Mutations of WHAT?
David Lynn Abel
ProtoBioCybernetics/Protocellular Metabolomics, The Gene Emergence Project, The Origin of Life Science Foundation, Inc USA.
*Corresponding author: David Lynn Abel, ProtoBioCybernetics/Protocellular Metabolomics, The Gene Emergence Project, The Origin of Life Science Foundation, Inc USA.
Received: 30 September 2025; Accepted: 09 October 2025; Published: 17 October 2025.
Article Information
Citation: David Lynn Abel. Mutations of WHAT? Journal of Bioinformatics and Systems Biology. 8 (2025): 83-98.
View / Download Pdf Share at FacebookAbstract
Typically ignored in evolutionary biology is the perfectly legitimate scientific question: “Mutations of WHAT?” What exactly is being altered? What programmed the initial genome? Are all polymorphisms pointless, or are some prescribed? The object acted upon by mutations was already programming and cybernetically executing extraordinary computational biofunction. Otherwise, no organism, let alone the fittest, would be alive to differentially survive and reproduce. What orchestrated the first symphony of homeostatic protometabolism far from equilibrium in a naturalistic, inanimate, prebiotic environment? Whatever this something is that is so subject to mutation even uses various superimposed, multi-dimensional sign/symbol systems to represent and process its semiotic commands. Shannon’s measure of Uncertainty is far from Szostak’s “functional information,” [1,2] and even further from Abel’s more refined “Prescriptive Information (PI)” [3-5]. We settle for measuring mere general, nonspecific statistical possibility in large phase spaces. We fail to appreciate that irreversible nonequilibrium thermodynamics lacks utilitarian direction, purpose, and efficaciousness. No explanation is provided for specific steering, control and the formal organization of life. The mere self-ordering phenomena of Prigogine does not begin to explain the integration of circuits or the formal orchestration of biosystems. What crafted initial protometabolic schemes? What was the source of Prescriptive Information (PI), recipe, commands and the execution of computational halting? What engineered the first subcellular nanocomputers and stunning molecular machines at the same time and place as the first programs? If mutations are so critical to evolution, why don’t we regard what is being mutated as being far more critical?.
Keywords
<p style="text-align:justify">Genetic information; genomic programming; Prescriptive Information (PI); Nonrandom mutations; Spontaneous mutations; Mutational Drift vs Genetic Drift; Directed polymorphisms; Rapid Adaptation; Population Genetics; Selection; Evolution; Prescribed Polymorphic Adaptation (PPA).</p>
Article Details
Introduction
The object of mutation (what mutations alter) had to have steered and controlled events toward biofunction and biosystems prior to alteration [6-8]. Some cause had to generate this effect of the integration of circuits and orchestration of the symphony of life [9-12]. Science’s responsibility is to pursue questions of causation and to elucidate the mechanisms explaining “How?” These are not philosophic questions. They are legitimate scientific questions. Historical science is greatly limited empirically. But that does not absolve us from seeking the elucidation of historical mechanisms of causation. The reductionism of life-origin science is extremely valuable in trying to define and understand life. Even protocellular metabolomics required steering toward biofunction and sustaining controls. Instructive and efficacious executable commands had to be issued and somehow be recorded and called up when needed. They had to be heritable. Metabolism-First models of life origin immediately suffer because of a lack of heritability. Of major interest, therefore, is how some form of The First Gene [13] was crafted and how biofunction was first prescribed [14].
The word "gene" was coined by Wilhelm Johannsen in 1909, He was a Danish botanist. He used the word “gene” to describe the Mendelian units of heredity. Its root meant to “give birth to” or to “beget.” The term “genome” was coined in 1920 by Hans Winkler, also a botanist. The German word "Gen" (meaning "gene") and the Greek suffix "-ome" (total) were combined to refer to the total set of controlling instructions and programming of the halting computations that are the essence of life [9,15,16]. Hugo de Vries used the term "mutation" to refer to distinct, sudden, heritable changes observed in the evening primrose plant, Oenothera lamarckiana. It wasn’t until the 1930’s that Thomas Hunt Morgan studied fruit fly mutations. This led to the widespread acceptance of the modern evolutionary synthesis. Watson and Crick refined the specific DNA role in 1953.
Stencel and Crespi in a paper entitled, “What is a genome” [17] employ George C. Williams’ conception of the evolutionary genome as:
“A set of genetic material, in a lineage that, due to common interests, tends to favor the same or similar phenotypes.”
Notice how “genetic material,” as usual, is just presupposed rather than explained. “Genetic material” cannot be used to define “genome.” The genome does not “favor” already-existing phenotypes. The environment does. The genome precedes, causes and programs life’s phenotypes. All known life is programmed and cybernetically processed. This alone produces living phenotypes.
Critiquing Stencel and Crespi’s definition, abiogenists might inquire, “What ‘common interests’ existed in an inanimate environment?” Or, from the perspective of a prebiotic environment, “What’s a ‘genome’ or a ‘phenotype’? What cared?” How could a prebiotic environment have “known” the difference? What naturalistic vector of instructional progress existed in an inanimate environment, prior to evolution (the differential survival and reproduction of already-living organisms)?
Work as defined by physics has nothing to do with “usefulness.” Inanimacy had no values or objectives. A prebiotic environment knew nothing about function. “Utility” is not a physicodynamic attribute; it is an abstract, conceptual, nonphysical formalism, the same as mathematics, logic theory, language and engineering. To achieve nontrivial function requires motive, intent, concept and goal. It is almost always a multistep process wherein the purposeful orchestration consists of a sequence of purposeful steps. Physical interactions “pursue” nothing, biofunction included. Inanimate nature had no interests or goals. Physico-chemical reactions “knew” only Chance and Necessity. In an inanimate environment, “function” was no better than nonfunction. Could sophisticated function have just spontaneously generated? If so, why have we never observed such “self-organization” and “emergence” since? Not even a simple piece of wire has ever “self-organized” or “emerged” from iron ore in the ground [18]. A long piece of metal alloy with constant diameter and the needed tensile strength and malleability arises only out of Choice Causation (CC), not Physicodynamic Causation (PC). If that is true of a simple piece of wire, where does that leave purely physicalistic models of abiogenesis? Nothing is more carefully programmed and engineered than life. No nontrivial halting computation has ever been programmed randomly. Laws and constraints cannot actively select from among real options in pursuit of function, either. Function is a formal concept requiring valuation, desire, purposeful pursuit and goal. Nontrivial function is not a physicodynamic effect. An inanimate environment is blind and indifferent to function. We cannot just presuppose pre-biotic preference for biofunction over no function in an inanimate cosmos ruled by nothing but physicodynamic law and constraint.
All known life is programmed and cybernetically processed
The programming commands of living organisms are empirically efficacious in prescribing homeostatic metabolism. The programming of life, when cybernetically processed, executes computational success (halting). Life is computed. It is fundamentally nonphysical. It is just instantiated into physicality using physical symbol vehicles in a formal Material Symbol System [19,20]. This is much the same as the game of Scrabble. Physical blocks of wood are used to spell abstract words with meanings. The physicality of the blocks of wood is not the point. Nucleosides, amino acids, lncRNAs, operons, enhancers, transcription factors, enzymes, methyl and acetyl groups are like blocks of wood in Scrabble. The genome is the immediate source of non-physical formal Prescriptive Information (PI) that is just instantiated into physicality [21]. PI consists of executable commands needed to compute formal function. PI instructs which choices from among real options to make, or it provides recordation of strings of efficacious choices already made [3]. The genome’s computations have already been empirically proven to halt (successfully compute), overcoming Turing’s “halting problem” that would have been encountered during initial programming. This raises the question of how an inanimate environment programmed the first gene in a presumed protocell. If highly intelligent human programmers cannot outsmart Turing’s “ halting problem,” how did inanimate nature accomplish it?
No computational halting—No living phenotypic organisms. Life is all about executable commands and the processing of those commands by some incredible nano equipment. What is the ultimate source of PI in an inanimate nature that programmed the first genome?.
Natural selection cannot explain initial programming.
Natural selection is always after-the-fact of already-programmed, already-cybernetically processed, already-living organisms. Evolution is nothing more than the differential survival and reproduction of the fittest already-living organisms. There is only one reason this simple fact is so easily lost from otherwise intelligent scientists’ consciousness. The starting philosophic axiom from which all traditional scientific reasoning proceeds precludes acknowledgement of the obvious. Choice Causation is real. Genomically, it preceded and caused all living systems, organismic agency, and life itself. Evolution and natural selection are irrelevant to abiogenesis science because they are both secondary—after-the-fact of already existing life. Organisms must already exist to differentially survive and reproduce. Organisms do not already exist without executable commands and the cybernetic processing of those executable commands [15,16].
No already-living organism programed itself into existence. That is a logical impossibility. Neither do physical laws and constraints program anything. Laws and constraints cannot make purposeful choices in pursuit of utility. Laws, constraints and forces cannot choose between a “1” and a “0” with controlling or computational intent. Whenever this realization crystalizes in the mind of philosophic naturalists, “molecular evolution” comes to the supposed rescue. Inanimate molecules are said to have evolved into living ones. The already indefinite definition of life is morphed into gradualistic concepts of life origin [22-24]. When the definition of “life” is blurred, “death” is also blurred to make protobiont and protocellular models seem more viable. Protocellular metabolomics becomes gradualistic. The problem is, way too many processes of life are interrelated and interdependent. An all-or-none reality exists at the subcellular level [25-32]. Nothing even close to life exists that does not manifest the extraordinary interdependence of pathways, cycles, feedbacks and biosystems. Holistic systems are formally organized and orchestrated entities.
We are very sloppy in our definitions. Definitions are critical to science. Weather and thermodynamic “systems,” for example, are not formal systems. They are self-ordered states (Prigogine’s “dissipative structures” [33-36]) requiring zero active selections. They are not organized. They are spontaneously-ordered. Protocellular metabolism, on the other hand, required innumerable active selections in the programming pursuit of biofunctional computations. Spontaneous mutations alter what they are mutating, almost always for the worse. Spontaneous mutations themselves cannot explain initial highly efficacious programming [15,16,37-39]. More than anything else, the answer to the question posed by this paper centers on efficacious active selections and executable commands that must be made prior to the realization of computational halting (integrated biofunction). This is something that so-called “natural” forces, laws and constraints cannot do. This is the point of failure of “molecular evolution” theory. Active selections in pursuit of utility would have been needed prior to function [39]. Active selections would have been needed at the genomic level, not just the secondary, passive, after-the-fact of living organism selections [39]. Mutations don’t create what they are mutating. Spontaneous purposeless mutations don’t program anything.
Spontaneous “Mutational Drift” vs. “Genetic Drift:
“Mutational Drift” is very different from “Genetic Drift.” Genetic Drift is related to a species’ general population and is the product of scientists’ sampling methods. Mutational Drift, on the other hand, accumulates within an individual cell line’s spontaneous mutation history. The effect of Mutational Drift is an increase in genomic randomness and a decrease in genomic Prescriptive Information from the equivalent of “typographical errors” and 2nd Law biochemical degradations. This is why spontaneous mutations have such an extremely low “beneficial” rate. Why would we expect anything different?. Genetic drift addresses organismal population genetic variability within a species. The frequency of alleles can change in a population of organisms as a result of random sampling of that population. Genetic drift is not caused by specific mutations or natural selection. Allelic frequency varies according to sampling differences. They are not caused by or even related to fitness or adaptation. Genetic drift is most affected by sampling within small populations. Within a population, some genes can be passed on to the next generation without genomic programming intent, and others are not. Drought or an unusually cold winter, for example, can wipe out a large portion of a certain species’ population. Sampling the surviving smaller population may give considerably different allelic frequencies from sampling of the original population. Some alleles can be lost altogether. Other can become “fixed” in an updated wild type, as most would say, “randomly.”
“Mutational drift,” on the other hand,” describes the result of accumulating spontaneous mutations within the genome of the same individual cell line. Both random and nonrandom mutations can be spontaneous. Nonrandom spontaneous mutations are the result of purely physicodynamic forces, laws and constraints. They are not directed toward bio-functionality or usefulness to that organism. In extremely rare cases, a truly spontaneous mutation, whether random or nonrandom, can result in a trivial genomically beneficial phenotypic trait. But this occurs without intent, by accident. They are extremely rare. These same spontaneous mutations are often accompanied by deleterious effects worse than the trivial benefit. Often the supposed benefits used as examples of beneficial spontaneous mutations are very contrived. A classic example is claiming that the sickle cell point mutation is beneficial. The erythrocytes of patients with the sickle cell anemia are so pathogenic that the malaria parasite has no ability to invade them. Any sickle cell anemia patient would gladly exchange their horrible hypoxic aches from lactic acid build-up and shortened lifespan for having to take the same anti-malarials that everyone else has to take. This spontaneous point mutation is not “beneficial” as claimed. The supposed benefit is extremely contrived. Many of the claimed beneficial spontaneous mutations are in fact prescribed polymorphisms called up into upper memory from pre-programmed modules waiting for rapid response to extreme environmental challenges [37]. Many supposedly “neutral” spontaneous mutations are only neutral in the context of one superimposed “language.” Down the line, for example, we may find that a supposedly neutral spontaneous mutation in triplet codon language was not so neutral in the superimposed sextet translational pausing code language [40]. This could lead to serious protein misfoldings, the subtle effects that may take decades to recognize. What was thought to be a neutral mutation may not be.
Prescriptive Information (PI) provides efficacious executable commands
Prescriptive Information cannot be quantified by mere Shannon Uncertainty measures. It is not just a phase space of thermodynamic possibilities. Statistics are descriptive only. Prescription is causative of real efficacious effects [41-43]. Choice Causation is real [44]. There is more to reality than Chance and Necessity, as proven by subcellular and supracellular life. Insisting on Chance OR Necessity is a false dichotomy [45]. A third fundamental category of reality exists [10]. Choice Causation exists in addition to Physicodynamic Causation [14,46-51]. This is known as the Universal Determinism Dichotomy (UDD) [44]. Not even the simplest concept of a heat engine can be explained without the intervention of Maxwell’s demon’s purposeful choices in deciding when to open and close the trap door [52].
Figure: Maxwell’s Demon has to purposefully choose when to open and close the trap door to accomplish his goal. The gas molecules are inert/ideal. No physicodynamic mechanism can explain spontaneous formal self-organization to the utilitarian end of creating even the simplest non-trivial heat engine equivalent. To create a heat differential between compartments requires purposeful choices. No Choice Causation—No Heat Engine. No Heat Engine—No Life far from equilibrium. (Figure from Chapter 10, “Moving ‘far from equilibrium’ in a prebiotic environment: The role of Maxwell’s Demon in life origin.” In Genesis - In the Beginning: Precursors of Life, Chemical Models and Early Biological Evolution Seckbach, J Gordon, R., Eds. Springer: Dordrecht).
In a 2012 book chapter entitled, “Moving ‘far from equilibrium’ in a prebiotic environment: The role of Maxwell’s Demon in life origin”[52], Abel posed the following question:
Can we falsify the following null hypothesis?
- • “A kinetic energy potential cannot be generated by Maxwell’s Demon from an ideal gas equilibrium without purposeful choices of when to open and close the partition’s trap door between compartments.”
- • If we can falsify this null hypothesis with an observable naturalistic mechanism, we have moved a long way towards modeling the spontaneous molecular evolution of life. Falsification is essential to discount teleology. But life requires a particular version of “far from equilibrium” that explains formal organization, not just physicodynamic self-ordering as seen in Prigogine’s dissipative structures. Life is controlled and regulated, not just constrained. Life obeys arbitrary rules of behavior, not just invariant physical laws. To explain life’s origin and regulation naturalistically, we must first explain the more fundamental question, “How can hotter, faster moving, ideal gas molecules be dichotomized from cooler, slower moving, ideal gas molecules without the Demon’s Choice Causation operating and closing the trap door?”
Since 2012, this null hypothesis has never been falsified in the literature. The latest genomic and molecular biological research repeatedly attests to the need for Maxwell’s Demon’s choices to orchestrate genomics. “Sustained Functional Systems (SFS) operating far from equilibrium exist only through the agency of Maxwell’s demon.” [50]. Unless we want to exclude the science of biology from the natural sciences, the survival of metaphysical naturalism as the starting axiom of science will require the falsification of this null hypothesis. Homeostatic metabolism must be steered by purposeful efficacious executable commands. These are clearly mediated by genomic programming, including the cybernetic processing/execution of that programming. To make matters even more formally “transcendent” to physicality, the RNA and protein products of that programing feed back to affect the programming itself, and conceptually complexify linear digital programming into not only a three-dimensional, but a multi-dimensionally-coded formal genome. Even the codon table is formal.
Is there any room for such “Choice Causation” in naturalistic science? If not, we had better be able to propose a purely physicodynamic mechanism for organizing potential energy, harnessing it, transducing it, calling it up only when needed, adapting it, and utilizing it without burning up the protocell in the process. Attempts have been made to reformat the Universal Determinism Dichotomy (UDD) into a dichotomy of physicodynamic determinism vs. evolutionary determinism. But evolutionary “determinism” is in actuality an effect, not a cause. The best programming produces the fittest organisms. Evolution occurs only secondary to alterations (algorithmic optimization) in genomic prescription. Differential survival of computed organisms is an effect. What causes the controls of genomic prescription? Certainly not pointless spontaneous mutations. The GS Principle and F > P Principle both obtain. The GS (Genetic Selection) Principle [41,53] states that biological selection must occur at the nucleotide-sequencing molecular-genetic level of 3'5' phosphodiester bond formation. After-the-fact differential survival and reproduction of already-living phenotypic organisms (ordinary natural selection) does not explain polynucleotide prescription and coding. All life depends upon literal genetic algorithms. “Fittest” means “optimized programming.” Even epigenetic and three-dimensional "genomic" factors such as coiling around histone proteins, regulation by long noncoding RNAs (lncRNAs), DNA methylations, histone acetylations and transcription factor protein regulation are ultimately instructed and prescribed by prior linear digital DNA programming. Biological control requires selection of particular configurable switch-settings and their integrated circuits to achieve potential function. Only controls make orchestrated biofunction possible, not mere constraints [54].
The F > P (Formalism > Physicality) Principle [49,51] states that “Formalism not only describes, but preceded, prescribed, organized, and continues to govern and predict Physicality.” The F > P Principle is an axiom that defines the ontological primacy of formalism in a presumed objective reality that transcends both human epistemology, our sensation of physicality, and physicality itself. The F > P Principle works hand in hand with the Law of Physicodynamic Incompleteness, which states that physicochemical interactions are inadequate to explain the mathematical and formal nature of physical-law relationships. Physicodynamics cannot explain formalisms such as quantum entanglement, teleportation and metrology. Physicodynamics cannot generate formal processes and procedures leading to nontrivial function[43,55]. Chance, necessity and mere constraints cannot steer, program or optimize algorithmic/computational success to provide desired nontrivial utility. Finally, physicality alone cannot explain life. The F > P Principle denies the notion of unity of Prescriptive Information (PI) with mass/energy. The F > P Principle distinguishes instantiation of formal choices into physicality from physicality itself. The arbitrary (not random, but free of physicodynamic determinism) setting of configurable switches and the selection of symbols in any Material Symbol System [19,56] is physicodynamically indeterminate—decoupled from physicochemical determinism. The above general scientific principles govern both code origin and efficacious genomic alterations leading to adaptation. Genomic alterations are either pointless spontaneous mutations or prescribed polymorphisms [37]. If they are directed polymorphisms, they are programmed by Choice Causation. They are purposefully steered with active selections at bona fide decision nodes. Even the feedback of such factors such as regulatory lncRNAs [57-62] and transcription factor proteins’ effect on DNA activation [63-67] are themselves ultimately prescribed by DNA formal coding. Even DNA methylations [68-71] and histone acetylations [72-75] are controlled by prior linear digital DNA coding. The multi-dimensional Genome is prescribed in part by linear digital formal coding of translational pausing [76], operons (activators, promotors and operators primarily in prokaryotes) [77-79], RNAs (e.g., lncRNAs) [80-83], widely dispersed enhancers [84-87], intrinsic disordered proteins (IDPs)[88,89,90, histone {Li, 2025 #20113,91-93] and chaperone proteins [94-98] correlated with prescriptive DNA nucleotide sequencing. All of this impressive functionality is formal, not merely physicodynamic. There is only one reason pointless spontaneous mutations are accredited with being regularly beneficial—they are confused with prescribed polymorphisms. These polymorphisms are programmed by the very thing that we claim gets mutated. What is it that directs polymorphisms so effectively toward efficacious RNA alternative splicing, micro and mini satellites, regulatory lncRNAs, operons, enhancers, altered tandem repeat units and the number of those units that cause immediate adaptations [99-103]. The changing nature of tandem repeat units and their number, for example, is no accident. While some represent pointless and deleterious mutations, others are clearly prescribed polymorphisms aimed at rapid adaptation to environmental challenges [99,104-107]. Each of these genomic phenomena, including crossing over, is no accident. The exchange of genetic material is intended to improve rubustness.
Engineering uses physicality; but physicality cannot generate engineering
Life itself originates from the far side of The Cybernetic Cut [42,43]. The Cybernetic Cut is an infinitely deep ravine that dichotomizes Physicodynamic Causation from Formalistic Choice Causation [43,55]. As a major corollary, physicodynamics cannot explain or generate life. Life is fundamentally formal. It is programmed, computational and invariably cybernetic. Configurable switch settings and the syntax of physical symbol vehicles in a material Symbol System (MSS) is how The Cybernetic Cut is traversed [42,55]. A one-way narrow bridge across the great ravine called “The Configurable Switch Bridge” makes possible the instantiation of formal choices into physicality. The Configurable Switch Bridge and Material Symbol Systems (MSSs) are the key to all design and engineering functions controlling physicality. Configurable switches and physical symbol vehicles are physical. But their setting and symbol selections are purely formal. Naturalistic abiogenesis models invariably violate the fundamental principle of The Cybernetic Cut [42,43,55]. Physicodynamics lies on the near side of this infinitely deep ravine known as The Cybernetic Cut. Formalisms like mathematics, logic theory, true organization (not Prigogine’s self-ordering), orchestration and language arise only on the far side of this ravine where Choice Causation originates [42,43,55]. Formalisms’ entry into the near physicodynamics side of the ravine requires traversing the narrow one-way Configurable Switch Bridge from the far side to the near physicodynamic side [42,43,55].
Physical configurable switches are “dynamically inert.” But the setting of these switches is altogether formal. For example, the light switch on your wall is never turned off by the force of gravity. It is only turned off by Choice Causation. Alternatively, Material Symbol Systems (MSS) [19,20] using representational physical symbol vehicles can also be used to introduce formal controls into physicality from the far side of The Cybernetic Cut. Genomics prescribes the computations of homeostatic metabolism far from equilibrium. These computations don’t just happen spontaneously. This is why our interest in nonrandom mutations is so quickened. Biologists want to elucidate the cause of the effect that we call “adaptation.” We have always known, at least subconsciously, that random mutations could not have collectively programmed such sophisticated “halting” computations. Thus, we must first address the source of the genomic programming and highly efficacious configurable switch-settings that pre-existed alteration. Pointless mutations could not have been initial programming’s source. They could not have chosen such sophisticated symbol and syntax active selections according to rules rather than laws. Mutations only modify programming, not create it. What caused the instantiation into physicality of formal Prescriptive Information (PI) [3-5] in the first place? Without it, there is nothing of value to mutate.
The orchestration of life makes use of physical factors such as Gibbs free energy in aminoacylation and protein folding [108]. But these physical factors are not what does the orchestrating. They are just used, the way an engineer uses steel and concrete to design and engineer a materialized suspension bridge. The idea of a suspension bridge is pure, abstract, formal concept. Prescription of function is far more conceptually complex than just recorded amino acid sequencing. 70% of proteins in the Protein Data Bank (PDB) contain unspecified regions with missing electron densities (IDPs/IDRs). X-ray crystallography represents averaged pictures of this conformational flexibility [109,110]. This is caused mostly by thermal fluctuations. But not even this “intrinsic disorder of proteins” (IDPs) explains their specific functionality. Such physical variability itself does not define the pointed controls of biofunction. The flexibility is just used by formal control mechanisms in the pursuit of homeostatic metabolism far from equilibrium. This control comes from the Prescriptive Informational commands of the genome. This is what mutates, whether pointlessly, or by prescribed polymorphisms intended to hasten adaptation. Eukaryotic proteomes have a higher fraction of intrinsic disorder than prokaryotic proteomes. So, there are at least two very different categories of proteins: polyamino acid sequences that fold naturally into pre-prescribed domains vs. sequences that yield IDPs/IDRS. The final models lack residue definition [111,112]. Unspecified regions account for considerable conformational flexibility. But programming and reprogramming need such contingency to allow choices from among real options, especially for adaptation. This is what programming means. IDPs/IDRs are programmed to have specific functions of regulation, recognition, and signaling. “Disordered” rather than “ordered” is good for flexibility of genomic control. Well-developed regulation networks exist in multicellular eukaryotes, especially, that employ IDPs/IDRs flexibility to perform impressive regulatory functions.
Material Symbol Systems are formal even though they use physical symbol vehicles
Life is prescribed with executable commands that employ physical symbol vehicles in Material Symbol Systems (MSSs) [19,20]. Instructions use representational symbols that have meaning beyond their mere physicality. Formal linguistic rules must be shared and voluntarily obeyed by source and recipient across a Shannon channel. The fixed laws of physicodynamics have nothing to do with meaning. Life is semiotic, cybernetic and computational. Signs, symbols and syntax must be actively selected in any form of semiosis [113-115]. Every time a nucleoside is actively selected to add to a polynucleotide string, a choice must be made from among four options. Each choice at each decision node must be made prior to any phenotypic benefit that the environment could favor. The fact that the symbol vehicles are physical does not change the fact that their representations in the codon table and transitional pausing table are formal rather than physicodynamic. The source of biological codes, especially when superimposed and multidimensional, presents a whole new can of worms for naturalistic attempts to explain life [116-120]. Most of the abiogenesis papers of the last four decades have focused solely on physicodynamic aspects with hardly any mention of the obvious formal controls of virtually all of life’s processes. No published naturalistic model thus far generates the needed steering and controls that define life. Many of the problems encountered in life-origin science are summarized in two books [13,14] and in very recent papers [9,15,16,37,38,46,47] The futility of most abiogeneic models stems from failure to acknowledge that life itself could only have originated from the far side of The Cybernetic Cut [42,43].
Symbolization in biosemiosis requires active selection of those symbols
Barbieri’s enumeration of multiple biological codes [121,122] has long-since been substantiated [3,115,117-120,123-146]. Symbolization is not possible without active selection of those symbols from among real symbol options. Similarly, syntax must be actively selected—not only the initial active selection of each nucleoside, but each codon, the sequencing of codons and the active selection of syntactical meaning according to rules, not laws. Each tandem repeat unit sequence and number of unit repeats has meaning and effect. Each kind of polymorphism, each transcription factor and its specific binding site, each RNA alternate splicing, methylation site, acetylation site in histones, phosphorylation, transposition of coding sequences, micro and minisatellite matter functionally. They are not purposeless and random. Their effects cannot be reduced to mere physicochemical Necessity, either. All of these functions result from formal representations of executable commands. The executable commands are messaged. They are semiotic. Semiosis is formal, even though it may use physical symbol vehicles in Material symbol systems (MSSs). Electromagnetism doesn’t issue executable commands or make programming choices. Gravity doesn’t steer events toward the goal of usefulness. Neither the strong nor the weak nuclear force can program. Quantum entanglement, irreversible nonequilibrium thermodynamics and mere probabilism are not specifically determinative of biofunction.
What about Central Dogma Doubts
Much conjecture has arisen over the seeming reverse flow of Prescriptive Information (PI) in opposition to the Central Dogma from various alternate splicing RNA controls, satellites, transcription factors, epigenetic switches, transpositions, tandem repeat (TR) polymorphisms, methylations, acetylations, phosphorylations, etc. But eventually all these “reverse flows of PI” will be proven to be directed from a central programming source that involves the call-up of reserve programming modules in response to environmental challenges. Ultimately, the Central Dogma is not as compromised as supposed. Unfortunately, many are confusing spontaneous "non-random mutations" with being "directed" ones. Non-random can be physicodynamically caused with no formal/conceptual/functional/prescriptive component at all.
Wong et al advocate replacing “prebiotic chemistry” with “protobiotic process” [147] Their point is that not all chemistry that happens before life on a planet must be directly or causally related to life's onset. When abiogenist authors wish to highlight certain processes that they assume to be relevant, they frequently subconsciously incorporate teleological factors without realizing it. Examples include explanatory phrases such as “in order to . . .” and “so that . . .” But we should also ask what a prebiotic environment knows of “processes”? “Process” presupposes a lot of teleological aspects within its own conception. Processes normally lead somewhere useful. They presumably accomplish some needed goal for which the process was undertaken. Unless the term is misused, it is formal. It is often misued in weather forecasting, thermodynamics and chaos theory. Unsteered physicodynamic sequences of events are not true processes. Can an inanimate environment undertake a useful process? How and why would “a useful process” be undertaken by Chance and Necessity? Neither is capable of launching such a quest. Our starting axiom of naturalistic science must be bogus. Physicodynamics is not sufficient. Clearly abundant empirical evidence exists in reality of all sorts of subcellular and supracellular formal processes. Demetrius points out that the origin of cellular life can be described in terms of the transition from inorganic matter to the emergence of cooperative assemblies of organic matter [148]. He emphasized the essential element of Directionality in life origin process. Missing from virtually all abiogenic papers, irreversible nonequilibrium thermodynamic ones in particular, is any answer to the question, “What exactly steered that directionality toward the goal of becoming alive, staying alive, and propagating offspring?”. Phenotypes don’t just happen. They are programmed and processed computations. If the programming improves, the phenotypes improve. But what optimizes the programming? More importantly, what generated any programming in the first place? Even when organisms already exist, evolution still doesn’t program. Programming requires active selections at bona fide decision nodes. These decision-node choices have to be made prior to computational halting—prior to any phenotypic existence or benefit. Only those organisms with the most optimized programming secondarily survive better.
The only acceptable answer to the question, “Mutations of WHAT?”
The answer to our primary question is that “Already-recorded efficacious choice-based executable programming commands in the wild type genome are being altered.” Based on universal empirical experience and reason, those commands could not plausibly [149-151] have been prescribed by nothing but spontaneous purposeless mutations [9,15,16,37,39]. The Universal Plausibility Metric and Principle come into play [149-151]. We must be more careful to start dichotomizing spontaneous pointless mutations from “prescribed polymorphisms (PPs)” [37].” PPs are programmed. Pointless mutations are not. Spontaneous “Mutational Drift” (different from “Genetic Drift”) only accumulates nonefficacious randomness into that cell-line’s genome. Rapidly adaptive polymorphisms, on the other hand, are prescribed by cybernetic modules held in reserve and called up into upper memory upon environmental challenge [15,16,37]. They are anything but pointless mutations. They are fully directed—dare we admit it—purposeful. They account for far more rapid adaptation than could ever be accredited to evolution. These programming modules are ready and waiting to respond as needed.
The essence of genomes is the multi-dimensional DNAome of switched on and off “efficacious executable commands.” The first of these many dimensions is almost always overlooked (fanatically “metaphysically/philosophically denied” might be a better term). It is the dimension of Choice Causation rather than Psychodynamic Causation. The Universal Determinism Dichotomy (UDD) must be acknowledged [10,44,152].
The next dimension is the choice of the specific nucleoside next to be polymerized in the linear digital string of nucleic acid. The next dimension after that is the syntax of individual quaternary choices. Next is the dimension of language and formal coding tables of rules, not laws, used to interpret the meaning across a Shannon channel of the syntactical sequencing of physical symbol vehicles. Next is the tertiary structure of folding prescribed by multiple causative factors including amino acid sequencing, the sextet translational pausing code superimposed on the triplet coding[40], followed by chaperone and foldamer directing of folding. Then there are the operons, widely dispersed enhancers. Still more dimensions are added with factors such as “intrinsic disorder of proteins (IDPs) that cannot be enumerated even with crystallography. Only averaged data of specifically prescribed controls can be identified. But the particular shape that goes into this average is specifically causative [153-155]. Next is the extensive role of purposeful alternate splicing of multiple forms of RNA, the lncRNAs especially. Then there are the transcription factors, the specific controlling sites of DNA methylations [156-161]. Then there are the three-dimensional coiling factors around the histone proteins regulated by acetylation choices [162-164]. The list of nonphysical formal controls instantiated into physicality goes on and on. These formal controls, not constraints, are the essence of life. Life is programmed computation [16].
Biosemiotics in particular compounds the problem of life origin. These already-existing executable commands are instantiated into physicality using physical symbol vehicles in a Material Symbol System (MSS) [19,122]. Physical objects are being used to semiotically represent meaning and instructions for how to do something useful. These initial abiogenic instructions would have been useless from day one if some equivalent of a Turing machine had not also been caused at the same place and time as the symbolically recorded executable commands in the Turing tape. The most basic cybernetic programming consists of a purposeful choice between a “zero” and a “one.” Will the formal logic gate be set to “Open” or set to “Closed”? Disallow that choice because of purely metaphysical prejudice, and no form of programming is logically possible. Yet all known life is programmed and cybernetically processed. No example of nontrivial programming or its execution independent of purposeful choice causation has ever been observed. Mere complexity is not conceptual complexity. Conceptual complexity is required to generate sophisticated functions. Conceptual complexity is formal, not merely physico-chemical. Fixed laws do the same thing the same way every time with very little statistical variation. Laws and constraints cannot actively select for formal function from among the contingent options that laws severely limit.
Normally we just presuppose the existence of the PI that mutates. We are not supposed to ask the perfectly legitimate scientific question of, “What naturalistic mechanism generated this PI?” We know full well what thin ice that would be to walk on for our careers. So, what we hear is, “You’re asking the wrong question.” No, we’re not! We are asking THE question that should have been better pursued by science 150 years ago. We subconsciously, if not consciously, sense it is a serious threat to our lifelong cherished purely metaphysical worldview. We have defined this religious belief into our very definition of science. Thomas Kuhn called it a “paradigm rut” [165]. It has rapidly become progressively more of an embarrassment as a result of genomic and molecular biological advances in particular. Even before that, Nobel laureates were troubled by “the unreasonable effectiveness of mathematics in the natural sciences” [166,167]. There is nothing physical about mathematics, life’s programming and computations, or the scientific method itself. Blind belief in physicalism has no place in science, the science of biology, especially. The prior programming that is being altered by mutations is abstract, conceptual, controlling (not merely constraining), nonphysical, formal, Prescriptive Information (instructions and commands). The most pressing questions relating to mutagenesis and its capabilities must be preceded by questions relating to the origin of what is being mutated.
Before we can discuss the cause of efficacious nonrandom mutations, we must first ask what caused the efficacious commands in the first place, prior to any mutation. No empirical or logical basis exists for believing that pointless spontaneous mutations generated the initial efficacious commands. Halting programing would have had to previously exist for polymorphisms to optimize. Neither Chance nor Necessity could have programmed the initial Prescriptive Information. Neither Chance nor Necessity could improve that prior programming except trivially and extremely infrequently. The initial halting computational commands of the wild type could only have been generated by efficacious Choice Causation. That Choice Causation is what is mutating. If that PI is mutating for the better, those mutations are almost certainly programmed by intended efficacious polymorphisms known as Prescribed Polymorphic Adaptation (PPA) [37]. Pointless mutations, even when nonrandom from physicodynamic causation, will almost always be found to be deleterious. Many may seem to be neutral initially. Superimposed coding languages may not become initially apparent. Mutations thought to be neutral in one superimposed language may not be neutral in others (e.g. translational pausing code superimposed on codon coding) [5,40] . The organism is likely to suffer down the road from many supposedly “neutral” mutations. Those purposeless mutations will have unforeseen deleterious effects not yet appreciated in other multidimensional semiotic codes and languages.
Failure to differentiate pointless spontaneous mutations from prescribed polymorphisms results in a grossly inflated attribution of functionality to pointless mutations. “Mutational drift” only worsens the cell line’s genomic random deterioration. A much higher beneficial rate is accredited to spontaneous purposeless mutations than they deserve. Simultaneously, appreciation of the importance of prescribed and programmed polymorphic adaptation is diminished. We blind ourselves, often for purely metaphysical reasons, to the fact of programming modules being called-up into upper memory when needed that direct polymorphisms toward rapid adaptation. But even before discussion of mutation begins, the most fundamental question of biological research is, “How were prior halting programmed computations written in an inanimate, prebiotic environment?” Mutations do not write programming. Mutations alter programming, almost invariably for the worse. Until we stop sweeping the question under the rug of programming origin, we are not going to progress very far with abiogenesis science, molecular evolution theory, or elucidation of the organism’s rapid adaptation capability.
References
- Szostak, J.W. Functional information: Molecular messages. Nature 2003, 423, 689.
- Hazen, R.M, Griffin, P.L, Carothers, J.M, Szostak, J.W. Functional information and the emergence of biocomplexity. Proc Natl Acad Sci U S A 2007, 104 Suppl 1, 8574-8581.
- Abel, D.L. The biosemiosis of prescriptive information Semiotica 2009, 2009, 1-19 (Available at www.DavidAbel.us Last accessed 10/2025), doi:10.1515/semi.2009.026.
- Abel, D.L. Prescriptive Information (PI) [Scirus SciTopic Page] Available online: www.DavidAbel.us (accessed on 10/2025).
- D'Onofrio, D.J, Abel, D.L, Johnson, D.E. Dichotomy in the definition of prescriptive information suggests both prescribed data and prescribed algorithms: biosemiotics applications in genomic systems (Last accessed 9/2024). Theor Biol Med Model 2012, 9, 8, doi:10.1186/1742-4682-9-8.
- Chatterjee, S, Yadav, S. The Coevolution of Biomolecules and Prebiotic Information Systems in the Origin of Life: A Visualization Model for Assembling the First Gene. Life (Basel) 2022, 12, doi:10.3390/life12060834.
- Higgs, P.G, Pudritz, R.E. A Thermodynamic Basis for Prebiotic Amino Acid Synthesis and the Nature of the First Genetic Code. Astrobiology 2009, 9, 483-490, doi:doi:10.1089/ast.2008.0280.
- Nelson, K.E, Levy, M, Miller, S.L. Peptide nucleic acids rather than RNA may have been the first genetic molecule. Proc Natl Acad Sci U S A 2000, 97, 3868-3871.
- Abel, D.L. What is Life? Archives of Microbiology and Immunology 2024, 8, 428-443, doi:10.26502/ami.936500189.
- Abel, D.L. The three fundamental categories of reality In The First Gene: The Birth of Programming, Messaging and Formal Control, Abel, D.L., Ed, LongView Press-Academic: Biolog. Res. Div.: New York, N.Y., 2011; pp. 19-54 (http://DavidAbel.us Last accessed in 19/2025).
- Abel, D.L. What is ProtoBioCybernetics? . In The First Gene: The Birth of Programming, Messaging and Formal Control, Abel, D.L., Ed, LongView Press-Academic: Biolog. Res. Div.: New York, N.Y., 2011; pp. 1-18 (http://DavidAbel.us Last accessed in 11/2024).
- Abel, D.L. Is life unique? Life 2011, 2, 106-134 (also available from www.DavidAbel.us Last accessed Dec, 2024), doi:DOI 10.3390/life2010106
- Abel, D.L., (Ed.) The First Gene: The Birth of Programming, Messaging and Formal Control. LongView Press-Academic: Biolog. Res. Div.: New York, NY, 2011; p. 389.
- Abel, D.L. Primordial Prescription: The Most Plaguing Problem of Life Origin Science LongView Press Academic: New York, N. Y., 2015.
- Abel, D.L. The Common Denominator of All Known Lifeforms. Journal of Bioinformatics and Systems Biology 2025, 8, 29-35 doi:10.26502/jbsb.5107099.
- Abel, D.L. Life is Programmed Computation. Journal of Bioinformatics and Systems Biology 2025, 8, 1-16, doi:10.26502/jbsb.5107097
- Stencel, A, Crespi, B. What is a genome? Molecular Ecology 2013, 22, 3437-3443, doi:10.1111/mec.12355
- Abel, D.L. The capabilities of chaos and complexity Int. J. Mol. Sci. 2009, 10, 247-291 (Also availabel at www.DavidAbel.us Last accessed 249/2024), doi:10.3390/ijms10010247.
- Rocha, L.M. Evolution with material symbol systems. Biosystems 2001, 60, 95-121.
- Abel, D.L. Linear Digital Material Symbol Systems (MSS) In The First Gene: The Birth of Programming, Messaging and Formal Control, Abel, D.L., Ed, LongView Press--Academic, Biol. Res. Div.: New York, N.Y., 2011; pp. 135-160 (http://DavidAbel.us Last accessed in 111/2024).
- Abel, D.L, Trevors, J.T. More than Metaphor: Genomes are Objective Sign Systems. In BioSemiotic Research Trends, Barbieri, M., Ed, Nova Science Publishers: New York, 2007; pp. 1-15 Downloadable from http://DavidAbel.us (Last accessed 11/2024).
- Malaterre, C. Is Life Binary or Gradual? Life 2024, 14, 564.
- Pavlinova, P, Lambert, C.N, Malaterre, C, Nghe, P. Abiogenesis through gradual evolution of autocatalysis into template-based replication. FEBS letters 2023, 597, 344-379, doi:10.1002/1873-3468.14507.
- Rosas, A, Morales, J.D. Cooperation and the Gradual Emergence of Life and Teleonomy. In Life and Evolution: Latin American Essays on the History and Philosophy of Biology, Baravalle, L., Zaterka, L., Eds, Springer International Publishing: Cham, 2020; pp. 85-101.
- Behe, M.J. Darwin's Black Box; Simon & Shuster: The Free Press: New York, 1996.
- Marks II, R.J, Behe, M.J, Dembski, W.A, Gordon, B.L, Sanford, J.C., (Eds.) Biological Information: New Perspectives. World Scientific: Cornell University Proceedings, 2013.
- Behe, M.J. Getting there first: An evolutionary rate advantage for adaptive loss-of-function mutations. In Biological Information: New Perspectives, Marks II, R.J., Behe, M.J., Dembski, W.A., Gordon, B.L., Sanford, J.C., Eds, World Scientific: Cornell Conference Proceedings, 2013; pp. 450-473.
- Behe, M.J. Experimental evolution, loss-of-function mutations, and "the first rule of adaptive evolution". Q Rev Biol 2010, 85, 419-445.
- Behe, M.J, Snoke, D.W. Simulating evolution by gene duplication of protein features that require multiple amino acid residues. Protein Science 2004, 13, 2651-2664, doi:https://doi.org/10.1110/ps.04802904.
- Meyer, S.C. Darwin's Doubt; Harper Collins: New York, NY, 2013.
- Meyer, S, Nelson, P. Can the origin of the genetic code be explained by direct DNA templating? BIO-Complexity 2011, 2011, 1-10, doi:doi:10.5048/BIO-C.2011.2.
- Meyer, S.C. Signature in the Cell; Harper Collins: New York, 2009.
- Prigogine, I. The End of Certainty; The Free Press: New York, 161-162, 1997.
- Nicolis, G, Prigogine, I. Exploring Complexity; Freeman: New York, 1989.
- Prigogine, I, Stengers, I. Order Out of Chaos; Heinemann: London, 285-287, 297-301, 1984.
- Prigogine, I. From Being to Becoming; W. H. Freeman and Co: San Francisco, 1980.
- Abel, D.L. Spontaneous Mutations vs. Prescribed Polymorphisms. Journal of Bioinformatics and Systems Biology 2025, 8, 68-82, doi:10.26502/jbsb.5107106
- Abel, D.L. “Assembly Theory” in life-origin models: A critical review. Biosystems 2025, 247, 105378, doi:10.1016/j.biosystems.2024.105378.
- Abel, D.L. Selection in molecular evolution. Stud Hist Philos Sci 2024, 107, 54-63, doi:10.1016/j.shpsa.2024.07.004.
- D'Onofrio, D.J, Abel, D.L. Redundancy of the genetic code enables translational pausing Front Genet 2014, 5, 140 (doi:10.3389/fgene.2014.00140.
- Abel, D.L. The GS (Genetic Selection) Principle. Frontiers in Bioscience 2009, 14, 2959-2969 Principle (Also availabel at www.DavidAbel.us Last accessed Nov 2024), doi:10.2741/3426.
- Abel, D.L. The ‘Cybernetic Cut’: Progressing from Description to Prescription in Systems Theory The Open Cybernetics and Systemics Journal 2008, 2, 252-262 (Also available at www.DavidAbel.us Last accessed 259/2025), doi:10.2174/1874110X00802010252.
- Abel, D.L. The Cybernetic Cut: Progressing from Description to Prescription in Systems Theory [Scirus SciTopic Page] (www.DavidAbel.us Last accessed 8/2025). Available online: www.DavidAbel.us (accessed on
- Abel, D.L. The Universal Determinism Dichotomy (UDD): Physicodynanamic Determinism (PD) vs Choice Determinism (CD). Scopus Sci Topic Paper 2005, doi:https://www.davidabel.us/papers/The%20Universal%20Determinism%20Dichotomy-UDD4.pdf.
- Trevors, J.T, Abel, D.L. Chance and necessity do not explain the origin of life Cell Biol Int 2004, 28, 729-739 (Also available at www.DavidAbel.us Last accessed 711/2024), doi:https://doi.org/10.1016/j.cellbi.2004.06.006.
- Abel, D.L. Selection in molecular evolution. Studies in History and Philosophy of Science, 2024, 107, 54-63, doi:10.1016/j.shpsa.2024.07.004.
- Abel, D.L. Why is Abiogenesis Such a Tough Nut to Crack? Archives of Microbiology and Immunology 2024, 8, 338-364, doi:10.26502/ami.936500182.
- Abel, D.L. Physicodynamic Incompleteness. Scirus Sci-Topic Paper (http://DavidAbel.us (last accessed 4/2025) 2022.
- Abel, D.L. Formalism > Physicality (F > P) Principle SciTopic Paper Available online: http://DavidAbel.us (accessed on 4/1/2025).
- Abel, D.L. Moving 'far from equilibrium' in a prebiotic environment: The role of Maxwell’s Demon in life origin In Genesis - In the Beginning: Precursors of Life, Chemical Models and Early Biological Evolution, Seckbach, J., Gordon, R., Eds.Seckbach, J., Ed, Springer: Dordrecht, 2012; pp. 219-236 (http://DavidAbel.us Last accessed in 211/2024).
- Abel, D.L. The Formalism > Physicality (F > P) Principle In The First Gene: The Birth of Programming, Messaging and Formal Control, Abel, D.L., Ed, LongView Press Academic: New York, N.Y., 2011; pp. 325-351 (http://DavidAbel.us (last accessed 311/2024).
- Abel, D.L. Moving “Far from Equilibrium” in a Prebiotic Environment: The Role of Maxwell’s Demon in Life Origin. In Genesis - In The Beginning: Precursors of Life, Chemical Models and Early Biological Evolution, Seckbach, J., Ed, Springer Netherlands: Dordrecht, 2012; pp. 219-236.
- Abel, D.L. The Genetic Selection (GS) Principle In The First Gene: The Birth of Programming, Messaging and Formal Control, Abel, D.L., Ed, LongView Press--Academic: New York, N.Y., 2011; pp. 161-188 (http://DavidAbel.us Last accessed in Nov 2024).
- Abel, D.L. Constraints vs. Controls: Progressing from description to prescription in systems theory Open Cybernetics and Systemics Journal 2010, 4, 14-27 (http://DavidAbel.us Last accessed 17/2025), doi:10.2174/1874110X01004010014
- Abel, D.L. The Cybernetic Cut and Configurable Switch (CS) Bridge In The First Gene: The Birth of Programming, Messaging and Formal Control, Abel, D.L., Ed, LongView Press--Academic, Biol. Res. Div. 55-74 New York, N.Y., 2011; pp. 55-74 (http://DavidAbel.us Last accessed in 58/2025).
- Rocha, L.M, Hordijk, W. Material representations: from the genetic code to the evolution of cellular automata. Artif Life 2005, 11, 189-214.
- Xingli, D, Gareev, I, Roumiantsev, S, Beylerli, O, Pavlov, V, Zhao, S, Wu, J. Identify Key Genes and Construct the lncRNA-miRNA-mRNA Regulatory Networks Associated with Glioblastoma by Bioinformatics Analysis. Curr Med Chem 2025, doi:10.2174/0109298673372488250530173615.
- Meng, C, Zang, A, Du, F. Prognostic value of LncRNA SH3BP5-AS1 in non-small cell lung cancer and its regulatory effect on tumor progression. Discov Oncol 2025, 16, 1126, doi:10.1007/s12672-025-02906-4.
- Ma, J.L, Bai, Y.L, Liang, Y.D, Zhang, T, You, L, Xu, D.P. Integrated analysis of lncRNA-miRNA-mRNA regulatory network revealing molecular mechanisms of immune response and ion transport in Eriocheir sinensis under alkalinity stress. Comp Biochem Physiol C Toxicol Pharmacol 2025, 297, 110262, doi:10.1016/j.cbpc.2025.110262.
- Liu, Y, Wang, Y, Cui, L, Li, R, Xiao, D. LncRNA OIP5-AS1 suppresses lung adenocarcinoma progression and modulates macrophage polarization through the miR-429/DOCK4 regulatory axis. Front Pharmacol 2025, 16, 1569644, doi:10.3389/fphar.2025.1569644.
- Liang, J, Niu, X, Wang, G, Wang, M. Regulatory Role and Mechanism of lncRNA RNF217-AS1 in the Proliferation and Migration of Esophageal Cancer Cells. Cancer Manag Res 2025, 17, 1329-1337, doi:10.2147/CMAR.S515036.
- Cao, M, Li, J, Zhang, J, Lu, W. Prognostic value of LncRNA PSMA3-AS1 in prostate cancer and its potential regulatory mechanism. Hereditas 2025, 162, 127, doi:10.1186/s41065-025-00485-6.
- Wlodarczyk, T, Lun, A, Wu, D, Shi, M, Ye, X, Menon, S, Toneyan, S, Seidel, K, Wang, L, Tan, J, et al. Epiregulon: Single-cell transcription factor activity inference to predict drug response and drivers of cell states. Nat Commun 2025, 16, 7118, doi:10.1038/s41467-025-62252-5.
- Liu, S, Heng, Y, Sun, M, Zhang, M, Zhang, F, Chen, S, Li, J, Zhou, H, Deng, X.W. The transcription factor AHL21 links phyB and PIF4 to promote photomorphogenesis in Arabidopsis. Plant Physiol 2025, doi:10.1093/plphys/kiaf341.
- Kim-Yip, R.P, Denberg, D, Faerberg, D.F, Nunley, H, Leite, I, Chalifoux, M, Joyce, B, Toettcher, J, Gu, B, Posfai, E. Live imaging endogenous transcription factor dynamics reveals mechanisms of epiblast and primitive endoderm fate segregation. Curr Biol 2025, doi:10.1016/j.cub.2025.07.031.
- Itoh, K, Ossipova, O, Sokol, S.Y. Myocardin-related transcription factor regulates actomyosin contractility and apical junction remodeling during vertebrate neural tube closure. Development 2025, doi:10.1242/dev.204681.
- de Villiers, R.M, Palfalvi, G, Kanai, A, Suzuki, Y, Hasebe, M, Ishikawa, M. STEMIN transcription factor drives selective chromatin remodeling for gene activation within a relaxed chromatin during reprogramming in the moss Physcomitrium patens. Plant J 2025, 123, e70386, doi:10.1111/tpj.70386.
- Baekgaard, C.H, Lester, E.B, Moller-Larsen, S, Lauridsen, M.F, Larsen, M.J. NanoImprint: A DNA methylation tool for clinical interpretation and diagnosis of common imprinting disorders using nanopore long-read sequencing. Ann Hum Genet 2024, 88, 392-398, doi:10.1111/ahg.12556.
- Mattei, A.L., Bailly, Nina and Meissner, Alexander. DNA methylation: a historical perspective. Trends in Genetics 2022, 38, 676-707, doi:https://doi.org/10.1016/j.tig.2022.03.010 1.
- Tang, A, Huang, Y, Li, Z, Wan, S, Mou, L, Yin, G. Analysis of a four generation family reveals the widespread sequence-dependent maintenance of allelic DNA methylation in somatic and germ cells. Sci Rep 2016, 6, doi:10.1038/srep19260.
- Subhash, S, Andersson, P.-O, Kosalai, S.T, Kanduri, C, Kanduri, M. Global DNA methylation profiling reveals new insights into epigenetically deregulated protein coding and long noncoding RNAs in CLL. Clinical epigenetics 2016, 8, 106, doi:10.1186/s13148-016-0274-6.
- Peleg, S, Feller, C, Ladurner, A.G, Imhof, A. The Metabolic Impact on Histone Acetylation and Transcription in Ageing. Trends Biochem Sci 2016, doi:10.1016/j.tibs.2016.05.008.
- Palladino, J, Mellone, B.G. The KAT's Out of the Bag: Histone Acetylation Promotes Centromere Assembly. Dev Cell 2016, 37, 389-390, doi:10.1016/j.devcel.2016.05.019.
- McBrian, M.A, al, e. Histone acetylation regulates intracellular pH. Mol Cell 2013, 49, 310-321.
- Turner, B.M. Histone acetylation and an epigenetic code. Bioessays 2000, 22, 836-845, doi:10.1002/1521-1878(200009)22:9<836::aid-bies9>3.0.co;2-x.
- D'onofrio, D.J, Abel, D.L. Redundancy of the genetic code enables translational pausing Frontiers in Genetics 2014, 5, 140 Open access doi:10.3389/fgene.2014.00140.
- Jagadeesan, R, Dash, S, Palma, C.S.D, Baptista, I.S.C, Chauhan, V, Makela, J, Ribeiro, A.S. Dynamics of bacterial operons during genome-wide stresses is influenced by premature terminations and internal promoters. Sci Adv 2025, 11, eadl3570, doi:10.1126/sciadv.adl3570.
- Kogay, R, Wolf, Y.I, Koonin, E.V. Horizontal Transfer of Bacterial Operons into Eukaryote Genomes. Genome Biol Evol 2025, 17, doi:10.1093/gbe/evaf055.
- Maniatis, T. From bacterial operons to gene therapy: 50 years of the journal Cell. Cell 2024, 187, 6417-6420, doi:10.1016/j.cell.2024.10.037.
- Chandhru, M, Ramamurthy, K, Sudhakaran, G, Choi, K.C, Valan Arasu, M, Arockiaraj, J. Aging-related epigenetic instability in lncRNAs and miRNAs mediates the development of hypertension. Eur J Pharmacol 2025, 178150, doi:10.1016/j.ejphar.2025.178150.
- Fu, X, Alkhamessi, R.N, Al-Aouadi, R.F.A, Kadham, M.J, Khasanova, S, Shree, M, Jassal, P, Sinha, A, Lakshmaiya, N, Wu, C. Inflammation-related lncRNAs in the regulation of kidney injuries; special emphasis on novel lncRNA-based delivery platforms. Naunyn Schmiedebergs Arch Pharmacol 2025, doi:10.1007/s00210-025-04516-x.
- Miao, P, Pan, C, Li, T, Li, Y. Potential role of immune-related LncRNAs in prognosis of hepatocellular carcinoma: an integrative study. Discov Oncol 2025, 16, 1661, doi:10.1007/s12672-025-03497-w.
- Wu, C, Liu, J, Li, Y, Yang, W, Wang, J, Liu, C. GreenCells: A comprehensive resource for single-cell analysis of plant lncRNAs. J Biol Chem 2025, 110678, doi:10.1016/j.jbc.2025.110678.
- Boa Ventura, P.V, Chaves, A.C, Pereira, M.S, de Carvalho, F.J.N, da Silva, B.F, Costa, R.A, Teixeira, R.S.C, Maciel, W.C, Carneiro, V.A. Exploring plant compounds as enhancers of ciprofloxacin activity against planktonic and biofilm cells of poultry-related enterobacteriaceae. Microb Pathog 2025, 208, 107995, doi:10.1016/j.micpath.2025.107995.
- Foutadakis, S, Bourika, V, Styliara, I, Koufargyris, P, Safarika, A, Karakike, E. Machine learning tools for deciphering the regulatory logic of enhancers in health and disease. Front Genet 2025, 16, 1603687, doi:10.3389/fgene.2025.1603687.
- Lai, X, Li, C, Tang, X, Luo, X, Wu, F, Liang, Y, Huang, B, Li, H. Super-enhancers in immune system regulation: mechanisms, pathological reprogramming, and therapeutic opportunities. Front Immunol 2025, 16, 1652398, doi:10.3389/fimmu.2025.1652398.
- Struhl, K. Distal enhancers loop to proximal enhancers, not to promoters. Nat Rev Mol Cell Biol 2025, doi:10.1038/s41580-025-00889-2.
- Lotthammer, J.M, Ginell, G.M, Griffith, D, Emenecker, R.J, Holehouse, A.S. Direct prediction of intrinsically disordered protein conformational properties from sequence. Nature methods 2024, 21, 465-476, doi:10.1038/s41592-023-02159-5.
- Struhl, K. Intrinsically disordered regions (IDRs): A vague and confusing concept for protein function. Molecular Cell 2024, 84, 1186-1187, doi:https://doi.org/10.1016/j.molcel.2024.02.023.
- Trivedi, R, Nagarajaram, H.A. Intrinsically Disordered Proteins: An Overview. Int J Mol Sci 2022, 23, doi:10.3390/ijms232214050.
- Li, H, Ogawa, H, Teng, D, Okame, Y, Namba, H, Honda, T. Human herpesvirus 6B U65 binds to histone proteins and suppresses interferon production. J Virol 2025, e0098425, doi:10.1128/jvi.00984-25.
- Tang, H, Zhang, K. Metabolic Tracing of Methyl Donor Utilization in Histone Methylation via Relative Quantification of Isotopomer Distribution Mass Spectrometry. ACS Chem Biol 2025, doi:10.1021/acschembio.5c00528.
- Zhang, Y, Zhang, Z, Chi, Y, Yun, H, Zhang, Q, Cao, W, Bai, H, Wei, B, Liu, H, Li, J, et al. Histone deacetylase inhibitors protect endothelial glycocalyx via suppressing hypoxia-inducible factor-1alpha-induced overexpression of matrix metalloproteinase 9 during hemorrhagic shock. J Trauma Acute Care Surg 2025, doi:10.1097/TA.0000000000004767.
- Bartuli, J, Jungwirth, S, Dixit, M, Okuda, T, Zimmermann, J.P, Erlacher, M, Pan, T, Volz, A, Huttenhofer, A, Warscheid, B, et al. tRNA as an assembly chaperone for a macromolecular transcription-processing complex. Nat Struct Mol Biol 2025, doi:10.1038/s41594-025-01653-y.
- Bhat, P.D, Nalli, A, Shoemaker, L, Kwong, J.Q, Qadota, H, Krishnamurthy, P, Benian, G.M. Age-related Decline of a Chaperone Complex in the Mouse Heart. bioRxiv 2025, doi:10.1101/2024.09.12.612759.
- Dantas, T.J, Abreu, D.M, De-Castro, M.J.G, De-Castro, A.R.G, Khobrekar, N.V, Rocha, S.A, Abreu, C.M.C. Dynein-2 requires HSP90 chaperone activity to ensure robust retrograde IFT and ciliogenesis. J Cell Sci 2025, doi:10.1242/jcs.264034.
- Urban, N.D, Gharat, K, Mattiola, Z.J, Scheutzow, A, Klaiss, A, Tabler, S, Huffaker, A.W, Grootveld, M, Skinner, M.E, Kirstein, J, et al. HSP-1-Specific Nanobodies Alter Chaperone Function in vitro and in vivo. bioRxiv 2025, doi:10.1101/2025.08.25.672099.
- Zhang, X, Young, C, Liao, X.H, Refetoff, S, Mutchler, S.M, Brodsky, J.L, Buck, T.M, Arvan, P. Thyroidal expression of ER molecular chaperone GRP170 is required for efficient TSH-mediated thyroid hormone synthesis. JCI Insight 2025, 10, doi:10.1172/jci.insight.191837.
- Tomkins, J.P. Genomic Tandem Repeats: Where Repetition is Purposely Adaptive. Acts and Facts 2025, 54, 14-17.
- Lujumba, I, Adam, Y, Ziaei Jam, H, Isewon, I, Monnakgotla, N, Li, Y, Onyido, B, Fredrick, K, Adegoke, F, Emmanuel, J, et al. A practical guide to identifying associations between tandem repeats and complex human traits using consensus genotypes from multiple tools. Nat Protoc 2025, doi:10.1038/s41596-025-01231-y.
- Homma, K, Anbo, H, Ota, M, Fukuchi, S. Longer internal exons tend to have more tandem repeats and more frequently experience insertions and deletions. Life Sci Alliance 2025, 8, doi:10.26508/lsa.202403148.
- Ershova, E.S, Umriukhin, P.E, Zinchenko, R.A, Vasilieva, T.P, Kostyuk, S.E, Shabalin, N.Y, Salimova, T.A, Malinovskaya, E.M, Veiko, N.N, Kostyuk, S.V. Variation in the Content of Three Tandem Repeats of the Human Genome (Ribosomal, Satellite III, and Telomere) in Peripheral Blood Leukocyte DNA of People of Different Ages (5-101 Years). J Aging Res 2025, 2025, 8847073, doi:10.1155/jare/8847073.
- Cho, S.T, Wright, E.S. Accurate detection of tandem repeats exposes ubiquitous reuse of biological sequences. Nucleic Acids Res 2025, 53, doi:10.1093/nar/gkaf866.
- Reinar, W.B.e.a. Length Variation in Short Tandem Repeats Affects Gene Expression in Natural Populations of Arabidopsis thaliana. Plant Cell 2021, 33, 2221–2234.
- Sawaya, S.e.a. Microsatellite tandem repeats are abundant in human promoters and are associated with regulatory elements. . PLoS ONE 2013, 8.
- Gemayel, R, Cho, J, Boeynaems, S, Verstrepen, K.J. Beyond junk-variable tandem repeats as facilitators of rapid evolution of regulatory and coding sequences. Genes (Basel) 2012, 3, 461-480, doi:10.3390/genes3030461.
- Ma, L, Jensen, J.S, Mancuso, M, Hamasuna, R, Jia, Q, McGowin, C.L, Martin, D.H. Variability of Trinucleotide Tandem Repeats in the MgPa Operon and Its Repetitive Chromosomal Elements in Mycoplasma genitalium. J Med Microbiol 2011, doi:jmm.0.030858-0 [pii] 10.1099/jmm.0.030858-0.
- Pawłowski, P.H, Zielenkiewicz, P. Determining the Identity Nucleotides and the Energy of Binding of tRNAs to Their Aminoacyl-tRNA Synthetases Using a Simple Logistic Model. Life 2024, 14, 1328.
- Mughal, F, Caetano-Anollés, G. Evolution of Intrinsic Disorder in Protein Loops. Life 2023, 13, 2055.
- Pawlowski, K, Zhang, B, Rychlewski, L, Godzik, A. The Helicobacter pylori genome: from sequence analysis to structural and functional predictions. Proteins 1999, 36, 20-30, doi:10.1002/(SICI)1097-0134(19990701)36:1<20::AID-PROT2>3.0.CO;2-X [pii].
- Uversky, V.N. A decade and a half of protein intrinsic disorder: biology still waits for physics. Protein Sci 2013, 22, 693-724, doi:10.1002/pro.2261.
- Djinovic-Carugo, K, Carugo, O. Missing strings of residues in protein crystal structures. Intrinsically Disord Proteins 2015, 3, e1095697, doi:10.1080/21690707.2015.1095697.
- Kull, K. Choosing and learning: Semiosis means choice. Sign Systems Studies 2018, 46, 452-466, doi:10.12697/SSS.2018.46.4.03.
- Kull, K. Choices by organisms: on the role of freedom in behaviour and evolution https://doi.org/10.1093/biolinnean/blac077. Biological Journal of the Linnean Society 2023, 139, 555–562.
- Kull, K. The aim of extended synthesis is to include semiosis. Theor Biol Forum 2022, 115, 119-132, doi:10.19272/202211402008.
- Di Giulio, M. The Origin of the Genetic Code: Matter of Metabolism or Physicochemical Determinism? Journal of Molecular Evolution 2013, 77, 131-133, doi:10.1007/s00239-013-9593-9.
- Kun, Á. The major evolutionary transitions and codes of life. Biosystems 2021, 210, 104548, doi:https://doi.org/10.1016/j.biosystems.2021.104548.
- Kull, K. Codes: Necessary, but not Sufficient for Meaning-Making. Constructivist Foundations 2020, 15, 137-139.
- Battail, G. Error-correcting codes and information in biology. Biosystems 2019, 184, 103987, doi:https://doi.org/10.1016/j.biosystems.2019.103987.
- Barbieri, M. A general model on the origin of biological codes. Biosystems 2019, 181, 11-19, doi:10.1016/j.biosystems.2019.04.010.
- Barbieri, M. The Organic Codes: An Introduction to Semantic Biology; Cambridge University Press: Cambridge, 2003.
- Rocha, L.M. The physics and evolution of symbols and codes: reflections on the work of Howard Pattee. Biosystems 2001, 60, 1-4.
- Rodríguez Higuera, C. Quantification and Realism: Locating Semiosis in the Description of Biological Systems. Biosemiotics 2021, 14, 1-12, doi:10.1007/s12304-021-09439-7.
- Alexander, V.N, Bacigalupi, J.A, Garcia, O.C. Living systems are smarter bots: Slime mold semiosis versus AI symbol manipulation. Biosystems 2021, 206, 104430, doi:10.1016/j.biosystems.2021.104430.
- Nomura, N, Muranaka, T, Tomita, J, Matsuno, K. Time from Semiosis: E-series Time for Living Systems. Biosemiotics 2018, 11, 65-83, doi:10.1007/s12304-018-9316-0.
- Kull, K. Choosing and learning: Semiosis means choice. Sign Systems Studies 2018, 46, 452, doi:10.12697/SSS.2018.46.4.03.
- Sharov, A, Maran, T, Tonnessen, M. Comprehending the Semiosis of Evolution. Biosemiotics 2016, 9, 1-6, doi:10.1007/s12304-016-9262-7.
- Affifi, R. The Semiosis of "Side Effects" in Genetic Interventions. Biosemiotics 2016, 9, 345-364, doi:10.1007/s12304-016-9274-3.
- Sharov, A.A, Vehkavaara, T. Protosemiosis: agency with reduced representation capacity. Biosemiotics 2015, 8, 103-123, doi:10.1007/s12304-014-9219-7.
- Kull, K. Semiosis stems from logical incompatibility in organic nature: Why biophysics does not see meaning, while biosemiotics does. Prog Biophys Mol Biol 2015, 119, 616-621, doi:10.1016/j.pbiomolbio.2015.08.002.
- Johnson, D.E. Biocybernetics and Biosemiosis. In Biological Information: New Perspectives, Marks II, R.J., Behe, M.J., Dembski, W.A., Gordon, B.L., Sanford, J.C., Eds, World Scientific: Cornell University Proceedings, 2013; pp. 402-414.
- Barbieri, M. Cosmos and History: Life is semiosis; The biosemiotic view of nature. The Journal of Natural and Social Philosophy 2008, 4, 29-51.
- Eslami-Mossallam, B, Schram, R.D, Tompitak, M, van Noort, J, Schiessel, H. Multiplexing Genetic and Nucleosome Positioning Codes: A Computational Approach. PLoS One 2016, 11, e0156905, doi:10.1371/journal.pone.0156905.
- Carter, J., Charles W. ; Wolfenden, R. tRNA acceptor stem and anticodon bases form independent codes related to protein folding. PNAS USA 2015, 112, 7489–7494, doi:10.1073/pnas.1507569112.
- Weatheritt, R.J, Babu, M.M. The Hidden Codes That Shape Protein Evolution. SCIENCE 2013, 342, 1325, doi:10.1126/science.1248425.
- Bystrykh, L.V. Generalized DNA barcode design based on Hamming codes. PLoS One 2012, 7, e36852, doi:10.1371/journal.pone.0036852.
- Ashlock, D, Houghten, S.K, Brown, J.A, Orth, J. On the synthesis of DNA error correcting codes. Biosystems 2012, 110, 1-8, doi:10.1016/j.biosystems.2012.06.005.
- Montañez, G, Marks II, R.J, Fernandez, J, Sanford, J.C. Multiple Overlapping Genetic Codes Profoundly Reduce the Probability of Beneficial Mutation. In Biological Information: New Perspectives, Marks II, R.J., Behe, M.J., Dembski, W.A., Gordon, B.L., Sanford, J.C., Eds, World Scientific: Cornell Conference Proceedings, 2011; pp. 139-167.
- Wallace, R. A scientific open season. Comment on 'A colorful origin for the genetic code: information theory, statistical mechanics and the emergence of molecular codes' by T. Tlusty. Phys Life Rev 2010, 7, 377-378; discussion 381-374, doi:10.1016/j.plrev.2010.07.007.
- Tlusty, T. A colorful origin for the genetic code: information theory, statistical mechanics and the emergence of molecular codes. Phys Life Rev 2010, 7, 362-376, doi:10.1016/j.plrev.2010.06.002.
- Takagi, H, Kaneko, K. Topological, statistical, and dynamical origins of genetic code. Comment on "A colorful origin for the genetic code: information theory, statistical mechanics and the emergence of molecular codes" by T. Tlusty. Phys Life Rev 2010, 7, 379-380; discussion 381-374, doi:S1571-0645(10)00091-6 [pii] 10.1016/j.plrev.2010.08.001.
- Wills, P.R. Informed generation: Physical origin and biological evolution of genetic codescript interpreters. Journal of Theoretical Biology 2009.
- Sternberg, R.V. DNA codes and information: formal structures and relational causes. Acta Biotheor 2008, 56, 205-232, doi:10.1007/s10441-008-9049-6.
- Jantzen, B, Danks, D. Biological Codes and Topological Causation. Philosophy of Science 2008, 75, 259-277, doi:doi:10.1086/592481.
- Escalera, S, Tax, D.M, Pujol, O, Radeva, P, Duin, R.P. Subclass problem-dependent design for error-correcting output codes. IEEE transactions on pattern analysis and machine intelligence 2008, 30, 1041-1054, doi:10.1109/TPAMI.2008.38.
- Barbieri, M., (Ed.) The Codes of Life: The Rules of Macroevolution (Biosemiotics). Springer: Dordrecht, The Netherlands, 2007.
- Wong, M.L, Christensen, M, Bartlett, S. Rethinking “Prebiotic Chemistry”. Perspectives of Earth and Space Scientists 2025, 6, e2025CN000275, doi:https://doi.org/10.1029/2025CN000275.
- Demetrius, L.A. Directionality theory and the origin of life. R. Soc. Open Sci. 2024, 11230623, doi:http://doi.org/10.1098/rsos.230623.
- Abel, D.L. The Universal Plausibility Metric (UPM) & Principle (UPP). Theoretical Biology and Medical Modelling 2009, 6, 27, doi:10.1186/1742-4682-6-27.
- Abel, D.L. The Universal Plausibility Metric and Principle In The First Gene: The Birth of Programming, Messaging and Formal Control, Abel, D.L., Ed, LongView Press--Academic: New York, N.Y., 2011; pp. 305-324 (http://DavidAbel.us (last accessed in Jan 2025)
- Abel, D.L. The Universal Plausibility Metric (UPM) & Principle (UPP) [Scirus SciTopic Page] (www.DavidAbel.us Last accessed 5/2025). Available online: http://DavidAbel.us (accessed on 5/2025).
- Abel, D.L, Trevors, J.T. Three subsets of sequence complexity and their relevance to biopolymeric information. Theoretical Biology and Medical Modeling 2005, 2, 29-45 www.DavidAbel.us (Last accessed Nov, 2023), doi:10.1186/1742-4682-2-29.
- Xie, J, Jin, X, Wei, H, Sun, S, Liu, Y. IDP-EDL: enhancing intrinsically disordered protein prediction by combining protein language model and ensemble deep learning. Briefings in bioinformatics 2025, 26, doi:10.1093/bib/bbaf182.
- Michael, K, Mohamad, J, Nasir, N, Tan, J. A Novel Fixed Dose Triple Combination Therapy (IDP-126) for Moderate to Severe Acne. Skin Therapy Lett 2025, 30, 5-7.
- Bugge, K, Sottini, A, Ivanovic, M.T, Buus, F.S, Saar, D, Fernandes, C.B, Kocher, F, Martinsen, J.H, Schuler, B, Best, R.B, et al. Role of charges in a dynamic disordered complex between an IDP and a folded domain. Nat Commun 2025, 16, 3242, doi:10.1038/s41467-025-58374-5.
- Ye, C, Zhao, Z, Lai, P, Chen, C, Jian, F, Liang, H, Guo, Q. Correction: Strategies for the detection of site-specific DNA methylation and its application, opportunities and challenges in the field of electrochemical biosensors. Anal Methods 2025, 17, 2365, doi:10.1039/d5ay90024g.
- Duckett, D, Vormittag-Nocito, E.R, Jamshidi, P, Sukhanova, M, Parker, S, Brat, D.J, Jennings, L.J, Santana-Santos, L. Accurate identification of primary site in tumors of unknown origin (TUO) using DNA methylation. NPJ Precis Oncol 2025, 9, 8, doi:10.1038/s41698-025-00805-z.
- Ding, L, Cao, S, Bai, L, He, S, He, L, Wang, Y, Wu, Y, Yu, S. Versatile fluorescence biosensors based on CRISPR/Cas12a for determination of site-specific DNA methylation from blood and tissues. Spectrochim Acta A Mol Biomol Spectrosc 2025, 329, 125520, doi:10.1016/j.saa.2024.125520.
- Zhu, M, Huang, Q, Li, H, Zhao, Y, Guo, H, Wang, T, Liu, X, Huang, Y, Hu, J, Fang, C, et al. The impact of site-specific DNA methylation in KCNJ11 promoter on type 2 diabetes. Heliyon 2024, 10, e39934, doi:10.1016/j.heliyon.2024.e39934.
- Ye, C, Zhao, Z, Lai, P, Chen, C, Jian, F, Liang, H, Guo, Q. Strategies for the detection of site-specific DNA methylation and its application, opportunities and challenges in the field of electrochemical biosensors. Anal Methods 2024, 16, 5496-5508, doi:10.1039/d4ay00779d.
- Tan, R, Bao, X, Han, L, Li, Z, Tian, N. [A two-site combined prediction model based on HOXA9 DNA methylation for early screening of risks of meningioma progression]. Nan Fang Yi Ke Da Xue Xue Bao 2024, 44, 2110-2120, doi:10.12122/j.issn.1673-4254.2024.11.07.
- Paz, N.E, Carneros, E, Pintos, B, Testillano, P.S. Targeting histone acetylation to enhance somatic embryogenesis in Quercus suber L. Tree Physiol 2025, doi:10.1093/treephys/tpaf111.
- Li, J, Fu, Y, Gu, X, Xie, Q, Liu, Z, Cao, Z, Li, L, Ren, J, Li, Y, Yang, H, et al. Aberrant histone acetylation and dysregulated synaptic plasticity in cognitive impairment induced by a high-methionine diet. Neural Regen Res 2025, doi:10.4103/NRR.NRR-D-25-00069.
- Garoffolo, G, Ferrari, S, De Martino, S, Pizzo, E, Candino, V, Curini, L, Macri, F, Kruithof, B.P.T, Mongelli, A, Grillo, M, et al. Reversion of aortic valve cells calcification by activation of Notch signalling via histone acetylation induction. Signal Transduct Target Ther 2025, 10, 311, doi:10.1038/s41392-025-02411-8.
- Kuhn, T.S. The Structure of Scientific Revolutions, 2nd 1970 ed, The University of Chicago Press: Chicago, 1970.
- Hamming, R.W. The unreasonable effectiveness of mathematics. The American Mathematical Monthly 1980, 87, 81-90.
- Wigner, E.P. The unreasonable effectiveness of mathematics in the natural sciences. Comm. Pure Appl. 1960, 13.

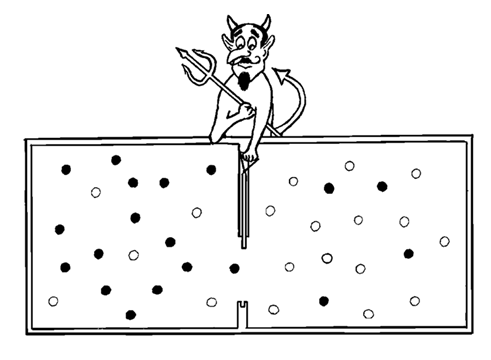

 Impact Factor: * 4.2
Impact Factor: * 4.2 Acceptance Rate: 77.66%
Acceptance Rate: 77.66%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 