Joint Secondary Transcriptomic Analysis of Non-Hodgkin’s B-Cell Lymphomas Predicts Reliance on Pathways Associated with the Extracellular Matrix and Robust Diagnostic Biomarkers
Naomi Rapier-Sharman1, Jeffrey Clancy1 and Brett E Pickett1*
1Department of Microbiology and Molecular Biology, Brigham Young University, Provo, UT 84602, USA
*Corresponding author: Brett E Pickett, Department of Microbiology and Molecular Biology, Brigham Young University, Provo, UT 84602, USA.
Received: 22 August 2022; Accepted: 21 September 2022; Published: 27 September 2022
Article Information
Citation: Naomi Rapier-Sharman, Jeffrey Clancy and Brett E Pickett. Joint Secondary Transcriptomic Analysis of Non-Hodgkin’s B-Cell Lymphomas Predicts Reliance on Pathways Associated with the Extracellular Matrix and Robust Diagnostic Biomarkers. Journal of Bioinformatics and Systems Biology 5 (2022): 119-135.
View / Download Pdf Share at FacebookAbstract
Approximately 450,000 cases of Non-Hodgkin’s lymphoma are annually diagnosed worldwide, resulting in ~240,000 deaths. An augmented understanding of the common mechanisms of pathology among larger numbers of B-cell Non-Hodgkin’s Lymphoma (BCNHL) patients is sorely needed. We consequently performed a large joint secondary transcriptomic analysis of the available BCNHL RNA-sequencing projects from GEO, consisting of 322 relevant samples across ten distinct public studies, to find common underlying mechanisms and biomarkers across multiple BCNHL subtypes and patient subpopulations; limitations may include lack of diversity in certain ethnicities and age groups and limited clinical subtype diversity due to sample availability. We found ~10,400 significant differentially expressed genes (FDR-adjusted p-value < 0.05) and 33 significantly modulated pathways (Bonferroni-adjusted p-value < 0.05) when comparing BCNHL samples to non-diseased B-cell samples. Our findings included a significant class of proteoglycans not previously associated with lymphomas as well as significant modulation of genes that code for extracellular matrix-associated proteins. Our drug repurposing analysis predicted new candidates for repurposed drugs including ocriplasmin and collagenase. We also used a machine learning approach to identify robust BCNHL biomarkers that include YES1, FERMT2, and FAM98B, which have not previously been associated with BCNHL in the literature, but together provide ~99.9% combined specificity and sensitivity for differentiating lymphoma cells from healthy B-cells based on measurement of transcript expression levels in B-cells. This analysis supports past findings and validates existing knowledge while providing novel insights into the inner workings and mechanisms of transformed B-cell lymphomas that could give rise to improved diagnostics and/or therapeutics.
Keywords
<p>B-cell; B-cell Non-Hodgkin Lymphoma; Biomarkers; Joint Analysis; Liquid Biopsy; RNA-seq; Transcriptomics</p>
Article Details
Abbreviations:
BCNHL- B-cell Non-Hodgkin Lymphoma; RNA-seq- RNA-sequencing; NCBI- National Center for Biotechnology Information; GEO- Gene Expression Omnibus; SRA- Sequence Read Archive; ARMOR-Automated Reproducible MOdular workflow for preprocessing and differential analysis of RNA-seq data; SPIA- Signaling Pathway Impact Analysis; GO- Gene Ontology; AUC- Area Under The Curve; DEGs- Differentially Expressed Genes; APOC1- Apolipoprotein C1; log2FC- log2 Fold Change; VCAM1- Vascular Adhesion Molecule 1; CCL18- C-C Motif Chemokine Ligand 18; CXCL9- C-X-C Motif Chemokine Ligand 9; LUM- Lumican; SLRPs- Small Leucine-Rich Proteoglycans; NS- Not Significant; NP- Not Present; C1QA- Complement C1q A chain; C1QB- Complement C1q B chain; C1QC- Complement C1q C chain; APOE- Apolipoprotein E; Lr- Likelihood ratio; COL1A1- Collagen type I Alpha 1 chain; COL27A1- Collagen type XXVII alpha 1 chain; psize- Number of Genes in a Pathway; NDE- Number of Differentially-Expressed Genes in Pathway; YES1- YES Proto-Oncogene 1 Src Family Tyrosine Kinase; FERMT2- FERM Domain Containing Kindlin 2; FAM98B- Family with Sequence Similarity 98 Member B; ROC- Receiver Operator Curve; DLBCL- Diffuse Large B-Cell Lymphoma; CTNNB1- Beta-Catenin/Beta Catenin; JNK- Janus Kinase; IGF- Insulin-like Growth Factor; IGFBP- Insulin-like Growth Factor Binding Protein.
1. Introduction
Lymphomas are the most common blood cancer, which primarily affects lymphocytes. There are three primary categories of lymphomas including Chronic Lymphocytic Leukemia/Small Lymphocytic Lymphoma, Hodgkin Lymphoma, and Non-Hodgkin Lymphoma. There are over 90 recognized types of Non-Hodgkin Lymphoma, which is diagnosed in ~450,000 patients worldwide annually, resulting in 240,000 deaths [1]. Among Non-Hodgkin lymphomas, only ~10-15% are T-cell lymphomas, while the remaining 85-90% are B-cell malignancies [2]. B-cell Non-Hodgkin Lymphomas (BCNHLs) pose a significant disease burden worldwide. BCNHL subtypes include Burkitt’s lymphoma, marginal-zone B-cell lymphomas, follicular lymphoma, diffuse large B-cell lymphoma, and mantle cell lymphoma [2]. B-cell lymphomas are dependent on their extracellular environment for activation and transformation into malignancies, including antigen activation of the B-cell receptor, canonical B-cell growth signals which are also essential to the maturation of healthy B cells, and signals delivered by other immune cells in the follicular/germinal center lymphoma microenvironment [3]. The research community has dedicated substantial effort to identify the attributes that characterize cancers across all types and subtypes—regardless of which tissue type first produces malignancies. Specifically, it has been suggested previously that all cancers share the following traits: selective proliferative advantage, altered stress response, vascularization, invasion and metastasis, metabolic rewiring, immune modulation, and an abetting microenvironment [4,5]. One example of a molecular mechanism that is common in cancer is malignant development through TP53 mutation, with multiple mutations in the TP53 being associated with hundreds of cancer subtypes [6]. Though not every gene-mechanism pairing will be widely found across malignant cells like TP53, identifying shared genes and mechanisms by performing joint secondary analysis on combined data from multiple previous research studies in a focused set of related cancer subtypes can be beneficial [7]. We can therefore leverage known mechanisms from well-studied subtypes to enable quicker, less expensive mechanism discovery for understudied subtypes. This approach could potentially enable researchers to identify shared mechanisms repurpose existing therapeutics to a wider swath of cancer types and subtypes. The widespread adoption of RNA-sequencing (RNA-seq) has opened new frontiers in disease research. Rather than identifying and characterizing individual cellular components, transcriptomic analyses can provide a mechanistic snapshot of the many genes that are upregulated or downregulated in response to a given stimulus or disease state, such as lymphoma. Characterizing these transcriptional patterns can aid in the identification of genes that could be worth further experimental investigation due to their selective modulation in diseased samples. Though the RNA-sequencing samples in the current study were previously published, analyzing them together in a joint secondary analysis can grant us new insights into disease mechanisms by increasing the signal of significant genes and reducing the statistical “noise” caused by outliers across patient subpopulations. The aim of this study was to perform a joint secondary analysis of transcriptomic data from de-identified publicly available B-cell Non-Hodgkin’s Lymphomas (BCNHLs) clinical samples to determine the shared underlying molecular mechanisms and biomarkers of B-cell lymphomas that are detectable after cellular transformation. We expect our analysis to validate past findings of B-cell cancer mechanisms and uncover mechanisms that have not been previously associated with BCNHL.
2. Methods
2.1 Collecting Samples
RNA-sequencing samples were acquired from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database [8] using the search term, “b-cell lymphoma” with the goal of finding B-cell non-Hodgkin’s lymphoma samples and healthy B-cell controls. The automatic GEO filters “Homo sapiens” and “high-throughput RNA-sequencing” were applied. Cell lines, formalin-fixed paraffin-embedded tissues, gene expression microarray experiments, single-cell (10X) RNA-sequencing experiments, xenografts, samples known to be infected with EBV and KSHV, and samples which contained more diverse cell types (i.e., whole blood, lymph node, PBMCs, brain, etc.) were excluded by hand. All samples that had one or more of these disqualifying attributes were excluded from the dataset prior to analysis, meaning that only a subset of the samples from an individual experiment may be represented in the joint secondary analysis. One study which matched all criteria was excluded due to inconsistent and unreliable sample labeling. Multiple myeloma, leukemia, and Hodgkin’s lymphoma samples were intentionally excluded in favor of focusing on B-cell non-Hodgkin’s lymphomas. Records were passed or failed against the standardized exclusion criteria detailed above by one team member, with some input from a second team member. To avoid inclusion bias, any sample that could not be excluded by our standardized exclusion criteria was included in the study. While a subset of the healthy control samples was obtained from the same RNA sequencing projects as the BCNHL samples, others were obtained from three lymphoma-unrelated B-cell datasets with healthy controls to create roughly equivalent-sized BCNHL and healthy groups. Final dataset assembly from GEO concluded on October 22, 2020, resulting in a dataset of 322 samples (134 BCNHL samples and 188 healthy B-cell controls) from ten studies [9-20]. The raw data for these experiments were previously collected by the primary authors and conform to the appropriate ethical oversight to protect patient autonomy and patient identity. All 10 primary RNA-sequencing datasets from which we gathered samples for our lymphoma joint secondary analysis have been published in the peer-reviewed literature, increasing overall confidence that each dataset has acceptable quality (Table 1, Figure 1).
2.2 Preprocessing of RNA-Sequencing Data
Following the manual curation of the RNA-seq samples, the fastq files were pre-processed as previously described [21]. In brief, fastq files containing RNA-sequencing data were downloaded from the Sequence Read Archive (SRA) using the sratools software package. The fastq files, the associated metadata file, and a configuration file for each dataset were then generated and used as input to the Automated Reproducible MOdular workflow for preprocessing and differential analysis of RNA-seq data (ARMOR) workflow [22]. A configuration file was used by ARMOR to appropriately set up a python-based Snakemake workflow [23]. In the ARMOR workflow, adapters and poor-quality regions of reads were trimmed with TrimGalore! [24], quality control metrics were calculated with FastQC [25], reads were mapped to the human GRCh38 transcriptome and total gene transcripts quantified with Salmon [26], significant differential gene expression was calculated using a negative binomial distribution implemented in edgeR [27], Gene Ontology (GO) enrichment was performed against terms from the MSigDB [28] while adjusting for inter-gene correlation using the Camera algorithm [29], and significant splice variants were predicted with DRIMseq [30]. The significant differentially expressed genes from the ARMOR workflow were then used as input to the signaling pathway impact analysis (SPIA) algorithm to enrich differentially expressed genes against intracellular signaling pathways from five databases including KEGG, Panther, BioCarta, Reactome, and NCI [31-35]. Differentially expressed genes outputted by ARMOR and DRIMSeq were evaluated by the effect measures log2 fold change and likelihood ratio respectively. Confidence in results was accomplished using false discovery-rate adjusted p-values.
2.3 Additional Analysis and Visualization of Differentially Expressed Genes and Gene Ontologies
The PRISMA flowchart template was used to generate figure 1, which is consistent with the accepted transparent reporting of joint secondary analysis generation and results [36]. The R package ggplot was used to construct the Fig 2 volcano plot from using FDRs and log2 fold change values for each gene from the edgeR output [37]. The KEGG ontology was extracted from the Brite Hierarchy using existing code [32]. Genes included in the Brite Hierarchy were then computationally matched to their corresponding edgeR log2 fold change values. A statistical enrichment of the KEGG gene ontologies was performed using the R package bc3net [38] prior to visualizing the bc3net enrichment results with the R package Treemap in Fig 3 [39].
2.4 Biomarker Prediction using Differentially Expressed Gene Data
Transcript-level read counts, generated by Salmon, were organized into a tabular format and samples were randomly assigned to either the testing set (30%) or the training set (70%). The R package randomForest was used to run a supervised classification analysis, with disease state (healthy or lymphoma) as the predictor, to determine biomarkers [40]. The initial results from the whole transcriptome were then reduced to the 3, 5, and 10 best-scoring transcriptional biomarkers, based on the mean Gini impurity decrease values for each of the features. These values were then sorted by size to determine the transcribed genes from the original dataset with the largest association. The area under the curve (AUC) was calculated from the receiver operator characteristic curves that were generated for each set of random forest results to determine the efficacy of the selected biomarkers for disease prediction.
2.5 Drug Prediction using Differentially Modulated Pathways
Drug prediction was conducted using the Pathways2Targets2.R algorithm [41]. Significantly modulated pathways (as determined by SPIA) were used as input for the Pathways2Targets algorithm to determine existing drugs that could potentially be repurposed for BCNHL. The Pathways2Targets algorithm takes the significantly affected pathways determined by SPIA, finds the members of those pathways, and searches the Open Targets drug database for drugs known to target the proteins from each pathway. The output table from this process was then summarized using a custom R script, most_common_treatments_2021_09_19.R [42].
3. Results
We acquired our BCNHL samples from publicly available projects on the NCBI Gene Expression Omnibus (GEO) database using the search term, “b-cell lymphoma” with the goal of finding B-cell non-Hodgkin’s lymphoma samples and healthy B-cell controls [8,21]. We excluded non-human samples, cell lines, formalin-fixed paraffin-embedded tissues, gene expression microarray experiments, single-cell (10X) RNA-sequencing experiments, xenografts, samples known to be infected with EBV and KSHV, and samples which contained more diverse cell types (i.e., whole blood, lymph node, PBMCs, brain, etc.). Study GSE142334 matched all study criteria, but contained file types incompatible with our bioinformatic workflow and was excluded due to inaccessibility. Study GSE93627, which seemed to match all of our criteria, was excluded in later stages of selection due to inconsistent and unreliable sample labeling. We also intentionally excluded multiple myeloma, leukemia, and Hodgkin lymphoma samples in favor of focusing on B-cell non-Hodgkin lymphomas. We then located additional healthy B-cell control samples from BCNHL-unrelated studies to even out case and control numbers, the final three studies cited in Table 1. In an effort to conform to best-practice PRISMA guidelines on transparent reporting of secondary joint analyses [36], we have included a detailed diagram on our sample selection process and PRISMA’s transparent reporting checklist (Fig 1, S1 File). Our final dataset included a total of 322 samples (134 BCNHL samples and 188 healthy B-cell controls) from ten studies (Table 1) [9-20]. Though the samples included in our joint secondary analysis were all clinical samples, the metadata provided by each original project varied greatly in both quantity and nature, making it difficult to discern the extent of sample heterogeneity or homogeny for variables other than lymphoma subtype. Given that the aim of this study was to generate a mechanistic profile for many BCNHL samples in comparison to healthy B-cells, the only evident source of heterogeneity is the distribution of BCNHL types across the included samples. We recognize that the included samples were largely skewed toward the diffuse large B-cell lymphoma subtype, which is the most common BCNHL subtype and resultantly has more data available on GEO than any other BCNHL subtype.
Figure 1: PRISMA flow diagram for transparent reporting of joint secondary analysis study selection. Contains a study-by-study breakdown of selection criteria. All studies included were retrieved from the Gene Expression Omnibus (GEO) database provided by NCBI.
|
Sample Phenotype |
Single End or Paired End Reads |
GEO Accession # |
Relevant Samples |
|
Large B-Cell Lymphoma |
Paired End |
GSE153437 [9] |
25 |
|
Diffuse Large B-Cell Lymphoma |
Paired End |
GSE130751 [10] |
63 |
|
B-Cell Lymphoma |
Single End |
GSE110219 [11] |
1 |
|
Diffuse Large B-Cell Lymphoma |
Paired End |
GSE95013 [12] |
28 |
|
Follicular Lymphoma |
Paired End |
GSE62241 [13,14] |
10 |
|
Diffuse Large B-Cell Lymphoma |
Paired End |
GSE50514 [15] |
7 |
|
Healthy |
Single End |
GSE110219 [11] |
1 |
|
Healthy |
Paired End |
GSE62241 [13,14] |
4 |
|
Healthy |
Paired End |
GSE45982 [16,17] |
8 |
|
Healthy |
Single End |
GSE92387 [18] |
12 |
|
Healthy |
Paired End |
GSE118254 [19] |
147 |
|
Healthy |
Paired End |
GSE110999 [20] |
16 |
Table 1: Study-based origin of samples included in the joint secondary analysis.
We began by trimming, mapping, and quantifying the RNA-sequencing reads prior to calculating the significant differential gene expression when comparing the Lymphoma samples to the non-diseased control samples. This comparison returned ~13,800 significant differentially expressed genes (DEGs) (Fig 2, Table 2, S2 and S3 Files). We then ranked this list by the FDR-corrected p-value for each gene. We observed that the top 20 DEGs include both novel and accepted differentially expressed genes associated with various Lymphomas. Specifically, we confirmed several genes that have previously been explored or characterized in various subtypes of BCNHL including Apolipoprotein C1 (APOC1; log2FC = 6.93, FDR = 8.55 × 10−117) and Vascular cell adhesion molecule 1 (VCAM1; log2FC = 7.85, FDR = 2.29 × 10−120) to be upregulated in BCNHLs. We also found two pathological BCNHL genes, C-C motif chemokine ligand 18 (CCL18; log2FC = 10, FDR = 3.74 × 10−123) and C-X-C motif chemokine ligand 9 (CXCL9; log2FC = 11, FDR = 4.31 × 10−141) to be upregulated in BCNHL as compared to healthy B-cells.
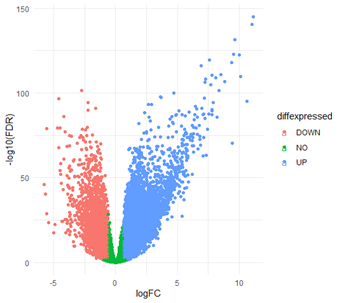
Figure 2: Visualization of Differentially Expressed Genes and Gene Ontologies. Differentially expressed gene volcano plot. Green dots represent genes which were not significantly differentially expressed between healthy B-cells and BCNHL, while the salmon and blue dots represent downregulated and upregulated genes respectively.
|
Gene Symbol |
Ensembl ID |
Log2 Fold Change (log2FC) |
FDR-corrected p-value |
|
|
1 |
LUM |
ENSG00000139329 |
11.1 |
1.11 × 10−145 |
|
2 |
CXCL9 |
ENSG00000138755 |
11 |
4.31 × 10−141 |
|
3 |
C1QC |
ENSG00000159189 |
9.65 |
2.70 × 10−132 |
|
4 |
C1QA |
ENSG00000173372 |
9.54 |
2.03 × 10−123 |
|
5 |
CCL18 |
ENSG00000278167 |
10 |
3.74 × 10−123 |
|
6 |
VCAM1 |
ENSG00000162692 |
7.58 |
2.29 × 10−120 |
|
7 |
C1QB |
ENSG00000173369 |
9.4 |
8.19 × 10−119 |
|
8 |
APOC1 |
ENSG00000130208 |
6.93 |
8.55 × 10−117 |
|
9 |
AL512646.1 |
ENSG00000203396 |
-15.6 |
2.24 × 10−115 |
|
10 |
CCL19 |
ENSG00000172724 |
8.48 |
1.27 × 10−111 |
|
11 |
SLAMF8 |
ENSG00000158714 |
7.77 |
4.01 × 10−111 |
|
12 |
COL3A1 |
ENSG00000168542 |
10.1 |
1.67 × 10−110 |
|
13 |
TCIM |
ENSG00000176907 |
8.07 |
7.86 × 10−110 |
|
14 |
RARRES2 |
ENSG00000106538 |
7.25 |
8.21 × 10−109 |
|
15 |
CXCL13 |
ENSG00000156234 |
8.8 |
1.72 × 10−107 |
|
16 |
SPARCL1 |
ENSG00000152583 |
7.24 |
6.42 × 10−107 |
|
17 |
PTGDS |
ENSG00000107317 |
7.69 |
1.07 × 10−105 |
|
18 |
COL1A2 |
ENSG00000164692 |
8.33 |
3.70 × 10−102 |
|
19 |
CXXC5 |
ENSG00000171604 |
-2.73 |
3.70 × 10−102 |
|
20 |
C1R |
ENSG00000159403 |
4.7 |
1.41 × 10−100 |
Table 2: Top 20 significant differentially expressed genes between BCNHL and healthy samples.
We then examined the highest-ranking novel differentially expressed genes from our joint secondary analysis to identify transcriptional mechanisms of disease, with transcripts reported at the gene level. The first gene we observed using this approach was Lumican (LUM), which is a member of the small leucine-rich proteoglycan family (SLRPs) [43], and was substantially upregulated in lymphoma (log2 fold change = 11.1, FDR p-value = 1.11 × 10−145). In addition, the larger family of SLRPs appears to play a role in BCNHL (Table 3). Specifically, our data show that 12/18 SLRPs are expressed in healthy and/or cancerous B-cells, and that 11/12 B-cell-expressed SLRPs are significantly differentially expressed in BCNHL samples. We found that overall, the SLRP fold changes substantially differed (9/12 expressed SLRPs are upregulated, 2/12 are downregulated, 1/12 had no significant change), with the genes encoding SLRPs (especially Classes I and V) being well represented in the B-cell lymphoma transcriptome.
|
SLRP Class |
Name |
Log2 Fold Change |
FDR-corrected p-value |
|
Class I |
DCN |
2.88 |
1.67 × 10−41 |
|
BGN |
7.88 |
3.22 × 10−95 |
|
|
ASPN |
3.09 |
2.27 × 10−25 |
|
|
ECM2 |
2.1 |
1.44 × 10−17 |
|
|
ECMX |
NP |
NP |
|
|
Class II |
FMOD |
5.71 |
3.86 × 10−61 |
|
LUM |
11.1 |
1.11 × 10−145 |
|
|
PRELP |
0.617 |
1.54 × 10−4 |
|
|
KERA |
NP |
NP |
|
|
OMD |
NP |
NP |
|
|
Class III |
EPYC |
NP |
NP |
|
OPTC |
NP |
NP |
|
|
OGN |
NS |
NS |
|
|
Class IV |
CHAD |
-3.49 |
1.33 × 10−24 |
|
NYX |
NP |
NP |
|
|
TSKU |
1.37 |
1.62 × 10−16 |
|
|
Class V |
PODN |
1.66 |
5.70 × 10−10 |
|
PODNL1 |
-1.49 |
3.15 × 10−11 |
Table 3: Novel identification of differential expression of Small Leucine-Rich Proteoglycans (SLRPs) in BCNHL.
*NS = not significant; NP = not present.
We found that genes encoding the Complement C1q A (C1QA; log2FC = 9.54, FDR = 2.03 × 10−123), Complement C1q B (C1QB; log2FC = 9.4, FDR = 8.19 × 10−119) and Complement C1q C (C1QC; log2FC = 9.65, FDR = 2.7 × 10−132) chains were all dramatically and significantly upregulated in BCNHL. Complement proteins are typically regarded as components of the innate immune system, which bind to antigen-antibody complexes to facilitate the formation of the membrane attack complex to kill invading bacteria. Our finding adds to the growing body of work indicating an association between complement C1q expression and lymphoma pathology. Additionally, we detected AL512646.1 (also known as LOC100128906 and as a WDR45-like pseudogene) as differentially expressed by B-cell non-Hodgkin’s lymphoma samples, a novel observation which was somewhat unexpected. Though AL512646.1 is annotated as a pseudogene and has not been previously associated with cancer, the RNA-sequencing data shows that it is uniformly expressed in healthy B-cells and downregulated in at least a subset of BCNHLs (log2FC = -15.1, FDR = 2.24 × 10−115). Next, we used the DRIMSeq algorithm to determine which genes had significant differences in the presence of splice variants between BCNHL and healthy control samples. This analysis returned 320 genes for which splice variants were significantly different (Table 4, S4 File). Apolipoprotein E (APOE) was the most statistically significant splice variant (Lr [likelihood ratio] = 4470, # of alternate splice variants = 4, adjusted p-value = 0). Specifically, we observed the expression of APOE transcripts ENST00000252486, ENST00000425718, ENST00000434152, ENST00000446996, and ENST00000485628 to significantly differ between non-Hodgkin’s lymphoma and non-diseased B-cells.
|
Gene symbol |
Ensembl ID |
Lr* |
# of Alternate Transcripts |
Adjusted P-value |
|
APOE |
ENSG00000130203 |
4470 |
4 |
0 |
|
COL1A1 |
ENSG00000108821 |
1520 |
12 |
5.56 × 10−315 |
|
COL27A1 |
ENSG00000196739 |
1060 |
7 |
6.71 × 10−220 |
|
RPL5 |
ENSG00000122406 |
1040 |
10 |
3.86 × 10−214 |
|
KLF6 |
ENSG00000067082 |
961 |
6 |
7.41 × 10−201 |
|
SRSF6 |
ENSG00000124193 |
954 |
5 |
1.56 × 10−200 |
|
CYBRD1 |
ENSG00000071967 |
931 |
6 |
2.17 × 10−194 |
|
PLEKHM1P1 |
ENSG00000214176 |
924 |
5 |
3.78 × 10−194 |
|
VCP |
ENSG00000165280 |
912 |
6 |
2.37 × 10−190 |
|
DDX6 |
ENSG00000110367 |
872 |
7 |
8.01 × 10−181 |
|
THRAP3 |
ENSG00000054118 |
846 |
3 |
6.63 × 10−180 |
|
FCGR2B |
ENSG00000072694 |
771 |
4 |
1.75 × 10−162 |
|
CHI3L1 |
ENSG00000133048 |
715 |
4 |
2.40 × 10−150 |
|
IFITM3 |
ENSG00000142089 |
691 |
3 |
2.53 × 10−146 |
|
ADAM28 |
ENSG00000042980 |
719 |
11 |
4.56 × 10−144 |
|
CIB1 |
ENSG00000185043 |
662 |
2 |
2.04 × 10−141 |
|
ZNF318 |
ENSG00000171467 |
645 |
3 |
1.88 × 10−136 |
|
RPS28 |
ENSG00000233927 |
621 |
3 |
3.10 × 10−131 |
|
CCDC124 |
ENSG00000007080 |
549 |
3 |
1.14 × 10−115 |
|
ZNF335 |
ENSG00000198026 |
545 |
3 |
8.02 × 10−115 |
Table 4: Top 20 most significant splice variants (sorted by gene).
*Lr = likelihood ratio
We also observed that Collagen type I alpha 1 chain (COL1A1) had significant splice variants (Lr = 1520, # of alternate splice variants = 12, adjusted p-value = 5.55999999807983 × 10−315). Interestingly, our study also found that the COL1A1 gene was significantly upregulated in BCNHL (log2FC = 3.73, FDR = 9.78 × 10−48). We also observed novel significant splice variants in Collagen type XXVII alpha 1 chain (COL27A1), which was found to be significant in BCNHL (Lr = 1060, # of alternate splice variants = 7, adjusted p-value = 6.71 × 10−220). We then wanted to determine which functional terms in the Gene Ontology (GO) were over-represented by the list of DEGs in BCNHL. The Camera algorithm evaluated 14,901 terms (including gene ontologies and human phenotypes) for statistical enrichment against the significant differentially expressed genes that we generated with edgeR. Although there were 482 results (p-value < 0.05), none remained significant after multiple hypothesis correction (S5 File). The lack of significant results is somewhat expected given the overall molecular heterogeneity of BCNHL subtypes and the stringency of the Camera algorithm. To visualize the Gene Ontology changes, we used a hypergeometric enrichment algorithm that applied a p-value cutoff of 0.05. We then averaged the edgeR fold-change values for the genes of each gene ontology in the KEGG Brite hierarchy and plotted the enrichment results using the R Treemap package to better understand the contribution of various GO terms to the overall list of DEGs (S3 File). To better understand the results of our analysis at a more mechanistic level, we used the signaling pathway impact analysis (SPIA) algorithm to identify intracellular signaling pathways that play important roles in various subtypes of lymphoma after transformation. Briefly, this pathway-analysis algorithm generates a null distribution through bootstrapping to identify pathways that are significantly modulated based on the DEGs. Our analysis revealed 33 significantly modulated pathways between lymphoma B-cells and non-diseased B-cells (Table 5, S6 File). Specifically, we observed ten pathways that were involved with the extracellular matrix and connective tissue, bolded below in Table 5. The upregulation of these pathways indicates that transformed BCNHL likely benefits from modulations to the extracellular matrix.
|
Name |
pSize |
NDE |
tA |
pGFWER |
Source Database |
|
|
1 |
Integrin signalling pathway |
99 |
86 |
114.394 |
2.39 × 10−5 |
Panther |
|
2 |
Extracellular matrix organization |
204 |
180 |
80.7398395 |
3.14 × 10−5 |
Reactome |
|
3 |
ECM-receptor interaction |
70 |
64 |
77.659 |
4.47 × 10−5 |
KEGG |
|
4 |
Staphylococcus aureus infection |
32 |
29 |
110.568396 |
0.000503428 |
KEGG |
|
5 |
Complement and coagulation cascades |
36 |
32 |
37.5461944 |
0.000674242 |
KEGG |
|
6 |
Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
28 |
25 |
130.821315 |
0.000740169 |
NCI |
|
7 |
Cytokine-cytokine receptor interaction |
168 |
140 |
98.294 |
0.000780995 |
KEGG |
|
8 |
Focal adhesion |
182 |
150 |
236.615459 |
0.001133449 |
KEGG |
|
9 |
PI3K-Akt signaling pathway |
271 |
221 |
260.046197 |
0.001344307 |
KEGG |
|
10 |
Complement cascade |
29 |
27 |
102.278583 |
0.001440498 |
Reactome |
|
11 |
Systemic lupus erythematosus |
17 |
15 |
67.4577222 |
0.001616635 |
KEGG |
|
12 |
b cell survival pathway |
22 |
19 |
26.576 |
0.00167109 |
BioCarta |
|
13 |
Small cell lung cancer |
78 |
64 |
121.170067 |
0.001930414 |
KEGG |
|
14 |
Integrins in angiogenesis |
52 |
41 |
146.143424 |
0.00285984 |
NCI |
|
15 |
Olfactory transduction |
93 |
74 |
-148.8965 |
0.002966626 |
KEGG |
|
16 |
integrin signaling pathway |
37 |
29 |
77.0156667 |
0.003336442 |
BioCarta |
|
17 |
erk and pi-3 kinase are necessary for collagen binding in corneal epithelia |
34 |
26 |
166.268917 |
0.003755165 |
BioCarta |
|
18 |
RNA Polymerase I Promoter Clearance |
85 |
72 |
-40.156 |
0.004307328 |
Reactome |
|
19 |
Initial triggering of complement |
15 |
14 |
44.508 |
0.004480759 |
Reactome |
|
20 |
RNA Polymerase I Promoter Opening |
39 |
34 |
-40.907 |
0.004675938 |
Reactome |
|
21 |
RHO GTPases activate PKNs |
67 |
57 |
39.779 |
0.004802382 |
Reactome |
|
22 |
DNA Damage/Telomere Stress Induced Senescence |
61 |
52 |
32.7708077 |
0.004980633 |
Reactome |
|
23 |
Creation of C4 and C2 activators |
7 |
7 |
27.365 |
0.005633091 |
Reactome |
|
24 |
Collagen formation |
66 |
63 |
26.4272897 |
0.006045837 |
Reactome |
|
25 |
Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 |
41 |
35 |
39.094 |
0.006675281 |
Reactome |
|
26 |
MET activates PTK2 signaling |
18 |
16 |
63.573 |
0.007079287 |
Reactome |
|
27 |
Collagen degradation |
17 |
15 |
131.8905 |
0.008037505 |
Reactome |
|
28 |
MET promotes cell motility |
28 |
24 |
97.5445 |
0.00808298 |
Reactome |
|
29 |
Regulation of IGF Activity by IGFBP |
11 |
10 |
25.958725 |
0.008404989 |
Reactome |
|
30 |
Classical antibody-mediated complement activation |
5 |
5 |
27.354 |
0.008619298 |
Reactome |
|
31 |
Serotonin Neurotransmitter Release Cycle |
11 |
9 |
-13.301889 |
0.015608196 |
Reactome |
|
32 |
Class A/1 (Rhodopsin-like receptors) |
81 |
77 |
5.908 |
0.026176599 |
Reactome |
|
33 |
Peptide ligand-binding receptors |
79 |
75 |
5.84 |
0.040928099 |
Reactome |
Table 5: Significant differentially modulated signaling pathways include extracellular matrix.
*Abbreviations: psize = number of genes in pathway. NDE = number of genes from pathway which were differentially expressed. tA = measure of change between healthy and lymphoma expression; directionality indicates up- or down-modulation. pGFWER = p-value with adjustments appropriate to a multiplexed interaction network [31].
We next used the Pathways2Targets algorithm to identify potentially novel drug targets for BCNHL from the signaling pathway results (S7 File). De novo drug development can require decades and billions of dollars, whereas drug repurposing, which is defined as finding new indications for existing drugs, is much cheaper and faster. Many existing drugs have undergone in-depth research to identify their target proteins, and this target information is stored in databases such as DrugBank and OpenTargets. In brief, the Pathways2Targets algorithm takes the significant pathways (as previously determined by SPIA), finds each protein member of those pathways, and searches the OpenTargets database [6] for all drugs known to directly interact with each protein, and generates an extensive table containing all drugs known to interact with protein members of the significant pathways (S8 File). We sorted the results so that drug targets present in multiple signaling pathways would be ranked higher (Table 6). Though Pathways2Targets results are in no way conclusive of drug efficacy for a novel indication, the algorithm provides a short-list of drugs for subsequent validation in the laboratory and has a track record of returning many drugs already in use for a given disease and several novel drug candidates [44-47]. Based on the Pathway2Targets output, we predicted the most relevant existing FDA-approved drugs for other indications that could affect the lymphoma phenotype are Doxycycline, Ocriplasmin, and Collagenase. We also identified ATN-161 as a candidate drug, but it has only been tested in phase-two trials. Doxycycline is currently in use for BCNHL subtypes [48]. The other drug candidates are promising based on drug targeting data but require follow-up validation experiments.
|
Drug Name |
Drug ID |
Significant Pathways Targeted |
Is FDA Approved for Human Use in > 1 Indication |
Highest Clinical Trial Phase |
Has Been Withdrawn |
|
|
1 |
OCRIPLASMIN |
CHEMBL2095222 |
13 |
TRUE |
4 |
FALSE |
|
2 |
ATN-161 |
CHEMBL4297456 |
10 |
FALSE |
2 |
FALSE |
|
3 |
DOXYCYCLINE |
CHEMBL1200699 |
10 |
TRUE |
4 |
FALSE |
|
4 |
DOXYCYCLINE |
CHEMBL1433 |
10 |
TRUE |
4 |
FALSE |
|
5 |
AS-1409 |
CHEMBL2109413 |
9 |
FALSE |
1 |
FALSE |
|
6 |
COLLAGENASE CLOSTRIDIUM HISTOLYTICUM |
CHEMBL2108709 |
9 |
TRUE |
4 |
FALSE |
|
7 |
FIRATEGRAST |
CHEMBL2104967 |
9 |
FALSE |
2 |
FALSE |
|
8 |
L19IL2 |
CHEMBL2109608 |
9 |
FALSE |
3 |
FALSE |
|
9 |
L19SIP 131I |
CHEMBL2109412 |
9 |
FALSE |
2 |
FALSE |
|
10 |
L19TNFA |
CHEMBL2109589 |
9 |
FALSE |
2 |
FALSE |
|
11 |
VOLOCIXIMAB |
CHEMBL2108061 |
9 |
FALSE |
3 |
FALSE |
|
12 |
ABITUZUMAB |
CHEMBL2109621 |
8 |
FALSE |
2 |
FALSE |
|
13 |
AL-78898A |
CHEMBL4594457 |
8 |
FALSE |
2 |
FALSE |
|
14 |
CILENGITIDE |
CHEMBL429876 |
8 |
FALSE |
3 |
FALSE |
|
15 |
EPTIFIBATIDE |
CHEMBL1174 |
8 |
TRUE |
4 |
FALSE |
|
16 |
ETARACIZUMAB |
CHEMBL1743014 |
8 |
FALSE |
2 |
FALSE |
|
17 |
HUMAN C1-ESTERASE INHIBITOR |
CHEMBL4297549 |
8 |
TRUE |
4 |
FALSE |
|
18 |
INTETUMUMAB |
CHEMBL1743032 |
8 |
FALSE |
2 |
FALSE |
|
19 |
PEGCETACOPLAN |
CHEMBL4298211 |
8 |
FALSE |
3 |
FALSE |
|
20 |
STX-100 |
CHEMBL2109623 |
8 |
FALSE |
2 |
FALSE |
Table 6: Predicted BCNHL drugs based on signaling pathways.
Rather than solely rely on the significant differential expression data to determine biomarkers, we applied a more robust random forest machine learning classification method to predict biomarkers of BCNHLs. Specifically, the DEG statistics focus on identifying genes that have a significantly large difference in expression between two states (lymphoma vs. healthy), while the random forest approach identifies genes that consistently show distinct profiles that are strongly dependent on disease state (lymphoma vs. healthy). Consequently, the random forest approach generates a model to identify gene products that are capable of most accurately classifying either disease or healthy states for the data analyzed. The top three genes identified by our random forest analysis included YES1, FAM98B, and FERMT2 (Table 7, Figs 3A and 3B, S9 File). We then calculated the area under the curve for the receiver-operator characteristic curve, which showed that when the expression values from these three genes have a combined specificity and sensitivity of 99.889% when classifying whether the patient samples had BCNHL (Fig 3C). The top three genes identified by our random forest biomarker prediction are high-fidelity biomarkers of BCNHL due to their consistent and extreme upregulation across our 134 clinical BCNHL samples as compared to our 188 healthy B-cell samples. YES proto-oncogene 1, Src family tyrosine kinase (YES1), FERM domain containing kindlin 2 (FERMT2), and family with sequence similarity 98 member B (FAM98B) have previous associations with cancers, but our finding that they are associated with BCNHL as high-potential biomarkers is novel.
Figure 3: Biomarker Prediction Yields a Three-Gene Signature with 99% Predictive Ability. Random forest analysis was conducted using the normalized read counts for all sequenced genes from each sample and the disease condition associated with each sample (healthy or lymphoma). A) Initial random forest biomarker results quantified with mean decrease in Gini impurity and mean decrease in permutation show YES1, FAM98B, and FERMT2 as the highest-ranked diagnostic biomarkers (ranked by mean decrease of Gini impurity score). B) Random forest results for the top three genes in isolation. C) Receiver-operator characteristic curve using only YES1, FAM98B, and FERMT2 shows these three genes have 99.889% specificity and sensitivity when predicting BCNHL status (healthy or diseased) based on B-cell transcripts.
|
Gene Symbol |
Mean Gini Impurity Decrease |
edgeR Log Fold Change |
edgeR FDR P-value |
Disease Status |
Mean (Read Counts) |
Standard Deviation (Read Counts) |
Median (Read Counts) |
|
YES1 |
0.77 |
2.38 |
1.98x |
Lymphoma |
1151.756 |
1246.946 |
629 |
|
Healthy |
38.87234 |
66.01043 |
11 |
||||
|
FAM98B |
0.68 |
1.58 |
1.48x |
Lymphoma |
1797.452 |
1174.797 |
1456 |
|
Healthy |
248.4202 |
954.1375 |
32 |
||||
|
FERMT2 |
0.67 |
2.83 |
1.46x |
Lymphoma |
1246.993 |
1200.669 |
841 |
|
Healthy |
32.46809 |
73.10299 |
4 |
Table 7: BCNHL biomarkers predicted from gene expression using machine learning.
4. Discussion
The goal of this study was to collect and analyze publicly available RNA-seq data from GEO to find differentially expressed genes, pathways, splice variants, and biomarkers that are relevant to BCNHL after the cells are initially transformed. We confirmed several biologically- and clinically-relevant biomarkers and pathologic mechanisms that were identified previously, as well as novel entities. We found several key genes that are significantly differentially expressed in BCNHL including LUM and other SLRPs, complement protein components, and the supposed pseudogene AL512646.1. We confirmed that previously characterized biomarkers such as APOC1, VCAM1, CCL18, and CXCL9 are overexpressed in BCNHL, and that 320 genes including APOE, COL1A1, and COL27A1 had differentially expressed splice variants. We additionally found a BCNHL reliance on the upregulation of pathways associated with the extracellular matrix. We also predicted three transcriptional biomarkers that perform well at differentiating patients who have BCNHL from those who do not. To our knowledge, this is the largest joint secondary transcriptomic analysis of primary human samples in the BCNHL field to-date. Two large-scale integrative multi-platform genomic profiling projects were previously completed on diffuse large B-cell lymphoma (DLBCL) with the aid of transcriptomic sequencing [49, 50]. However, their applications of the RNA-sequencing data are distinct from the approach and purpose reflected in this project. Specifically, these prior studies used RNA-seq data to identify causative gene fusions as well as to predict subtypes, and to determine the location and frequency (respectively) of genetic aberrations that initiate disease. In contrast, we utilized RNA-seq data to better characterize the gene expression profiles of BCNHL after transformation had occurred, which provides a high-level view of characteristics that are shared across multiple BCNHL types. Though our applications of RNA-seq data were distinct, we were interested to find some shared results. Reddy et al. found that extracellular matrix and lymphatic vessel gene sets were important differentiators between their 33 gene expression-based proposed subtypes [49], while we noted an overall trend of extracellular matrix-associated pathway upregulation. Schmitz et al. noted the gain-of-function of multiple crucial genes along the PI3K pathway [50], which seems consistent with our data where we found two PI3K pathways to be upregulated. We believe that including representative samples from multiple BCNHL subtypes augments the signal(s) that are shared among the represented subtypes and could aid in the identification of shared mechanistic insights with reduced bias. Given our intentional focus on BCNHL, we did not include multiple myelomas, B-cell leukemias, or Hodgkin’s B-cell lymphomas. Promising future directions may include querying multiple databases for sequencing data and perhaps expanding the scope of future joint secondary analysis to include all B-cell malignancies. Since we used only publicly available data, there may be biases in age, gender, or ethnicity. Though there is previous evidence in the literature that directly associates BCNHL with some of our results, some of our findings are novel to BCNHL. We will therefore appeal to other models in cancer (i.e., other blood cancers, other non-blood cancers) in cases where no previously published research indicates the relationship between BCNHL and our results. Comparing our results against those from cancers outside of the BCNHL family is a direct appeal to the Hallmarks of Cancer [4]. Given that underlying mechanisms for cell growth, vascularization, disruption of the cell cycle, and other cellular attributes have the potential to be common across cancer subtypes, we expect that including research from different cancer models will help to accelerate research into shared cancer mechanisms. We therefore first pull on any research available in BCNHL, followed by research in other B-cell malignancies, other blood cancers, and finally all other cancers. We believe that identifying a possible mechanism for a gene that is associated with other cancers, but unresearched in BCNHL, is still relevant. We expect that a subset of these findings will justify additional wet lab experimentation.
4.1 Differentially Expressed Genes Suggest Shared Underlying Mechanisms for Lymphomas
Lumican (LUM) seems to play a role in the progression or non-progression of several different cancer types. Mahadevan et al. previously reported upregulated LUM in both T- and B-cell lymphomas, but offered no insights on potential mechanisms [51]. A literature search of parallel systems revealed that in breast cancer, high stromal-cell expression of LUM adjacent to the tumor stalls tumor growth, and lowered stromal expression of LUM correlates with higher breast cancer mortality rates and increased severity [52]. In melanoma, LUM in the extracellular matrix halts metastasis through direct interaction with alpha-2-beta-1 integrin [53]. Both breast cancer and pancreatic cancer cells have been documented to upregulate LUM, along with many other cancer types [43]. Overall, LUM expression by cancer cells seems to correlate with more aggressive cancers and poorer patient outcomes. The massive LUM upregulation illustrated in our samples may be because the BCNHL samples available on GEO were mostly from advanced or refractory cases of BCNHL. The prior finding that high LUM expression around tumors is protective against metastasis in several cancer subtypes indicates the potential for LUM as a cancer-stalling therapy. Interestingly, a subset of the members in the SLRP protein family have been previously identified in B-cell Non-Hodgkin’s lymphomas including DCN [54], BGN [54], ASPN [55], FMOD [56], LUM [51], PRELP [56], and TSKU [57]. However, other members within the SLRP family have not been previously considered as lymphoma biomarkers or potential pathology-inducing molecules. Our novel finding is that the SLRPs ECM2, CHAD, PODN, and PODNL1 are differentially expressed in BCNHL. Proteoglycans have been shown to be associated with pro-cancer mechanisms in prostate, breast, colon, lung, ovary, mesothelium, pancreatic, lymphoma, and esophageal cancers [43]. Our results show two upregulated pathways in BCNHL that were previously shown to be mechanistically intertwined with proteoglycans in cancer, which are the Focal Adhesion pathway [58] and the PI3K-Akt signaling pathway [59]. Taken together, these data may suggest a connection between previously established proteoglycan cancer mechanisms and B-cell non-Hodgkin’s lymphomas. Additional work is still required to elucidate the role(s) that these entities play in BCNHL.
Discussing our other top DEGs in the context of other cancers, increased expression of complement genes C1QA and C1QB at week 16 of mantle cell lymphoma treatment by Venetoclax and Ibrutinib was significantly associated with a worse prognosis [60], illustrating that C1QA and C1QB may be associated with resistance to cancer drugs. Jiang et al. showed via immunohistochemistry that C1QB localizes to the nuclei of gastric cancer cells [61]. C1QB’s nuclear localization suggests that C1QB may have additional function(s). Upregulation of C1QA, C1QB, and C1QC in peripheral T-cell lymphoma [62] and upregulation of C1QC in Epstein-Barr Virus-positive diffuse large B-cell lymphoma [63] have been reported previously. Though it is possible that BCNHL is upregulating expression of C1q chains A, B, and C in response to underlying patient-cohort deficiency in complement function, the whole C1q protein has been shown to mediate metastasis, motility, growth and proliferation, and adhesion in multiple other in-vitro and in-vivo cancer models [64].We consider this C1q research across multiple cancer types to indicate that an alternate, cancer-associated C1q function in BCNHL merits further investigation. Our results add to the growing body of work suggesting a potential alternate function of complement proteins in cancer that warrants further investigation.
In addition to our novel findings on differentially expressed genes, we were also able to detect statistically significant genes that were previously characterized in at least one subtype of BCNHL. The first of these proteins is Apolipoprotein C1 (APOC1), which we observed to be upregulated in BCNHL. APOC1 is one of three genes whose expression levels are predictive of diffuse large B-cell lymphoma severity [65], and it is also upregulated in late stage lung cancers as compared to early stage lung cancers [66]. This suggests that APOC1 may be contributing to cancer pathology across diverse cancers in multiple cell types. Our observation that C-C motif chemokine ligand 18 (CCL18), which has a well-recognized role in lymphoma, was upregulated in our BCNHL analysis is relevant since this gene assists large B-cell lymphoma in cell proliferation, the NF-Kappa-B pathway, and the PI3K-AKT pathway [67]. Its upregulation in macrophages and dendritic cells from cutaneous T-cell lymphoma lesions was associated with a negative prognosis [68]. Our finding of C-X-C motif chemokine ligand 9 (CXCL9) to be significantly upregulated in our analysis of B-cells is interesting since this gene has been shown to promote the progression of diffuse large B-cell lymphoma by halting degradation of beta-catenin (CTNNB1) and upregulating its initial expression [69]. Our findings support this proposed mechanism with CTNNB1 being upregulated in lymphoma (log2FC = 1.1, FDR = 1.54 × 10−33), while other elements of the CTNNB1 “destruction complex” were mostly downregulated. Specifically, several of the known components of the destruction complex that were detected in our analysis include APC (log2FC = -0.755, FDR = 3.51 × 10−11), GSK3B (log2FC = -0.692, FDR = 2.62 × 10−3), CSNK1A1 (not significant), AXIN1 (log2FC = 0.533, FDR = 3.96 × 10−10), BTRC (not significant), and FBW11 (log2FC = -0.692, FDR = 5.60 × 10−20). We identified several other genes that may be relevant to cancer pathology. Small but significant upregulation of AXIN1 is of interest for additional investigation due to its ties to CXCL9, and is not known to have multiple heterogenous functions [70]. AXIN1 regulates the Wnt and Janus Kinase (JNK) signaling pathways [71], and it regulates the Wnt pathway by degrading CTNNB1 [50]. If CTNNB1 is not degraded by AXIN1, CTNNB1 translocates to the nucleus and interacts with LEF1, which we found to be significantly upregulated, and TCF7 (not significant in this study), causing transcription of Wnt pathway target genes to occur [72, 73]. Wnt helps to regulate cell cycle and contributes to the increased growth rate of many cancer types [74]. AXIN1 activates the JNK signaling pathway by binding to MAP3K1, which we found to be significantly downregulated, or to MAP3K4, which was significantly upregulated [75]. Since CTNNB1 has been shown to contribute to apoptosis resistance in multiple myeloma cells [76], it is possible that BCNHL’s decreased ability to destroy CTNNB1 in may contribute to a similar pathogenic mechanism. Finally, VCAM1 upregulation is associated with a poor prognosis for patients with non-Hodgkin's lymphomas, and VCAM1 is under investigation as a potential serum biomarker for assessing disease progression [77]. Adhesion molecules such as VCAM1 promote cancer metastasis, or in the case of blood cancers, extravasation, by allowing cancer cells to exit the bloodstream and integrate with healthy tissues throughout the body [78].
4.2 Splice Variants Suggest Relevance to Lymphomas
To better understand the contribution of differentially expressed splice variants to disease, we examined the highest-ranked DRIMseq results. Our observation that Apolipoprotein E (APOE) was the highest-ranking splice variant result for BCNHL resonates with previous findings that associate this gene with pancreatic cancer pathology [79]. In addition, pediatric patients with malignant lymphoma and acute lymphoblastic leukemia who express isoforms E3 and E4 of APOE are at higher risk of developing extreme hypertriglyceridemia [80]. Though little research has been done concerning the mechanisms of APOE in BCNHL, we believe that APOE may be contributing to disease by participating in the Regulation of Insulin-like Growth Factor (IGF) activity by Insulin-like Growth Factor Binding Protein (IGFBP) Pathway, which is we found to be a significantly modulated pathway that includes APOE. The significance of APOC1 as a DEG in BCNHL, paired with the evidence of significant APOE splice variants suggest that apolipoproteins may be useful targets for future BCNHL treatments. Our observation of Collagen type I alpha 1 chain (COL1A1) as a highly ranked splice variant result is novel to the best of our knowledge. However, the literature indicates that the COLA1A-014 transcript regulates the CXCL12-CXCR4 axis in gastric cancer, leading to tumor progression [65]. In addition to displaying significant differences in splice variant expression, we also found COL1A1 to be significantly upregulated in BCNHL. COL1A1 has previously been reported to be upregulated in peripheral T-cell lymphoma [32]. In Hodgkin’s lymphoma, COL1A1 overexpression is associated with epigenetic silencing of the RNA demethylase ALKBH3 and reduced survival [81]. COL1A1 is a member of several of our significant upregulated pathways involving the extracellular matrix (ECM-receptor interaction, Focal adhesion, Extracellular matrix organization, and Collagen formation). This involvement in extracellular matrix-related pathways strengthens the case that the mechanism of COL1A1 may involve tumor cell interaction with its outer environment. Collagen type XXVII alpha 1 chain (COL27A1) having significant changes among its expressed splice variants in BCNHL is interesting since it was recently reported as being overexpressed in adenoid cystic carcinoma [82]. Like COL1A1, COL27A1 is a member of the upregulated Extracellular matrix organization and Collagen formation pathways, suggesting that COL27A1 could play a role in BCNHL extravasation.
4.3 Extracellular Matrix-Related Pathways may contribute to Disease
Our signaling pathway enrichment analysis broadened the scope of our analysis and interpretation. Many of our findings supported an interesting reliance of BCNHL on pathways associated with the extracellular matrix. Recent research has suggested the importance of extracellular matrix components in reactivating quiescent cancer cells through the β1-integrin signaling pathway [83]. It would follow that interaction with extracellular matrix components also plays a role in regulating cancer cells. To our knowledge, no studies have reported the integrin signaling pathway to be activated in BCNHL, though it has been reported as activated in the closely-related cancer NK/T-cell lymphoma [51]. The activation of these pathways suggests that malignant BCNHL cells may have an advantage by interacting with the extracellular matrix. Such interactions with the extracellular matrix are typically considered to be an important part of metastasis [78]. We found this result to be interesting since lymphomas are liquid tumors, unbound by extracellular matrix. This upregulation of pathways allowing interaction with the extracellular matrix may suggest that BCNHL could be invading non-lymphatic and/or non-circulatory tissues. The trend of extracellular matrix interaction is also seen in the DEG results, adding support to the idea that interaction with the extracellular matrix is important for BCNHL growth and survival. Additionally, COL1A1 and COL27A1, which are members of extracellular matrix-related pathways, are two of the genes with the most significantly differential expression of splice variants.
4.4 Drug Prediction Algorithm Returned both Tested and Novel Candidates
Of our top drug results, doxycycline is currently in use for ocular B-cell lymphomas [84, 85]. It is additionally under investigation for diffuse large B-cell lymphoma; recent work found doxycycline suppresses diffuse large B-cell lymphoma growth in vitro and in vivo via CSN5 inhibition [48]. ATN-161 is a novel drug candidate for BCNHL. Though it has only been tested in phase two of clinical trials, it has been a successful drug against refractory solid tumors, making it a promising drug candidate for other susceptible malignancies [86]. ATN-161 suppresses cancer via integrin beta1 alpha5 antagonism, disabling invasion and metastasis [87]. Ocriplasmin reverses vitreomacular adhesion via interaction with fibronectin and laminin [88]. Though ocriplasmin has never been used in cancer before, it may be a promising drug candidate due to its ability to modulate adhesion. Collagenase clostridium histolyticum is under investigation for treating collagen-rich uterine fibroids and was successful at reducing the stiffness of the tumors [89]. These predictions justify further validation experiments to determine their relevance in human BCNHL.
4.5 Machine Learning Predicts Novel Biomarkers of BCNHL
YES1, FERMT2, and FAM98B are novel biomarkers not previously associated with BCNHL. However, each has well-documented cancer associations. YES1 is a tyrosine kinase which regulates cell cycle and apoptosis in vitro and cell growth in vivo of tumors with YES1 amplification [90]. YES1 has been previously identified as a biomarker for non-small cell lung cancer and esophageal adenocarcinoma [91, 92] and may be a potential membrane biomarker. YES1 can anchor to the inner membrane with help from peptide SMIM30 [93], but whether it can flip to the outer leaflet has not been investigated. The role of YES1 in BCNHL pathology also needs additional investigation. FERMT2 has been pinpointed as a biomarker for other cancers previously including non-small cell lung cancer and prostate cancer [94, 95], but not for BCNHL. FERMT2 stabilizes CTNNB1, which is a well-documented activator of oncogene transcription, and is implicated in Wnt pathway regulation [96]. Additionally, FERMT2 enhances integrin signaling and mediates migration, invasion, and focal adhesion [97, 98]. Though FAM98B has been shown to play an important role in the development of multiple cancers, it has not previously been identified as a biomarker for any cancer. FAM98B is an arginine methyltransferase utilized in tumorigenesis and works in tandem with DDX1, a pan-cancer marker, in RNA metabolism/processing [7, 80]. Like YES1 and FERMT2, FAM98B has not been previously identified as a biomarker for BCNHL. These three genes have substantial diagnostic potential as a liquid biopsy that could be generalizable across B-cell non-Hodgkin’s lymphoma subtypes. Further experimental validation is needed to determine whether these are suitable as diagnostic or prognostic biomarkers.
5. Conclusions
In summary, our joint secondary analysis identified many significant differentially expressed genes and pathways that play a role in B-cell non-Hodgkin’s lymphomas. Our findings confirm results of previous BCNHL research, indicating that the statistical analyses applied within our computational workflow pipeline are effective at accurately identifying statistically significant genes, splice variants, and pathways with clinical and pathological relevance. Additionally, several of our results are novel, which need additional validation in future experiments. It is likely that at least some of these novel findings were detected due to the ability of our joint secondary analysis to reduce the statistical “noise” produced by outliers from individual studies and increase the biologically-relevant signal. Specifically, our preliminary findings suggest that LUM and 10 other small leucine-rich proteoglycans are significantly differentially expressed in BCNHL, that AL512646.1 is not a pseudo-gene, that APOE, COL1A1, and COL27A1 have significant differentially expressed splice variants in BCNHL, and that BCNHL is strongly reliant on the overexpression of extracellular matrix-associated pathways. The predominant drug prediction results nearly universally targeted extracellular matrix-associated mechanisms and has yielded several promising new potential drug candidates including ocriplasmin and ATN-161. Our random forest biomarker discovery pinpointed three novel biomarker genes not previously associated with BCNHL, YES1, FERMT2, and FAM98B, which show high fidelity in predicting lymphoma presence based on transcriptional levels in B-cells. We believe that additional experiments are needed to validate our results. These findings shed additional light on the underlying intracellular mechanisms of BCNHL and could be used in the development of improved diagnostics and therapeutics to further improve human health. We anticipate that future directions after wet-lab validation could include evaluating FAM98B, FERMT2, and YES1 expression in liquid biopsy as a diagnostic tool, investigating the utility of the predicted drugs against BCNHL, and determining the roles of the genes identifying by our analysis in BCNHL pathology.
Other Information
This joint secondary analysis was not registered. The review protocol was not prepared separately but is described in detail in the methods section. This joint secondary analysis received no specific financial support and was supported by general funding from Brigham Young University. Brigham Young University played no role in the ideation, synthesis, or analysis of this joint secondary transcriptomic analysis. The authors each declare that they have no competing interests. All raw data analyzed in this study can be found in the NCBI GEO database. Analytical codes can be found as cited in materials and methods. All processed data are available in the supplementary materials or at DOI: 10.5281/zenodo.4757764.
Acknowledgments
We thank the high-performance computing resources hosted by the BYU Research Computing Center. We also gratefully acknowledge those who generated, provided, and submitted the original data.
Conflicts of Interest
The authors declare no conflicts of interest.
Supplementary Information
Download the additional supplementary files from the below link
https://www.fortunejournals.com/supply/JBSB_6499.zip
References
- Global Burden of Disease Cancer Collaboration. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2016: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncology 4 (2018): 1553-1568.
- Shankland KR, Armitage JO, Hancock BW. Non-Hodgkin lymphoma. The Lancet 380 (2012): 848-857.
- Küppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer 5 (2005): 251-262.
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 100 (2000): 57-70.
- Fouad YA, Aanei C. Revisiting the hallmarks of cancer. Am J Cancer Res 7 (2017): 1016-1036.
- Ochoa D, Hercules A, Carmona M, et al. Open Targets Platform: supporting systematic drug-target identification and prioritisation. Nucleic Acids Res 49 (2021): D1302-D1310.
- Gao B, Li X, Li S, et al. Pan-cancer analysis identifies RNA helicase DDX1 as a prognostic marker. Phenomics 2 (2022): 33-49.
- Clough E, Barrett T. The Gene Expression Omnibus Database. In Statistical Genomics: Methods and Protocols. Methods in Molecular Biology E. Mathé, and S. Davis, eds. Springer, New York, NY (2016): 93-110.
- Faramand R, Jain M, Staedtke V, et al. Tumor Microenvironment Composition and Severe Cytokine Release Syndrome (CRS) Influence Toxicity in Patients with Large B-Cell Lymphoma Treated with Axicabtagene Ciloleucel. Clin Cancer Res 26 (2020): 4823-4831.
- Li M, Chiang YL, Lyssiotis CA, et al. Non-oncogene Addiction to SIRT3 Plays a Critical Role in Lymphomagenesis. Cancer Cell 35 (2019): 916-931.e9.
- Porpaczy E, Tripolt S, Hoelbl-Kovacic A, et al. Aggressive B-cell lymphomas in patients with myelofibrosis receiving JAK1/2 inhibitor therapy. Blood 132 (2018): 694-706.
- Teater M, Dominguez PM, Redmond D, et al. AICDA drives epigenetic heterogeneity and accelerates germinal center-derived lymphomagenesis. Nat Commun 9 (2018): 222.
- Raju S, Kretzmer LZ, Koues OI, et al. NKG2D-NKG2D Ligand Interaction Inhibits the Outgrowth of Naturally Arising Low-Grade B Cell Lymphoma In Vivo. J Immunol 196 (2016): 4805-4813.
- Koues OI, Kowalewski RA, Chang LW, et al. Enhancer sequence variants and transcription-factor deregulation synergize to construct pathogenic regulatory circuits in B-cell lymphoma. Immunity 42 (2015): 186-198.
- Rouhigharabaei L, Finalet Ferreiro J, Tousseyn T, et al. Non-IG aberrations of FOXP1 in B-cell malignancies lead to an aberrant expression of N-truncated isoforms of FOXP1. PLoS One 9 (2014): e85851.
- Béguelin W, Popovic R, Teater M, et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 23 (2013): 677-692.
- Verma A, Jiang Y, Du W, et al. Transcriptome sequencing reveals thousands of novel long non-coding RNAs in B cell lymphoma. Genome Med 7 (2015): 110.
- Jenks SA, Cashman KS, Zumaquero E, et al. Distinct Effector B Cells Induced by Unregulated Toll-like Receptor 7 Contribute to Pathogenic Responses in Systemic Lupus Erythematosus. Immunity 49 (2018): 725-739.e6.
- Scharer CD, Blalock EL, Mi T, et al. Epigenetic programming underpins B cell dysfunction in human SLE. Nat Immunol 20 (2019): 1071-1082.
- Wang S, Wang J, Kumar V, et al. IL-21 drives expansion and plasma cell differentiation of autoreactive CD11chiT-bet+ B cells in SLE. Nat Commun 9 (2018): 1758.
- Rapier-Sharman N, Krapohl J, Beausoleil EJ, et al. Preprocessing of Public RNA-Sequencing Datasets to Facilitate Downstream Analyses of Human Diseases. Data 6 (2021): 75.
- Orjuela S, Huang R, Hembach KM, et al. ARMOR: An Automated Reproducible MOdular Workflow for Preprocessing and Differential Analysis of RNA-seq Data. G3 (Bethesda) 9 (2019): 2089-2096.
- Köster J, Rahmann S. Snakemake--a scalable bioinformatics workflow engine. Bioinformatics 28 (2012): 2520-2522.
- Babraham Bioinformatics - Trim Galore! .
- Babraham Bioinformatics - FastQC A Quality Control tool for High Throughput Sequence Data. .
- Patro R, Duggal G, Love MI, et al. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14 (2017): 417-419.
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 (2010): 139-140.
- Liberzon A, Subramanian A, Pinchback R, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27 (2011): 1739-1740.
- Wu D, Smyth GK. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res 40 (2012): e133.
- Nowicka M, Robinson MD. DRIMSeq: a Dirichlet-multinomial framework for multivariate count outcomes in genomics. F1000Res 5 (2016): 1356.
- Tarca AL, Draghici S, Khatri P, et al. A novel signaling pathway impact analysis. Bioinformatics 25 (2009): 75-82.
- Ogata H, Goto S, Sato K, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 27 (1999): 29-34.
- Mi H, Muruganujan A, Casagrande JT, et al. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8 (2013): 1551-1566.
- Nishimura D. BioCarta. Biotech Software & Internet Report 2 (2001): 117-120.
- Fabregat A, Jupe S, Matthews L, et al. The Reactome Pathway Knowledgebase. Nucleic Acids Research 46 (2018): D649-D655.
- Page MJ, McKenzie JE, Bossuyt PM, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 372 (2021): n71.
- Wickham H. ggplot2. WIREs Computational Statistics 3 (2011): 180-185.
- Simoes R. de M, Emmert-Streib F. Bagging Statistical Network Inference from Large-Scale Gene Expression Data. PLOS ONE 7 (2012): e33624.
- Tennekes M, de Jonge E. Top-Down Data Analysis with TreemapS. 6 (2011).
- Liaw A, Wiener M. Classification and Regression by randomForest. R News 2 (2002): 18-22.
- Scott TM, Jensen S, Pickett BE. A signaling pathway-driven bioinformatics pipeline for predicting therapeutics against emerging infectious diseases. F1000Res 10 (2021): 330.
- naomi-rapier-sharman. summarize_Pathways2Targets2_output (2022).
- Appunni S, Anand V, Khandelwal M, et al. Small Leucine Rich Proteoglycans (decorin, biglycan and lumican) in cancer. Clin Chim Acta 491 (2019): 1-7.
- Scott TM, Jensen S, Pickett BE. A signaling pathway-driven bioinformatics pipeline for predicting therapeutics against emerging infectious diseases. F1000Res 10 (2021): 330.
- Krapohl J, Pickett BE. Meta-Analysis of Transcriptomic Datasets Quantifies Differential Gene Expression, Affected Pathways, and Predicted Drugs from Hantavirus-Infected Human Material,. In Review (2022).
- Moreno C, Bybee E, Tellez Freitas CM, et al. Meta-Analysis of Two Human RNA-seq Datasets to Determine Periodontitis Diagnostic Biomarkers and Drug Target Candidates. International Journal of Molecular Sciences 23 (2022): 5580.
- Gray M, Guerrero-Arguero I, Solis-Leal A, et al. Chikungunya virus time course infection of human macrophages reveals intracellular signaling pathways relevant to repurposed therapeutics. PeerJ 10 (2022): e13090.
- Pulvino M, Chen L, Oleksyn D, et al. Inhibition of COP9-signalosome (CSN) deneddylating activity and tumor growth of diffuse large B-cell lymphomas by doxycycline. Oncotarget 6 (2015): 14796-14813.
- Reddy A, Zhang J, Davis NS, et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell 171 (2017): 481-494.e15.
- Schmitz R, Wright GW, Huang DW, et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N Engl J Med 378 (2018): 1396-1407.
- Mahadevan D, Spier C, Croce KD, et al. Gene Expression Profiling of Peripheral T-Cell Lymphoma (PTCL, NOS) and Comparison to Diffuse Large B-Cell Lymphoma (DLBCL). Blood 104 (2004): 2277-2277.
- Troup S, Njue C, Kliewer EV, et al. Reduced Expression of the Small Leucine-rich Proteoglycans, Lumican, and Decorin Is Associated with Poor Outcome in Node-negative Invasive Breast Cancer. Clin Cancer Res 9 (2003): 207-214.
- Brézillon S, Pietraszek K, Maquart FX, et al. Lumican effects in the control of tumour progression and their links with metalloproteinases and integrins. FEBS J 280 (2013): 2369-2381.
- Kotlov N, Bagaev A, Revuelta MV, et al. Clinical and Biological Subtypes of B-cell Lymphoma Revealed by Microenvironmental Signatures. Cancer Discov 11 (2021): 1468-1489.
- Sarkozy C, Chong L, Takata K, et al. Gene expression profiling of gray zone lymphoma. Blood Advances 4 (2021): 2523-2535.
- Mikaelsson E, Jeddi-Tehrani M, ÖSterborg A, et al. Small Leucine Rich Proteoglycans as Novel Tumor Markers In Chronic Lymphocytic Leukemia. Blood 116 (2010): 694.
- Huang H, Zhang D, Fu J, et al. Tsukushi is a novel prognostic biomarker and correlates with tumor-infiltrating B cells in non-small cell lung cancer. Aging (Albany NY) 13 (2021): 4428-4451.
- Munesue S, Kusano Y, Oguri K, et al. The role of syndecan-2 in regulation of actin-cytoskeletal organization of Lewis lung carcinoma-derived metastatic clones. Biochem J 363 (2002): 201-209.
- Mahtouk K, Tjin EPM, Spaargaren M, et al. The HGF/MET pathway as target for the treatment of multiple myeloma and B-cell lymphomas. Biochim Biophys Acta 1806 (2010): 208-219.
- Davis J, Handunnetti SM, Sharpe C, et al. Long Term Responses to Venetoclax and Ibrutinib in Mantle Cell Lymphoma Are Associated with Immunological Recovery and Prognostic Changes in Inflammatory Biomarkers. Blood 134 (2019): 2791-2791.
- Jiang J, Ding Y, Wu M, et al. Identification of TYROBP and C1QB as Two Novel Key Genes with Prognostic Value in Gastric Cancer by Network Analysis. Front Oncol 10 (2020): 1765.
- Gao HX, Wang MB, Li SJ, et al. Identification of Hub Genes and Key Pathways Associated with Peripheral T-cell Lymphoma. Curr Med Sci 40 (2020): 885-899.
- Yoon H, Park S, Ju H, et al. Integrated copy number and gene expression profiling analysis of Epstein-Barr virus-positive diffuse large B-cell lymphoma. Genes Chromosomes Cancer 54 (2015): 383-396.
- Bulla R, Tripodo C, Rami D, et al. C1q acts in the tumour microenvironment as a cancer-promoting factor independently of complement activation. Nat Commun 7 (2016): 10346.
- Zamani-Ahmadmahmudi M, Nassiri SM. Development of a Reproducible Prognostic Gene Signature to Predict the Clinical Outcome in Patients with Diffuse Large B-Cell Lymphoma. Sci Rep 9 (2019): 12198.
- Ko HL, Wang YS, Fong WL, et al. Apolipoprotein C1 (APOC1) as a novel diagnostic and prognostic biomarker for lung cancer: A marker phase I trial. Thorac Cancer 5 (2014): 500-508.
- Zhou Q, Huang L, Gu Y, et al. The expression of CCL18 in diffuse large B cell lymphoma and its mechanism research. Cancer Biomark 21 (2018): 925-934.
- Miyagaki T, Sugaya M, Suga H, et al. Increased CCL18 expression in patients with cutaneous T-cell lymphoma: association with disease severity and prognosis. J Eur Acad Dermatol Venereol 27 (2013): e60-67.
- Ruiduo C, Ying D, Qiwei W. CXCL9 promotes the progression of diffuse large B-cell lymphoma through up-regulating β-catenin. Biomedicine & Pharmacotherapy 107 (2018): 689-695.
- Mani M, Chen C, Amblee V, et al. MoonProt: a database for proteins that are known to moonlight. Nucleic Acids Research 43 (2015): D277-D282.
- Kusano S, Raab-Traub N. I-mfa domain proteins interact with Axin and affect its regulation of the Wnt and c-Jun N-terminal kinase signaling pathways. Mol Cell Biol 22 (2002): 6393-6405.
- Bienz M. TCF: transcriptional activator or repressor? Current Opinion in Cell Biology 10 (1998): 366-372.
- Escobar G, Mangani D, Anderson AC. T cell factor 1: A master regulator of the T cell response in disease. Sci Immunol 5 (2020): eabb9726.
- Bugter JM, Fenderico N, Maurice MM. Mutations and mechanisms of WNT pathway tumour suppressors in cancer. Nat Rev Cancer 21 (2021): 5-21.
- Luo W, Ng WW, Jin LH, Z. et al. Axin Utilizes Distinct Regions for Competitive MEKK1 and MEKK4 Binding and JNK Activation *. Journal of Biological Chemistry 278 (2003): 37451-37458.
- Su N, Wang P, Li Y. Role of Wnt/β-catenin pathway in inducing autophagy and apoptosis in multiple myeloma cells. Oncol Lett 12 (2016): 4623-4629.
- Shah N, Cabanillas F, McIntyre B, et al. Prognostic value of serum CD44, intercellular adhesion molecule-1 and vascular cell adhesion molecule-1 levels in patients with indolent non-Hodgkin lymphomas. Leuk Lymphoma 53 (2012): 50-56.
- Zetter BR. Adhesion molecules in tumor metastasis. Semin Cancer Biol 4 (1993): 219-229.
- Omenn GS, Yocum AK, Menon R. Alternative splice variants, a new class of protein cancer biomarker candidates: findings in pancreatic cancer and breast cancer with systems biology implications. Dis Markers 28 (2010): 241-251.
- Tozuka M, Yamauchi K, Hidaka H, et al. Characterization of hypertriglyceridemia induced by L-asparaginase therapy for acute lymphoblastic leukemia and malignant lymphoma. Ann Clin Lab Sci 27 (1997): 351-357.
- Esteve-Puig R, Climent F, Piñeyro D, et al. Epigenetic loss of m1A RNA demethylase ALKBH3 in Hodgkin lymphoma targets collagen, conferring poor clinical outcome. Blood 137 (2021): 994-999.
- Arolt C, Meyer M, Hoffmann F, et al. Expression Profiling of Extracellular Matrix Genes Reveals Global and Entity-Specific Characteristics in Adenoid Cystic, Mucoepidermoid and Salivary Duct Carcinomas. Cancers 12 (2020): 2466.
- Barkan D, El Touny LH, Michalowski AM, et al. Metastatic growth from dormant cells induced by a col-I-enriched fibrotic environment. Cancer Res 70 (2010): 5706-5716.
- Kim TM, Kim KH, Lee MJ, et al. First-line therapy with doxycycline in ocular adnexal mucosa-associated lymphoid tissue lymphoma: A retrospective analysis of clinical predictors. Cancer Science 101 (2010): 1199-1203.
- Han JJ, Kim TM, Jeon YK, et al. Long-term outcomes of first-line treatment with doxycycline in patients with previously untreated ocular adnexal marginal zone B cell lymphoma. Ann Hematol 94 (2015): 575-581.
- Cianfrocca ME, Kimmel KA, Gallo J, et al. Phase 1 trial of the antiangiogenic peptide ATN-161 (Ac-PHSCN-NH2), a beta integrin antagonist, in patients with solid tumours. Br J Cancer 94 (2006): 1621-1626.
- Barkan D, Chambers AF. β1-Integrin: A Potential Therapeutic Target in the Battle against Cancer Recurrence. Clinical Cancer Research 17 (2011): 7219-7223.
- Baldo BA. Enzymes Approved for Human Therapy: Indications, Mechanisms and Adverse Effects. BioDrugs 29 (2015): 31-55.
- Jayes FL, Liu B, Moutos FT, et al. Loss of stiffness in collagen-rich uterine fibroids after digestion with purified collagenase Clostridium histolyticum. American Journal of Obstetrics and Gynecology 215 (2016): 596.e1-596.e8.
- Hamanaka N, Nakanishi Y, Mizuno T, et al. YES1 Is a Targetable Oncogene in Cancers Harboring YES1 Gene Amplification. Cancer Research 79 (2019): 5734-5745.
- Garmendia I, Pajares MJ, Hermida-Prado F, et al. YES1 Drives Lung Cancer Growth and Progression and Predicts Sensitivity to Dasatinib. Am J Respir Crit Care Med 200 (2019): 888-899.
- Liu Z, Jiang F, Tian G, et al. Sparse Logistic Regression with Lp Penalty for Biomarker Identification. Statistical Applications in Genetics and Molecular Biology 6 (2007).
- Pang Y, Liu Z, Han H, et al. Peptide SMIM30 promotes HCC development by inducing SRC/YES1 membrane anchoring and MAPK pathway activation. Journal of Hepatology 73 (2020): 1155-1169.
- Sun W, Guo J, Cheng Z, et al. Identification of the Dysregulated Pathways and Key Gene in Prostate Cancer by Transcriptome Analysis and Cell Biology Experiments,. In Review (2022).
- Su X, Liu N, Wu W, et al. Comprehensive analysis of prognostic value and immune infiltration of kindlin family members in non-small cell lung cancer. BMC Med Genomics 14 (2011): 119.
- Yu Y, Wu J, Wang Y, et al. Kindlin 2 forms a transcriptional complex with β-catenin and TCF4 to enhance Wnt signalling. EMBO Rep 13 (2012): 750-758.
- Kawamura E, Hamilton GB, Miskiewicz EI, et al. Fermitin family homolog-2 (FERMT2) is highly expressed in human placental villi and modulates trophoblast invasion. BMC Developmental Biology 18 (2018): 19.
- Yasuda-Yamahara M, Rogg M, Frimmel J, et al. FERMT2 links cortical actin structures, plasma membrane tension and focal adhesion function to stabilize podocyte morphology. Matrix Biology 68–69 (2018): 263-279.


 Impact Factor: * 4.2
Impact Factor: * 4.2 Acceptance Rate: 77.66%
Acceptance Rate: 77.66%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 