylogenetic Analysis
The result of the phylogenetic analysis is presented in Figure 1. The phylogenetic tree showed the most closely related organisms to our isolates on GenBank. The distance between the branches (containing the isolates) is represented by the vertical lines. The more the distance, the more the isolates are far apart in evolution i.e. the more the length of the vertical line, the more the difference between the isolates. The phylogenetic analysis showed that the isolates are closely-related to Aspergillus tamarii, Lasiodiplodia iranensis, Macrophomina phaseolina, Penicillium citrinum, Aspergillus oryzae and Aspergillus penicilliodes. These organisms are in turn closely related to Lasiodiplodia pseudotheobromae, Rhizoctonia bataticola, Penicillium griseofulvum, Aspergillus flavus and Pediastrum duplex. Isolates 1 (Aspergillus tamarii) and 5 (Aspergillus oryzae), and 2 (Lasiodiplodia iranensis) and 3 (Macrophomina phaseolina) were found to be more closely related to each other.
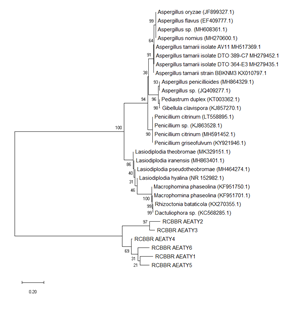
Figure 1: Phylogenetic tree showing fungal species of Arachis hypogaea using maximum composite likelihood method.
4. Discussion
The aim of this study was to determine the species identification of the fungal organisms associated with Arachis hypogaea using molecular techniques. Molecular techniques used in this study were successful in determining the fungal species associated with groundnut. The nucleotide lengths of the ITS 1 -2 genes obtained were: 562 base pairs, 445 bp, 580 bp, 507 bp, 523 bp and 620 bp for Aspergillus tamarii, Lasidioplodia iranensis, Macrophomina phaseolina, Penicillium citrinum, Aspergillus oryzae and Aspergillus penicilliodes respectively. Nuts are vulnerable to fungal infestations at various stages and periods. They can infiltrate the nuts while they are still on the trees. This usually happens when the hard shells or pods of the nuts are broken apart and the seeds are damaged by insects or pests, allowing the fungal spores to access the growing seeds. Harvesting, sorting, and washing the nuts before storage are further sources of infection. If the nuts are not handled properly at this stage, mould growth may develop, especially if the seeds are not properly dried to the recommended moisture level before storage [8,9].
In tropical and subtropical climates, Aspergillus tamarii is found in nuts (peanuts, pistachios, pecans, hazelnuts, walnuts, kola nuts, and betel nuts) and oilseeds. It can be isolated from a variety of foods, including wheat and other small grains, coffee beans, soybeans, spices, dried meat and fish products [10]. A. tamarii is allergic and capable of producing a number of poisonous secondary metabolites, it is not regarded hazardous and is rarely found as a human pathogen [11]. It is also known to cause plant dry rot. A. tamarii has been identified as a postharvest pathogen in guava [12] and has been isolated from diseased banana fruits [13]. Lasiodiplodia iraniensis is an endophytic fungus. It was first isolated from Mangifera indica, Salvadora persica, Terminalia catappa, Eucalyptus, Citrus and Juglans species in Iran. It has since been isolated from other plants in other continents, and is considered a plant pathogen. Symptoms of Lasiodiplodia spp. infection include branch dieback, stem cankers, gum exudation, necrotic lesions, neck rot, seed and fruit decay, and foliage yellowing, all of which are caused by vascular tissue obstruction and ultimately lead to plant death [14-18]. Macrophomina phaseolina is a soil-borne fungus that is widely distributed. Stem and root rot, charcoal rot, and seedling blight are diseases caused by M. phaseolina. The fungus has a large host range and is responsible for the deaths of over 500 cultivated and wild plants [19]. M. phaseolina can cause significant yield losses in crops including soybean, sorghum, and peanuts when temperatures are high and soil moisture is low.
Penicillium citrinum is a filamentous fungus that has a worldwide distribution and is likely one of the most widespread eukaryotic life forms on the planet. P. citrinum is a biotechnologically important microorganism due to its metabolite or enzyme production [20,21]. This species has been isolated from soil, (tropical) grains, spices, and indoor settings, among other things [22]. Citrinin, a nephrotoxin mycotoxin, has been reported to be consistently produced by P. citrinum [23]. Aspergillus oryzae is a filamentous fungus widely used in traditional fermentation and food processing industries such as soy sauce, soybean paste, and sake brewing. Aspergillus oryzae, is generally regarded as safe according to [24] and no strains of A. oryzae are known to produce aflatoxin.
Aspergillus penicillioides is a xerophilic fungus found in a variety of environments, including house dust. A. penicillioides is a key early colonizer of stored commodities and has been found in a variety of low aw habitats, including the solar salterns, mangroves, and estuaries [25-28] , grains, dried fruit, baked goods, salted fish, and spices [29,30]. Groundnut contamination with these fungal species will reduce germination ability of groundnut seeds during storage. All the fungi identified in this study have been known to infect groundnut either in the field or in storage causing different diseases. [31] reported the presence of Lasidioplodia theobromae in groundnut seeds causing collar rot disease.[3,32,33] reported that Fusarium spp., Rhizopus spp., Mucor spp. and Penicillum spp. were the most abundant fungi encountered in groundnut seeds. [33], isolated Aspergillus niger, Aspergillus flavus, Fusarium oxysporum, Macrophomina phaseolina and Rhizoctonia solani from seven (7) samples of groundnut collected from different localities in Pakistan. Most of these works were based on the use of cultural techniques to identify the fungal organisms associated with groundnuts. The findings from this study revealed that molecular techniques can be employed to determine the species identification of fungi associated with plant seeds. This study will increase the information on the fungal organisms associated with Arachis hypogaea seeds which is the benchmark of disease prevention and control in plant pathology.
5. Conclusion
Traditionally, the species composition of fungal communities has been determined using a combination of media culture and macro or microscopic characteristics. These methods of fungi identification can be extremely challenging, and they may result in a species list that is inaccurate. Molecular approaches on the other hand have proven to be less tedious, faster and efficient in providing information about the organisms associated with food crops. Therefore, the use of molecular techniques in species identification should be greatly encouraged in our Universities and Research Institutes and also in the field of plant pathology and fungal ecology as this allows for the comparison of DNA sequence information across known and new fungal species.
References
- Farid M. Toma, Nareen Q, et al. Isolation and Identification of Fungi from Spices and Medicinal Plants. Research Journal of Environmental and Earth Sciences 5 (2013): 131-138.
- Surendranatha Reddy EC, Sudhakar C, Eswara Reddy Aflatoxin contamination in groundnut induced by Aspergillus flavus fungi. A critical review. International Journal of Applied Biology and Pharmaceutical Technology 2 (2011):180-192.
- Syed DYN, Mehrel. Identification of Seed Borne Fungi on Farmer Saved Sorghum (Sorghum bicolor L.), Pearl Millet (Pennisetum glaucum L.) and Groundnut (Arachis hypogaea L.) Seeds. Agricultural Science Research Journals 3 (2013): 107-144.
- Chavan AM. Kakde BB. Studies of Abnormal Oilseeds Mycoflora from Marathwada Region. Bionana Frontier 2 (2008): 101-104.
- ISTA (International Seed Testing Association). International Rules for Seed Testing. Rules Amendment. Seeds Science Technology 29 (2016): 1-127.
- Ataga AE, Akueshi CO. Fungi Associated with Sunflower Seeds in Nigeria. Seed Research 1 (1986): 64-65.
- Kumar A, Kumar A, Lata C, et al. Effect of salinity and alkalinity on responses of halophytic grasses Sporobolus marginatus and Urochondra setulosa. Indian Journal of Agricultural Science 88 (2018): 149-157.
- Abdulla NQF. Evaluation of fungal flora and mycotoxin in some important nut products in Erbil local markets. ResearchJournal of Environmental and Earth Sciences 5 (2013): 330-336
- Adetunji M, Atanda O, Ezekiel CN. Fungal and bacterial metabolites of stored maize (Zea mays, L.) from five agro-ecological zones of Nigeria. Mycotoxin Research 30 (2014): 89-102
- Ogbole OO, Adebayo-Tayo BC, Salami KM, et al. Molecular identification and antimicrobial activity of endophytic fungi Aspergillus tamarii (Trichomaceae). Nigerian Journal of Pharmaceutical Sciences 16 (2017): 41-48
- Valentino MJG, Pineda FG, Fandialan MF. Phytopathogenicity of fungi associated with crown rot of guava (Psidium guajava). Plant Pathology and Quarantine 5 (2015): 7-13.
- Talha Azeem M, Saleem S, Nasreen S. Prevalence and detection of fungi associated with post harvest rots of banana in Karachi. International Journal of Biology and Biotechnology 13 (2016): 587-592.
- Freire FCO, Cardoso JE, Viana FMP, et al. Status of Lasiodiplodia theobromae as a plant pathogen in Brasil. Essentia 12 (2011): 53-71.
- Lima JS, Moreira RC, Cardoso JE, et al. Cultural, morphological and pathogenic characterization of Lasiodiplodia theobromae associated with tropical fruit plants. Summa Phytopathologica 39 (2013): 81-88.
- Marques MW, Lima NB, Morais JRMA. Species of Lasiodiplodia associated with mango in Brasil. Fungal Diversity 61 (2013): 181-193.
- Machado AR, Pinho DB, Pereira OL. Phylogeny, identification and pathogenicity of the Botryosphaeriaceae associated with collar and root rot of the biofuel plant Jatropha curcas in Brasil, with a description of new species of Fungal Diversity 67 (2014a): 231-247.
- Netto MSB, Assunção IP, Lima GSA. Species of Lasiodiplodia associated with papaya stem-end rot in Brasil. Fungal Diversity 67 (2014): 127-141.
- Khan SN. Macrophomina phaseolina as causal agent for charcoal rot of sunflower. Mycopath 5 (2007): 111-118.
- Tashiro Y, Ueno H, Takaba M, et al. Production of Functional Inulin-Type Fructooligosaccharides by an Enzyme from Penicillium citrinum Current Microbiology 74 (2017): 1114.
- Gu Y, Ding P, Liang Z, et al. Activated production of silent metabolites from marine-derived fungus Penicillium citrinum Fitoterapia 127 (2018): 207
- Samson RA, Frisvad JC. Penicillium subgenus Penicillium: new taxonomic schemes and mycotoxins and other extrolites. Stud Mycol 49 (2004): 1-266
- Malmstrøm J, Christophersen C, Frisvad JC. Secondary metabolites characteristic of Penicillium citrinum, Penicillium steckii and related species. Phytochemistry 54 (2000): 301-309.
- Machida M, Asai K, Sano M. Genome sequencing and analysis of Aspergillus oryzae, Nature, 438 (2005): 1157-1161.
- Butinar L, Frisvad JC, Gunde-Cimerman N. Hypersaline waters-a potential source of foodborne toxigenic Aspergilli and Penicillia. FEMS Microbiol. Ecol 77 (2011): 186-199.
- Gonsalves V, Nayak S, Nazareth S. Halophilic fungi in a polyhaline estuarine habitat. J. Yeast Fungal Res 3 (2012): 30-36.
- Nayak S, Gonsalves V, Nazareth S. Isolation and salt tolerance of halophilic fungi from mangroves and solar salterns in Goa – India. Indian J Mar Sci 41 (2012):164-172.
- Nazareth S, Gonsalves V, Nayak S. A first record of obligate halophilic aspergilli from the Dead Sea. Indian J. Microbiol 52 (2012): 22-27.
- Tamura M, Kawasaki H, Sugiyama J. Identity of the xerophilic species Aspergillus penicillioides: integrated analysis of the genotypic and phenotypic. J Gen Appl Microbiol 45 (1999): 29-37.
- Pitt JI, Hocking AD. Fungi and Food Spoilage (2009).
- Philips PM, Porter DM. Plant Diseases. American Phytopathological Society 82 (1998): 1205-1209.
- Akinseye OF, Ataga AE. Aqueous and Ethanolic Extracts of Vernonia amygdalinain the Control of Fungi Associated with Arachis hypogeae L. Scientia Africana 13 (2014): 281-289
- Akinnibosun FI, Osawuru EE. Quality Assessment of Peeled and Unpeeled Roasted Groundnut (Arachis hypogaea ) sold in Benin City, Nigeria. International Research Journal of Natural and Applied Sciences 2 (2015): 1-32.
- Rasheed S, Dawar S, Ghafar, et al. Seed Borne Mycoflora Groundnut. Pakistan Journal of Botany 36 (2004): 199-202.


 Impact Factor: * 4.2
Impact Factor: * 4.2 Acceptance Rate: 77.66%
Acceptance Rate: 77.66%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 