Oncogenic HPV Infection Dysregulates Histone H3 Clipping
Jorge Sandoval-Basilio1*, Sofia L. Alcaraz-Estrada2, Nicolás Serafín-Higuera3, Octavio D. Reyes-Hernández4, Gabriela Figueroa-González5, Magali Blanco-Morales1, Loreli Zamudio-Rodríguez1, Jessica Sanchez-Briones6
1Laboratorio de Biología Molecular, Universidad Hipócrates, Acapulco de Juárez, Gro., México
2November 20 National Medical Center, Instituto de Seguridad y Servicios Sociales de los Trabajadores del Estado, Mexico City, Mexico
3 Laboratorio de Biología Celular, Unidad Ciencias de la Salud, Facultad de Odontologia, Universidad Autonoma de Baja California Mexicali, BC, México
4Cancer Molecular Biology Laboratory, Zaragoza Multidisciplinary Experimental Research Unit, Zaragoza Faculty of Higher Studies, National Autonomous University of Mexico, Mexico City, Mexico
5Pharmacogenetics Laboratory, Zaragoza Multidisciplinary Experimental Research Unit, Zaragoza Faculty of Higher Studies, National Autonomous University of Mexico, Mexico City, Mexico
6Istituto Mexicano del Seguro Social 29, Mexico City, Mexico
*Corresponding author: Jorge Luis Sandoval Basilio, Molecular Biology Laboratory, Tower "A" third floor, Hippocrates University, Av. Andres de Urdaneta # 360, Fractionation Ovens, Acapulco, Gro. Zip 39355, Mexico.
Received: 03 October 2022; Accepted: 10 October 2022; Published: 31 October 2022
Article Information
Citation: Jorge Sandoval-Basilio, Sofia L. Alcaraz- Estrada, Octavio D. Reyes-Hernández, Gabriela Figueroa-González, Magali Blanco-Morales, Loreli Zamudio-Rodríguez, Jessica Sanchez-Briones. Oncogenic HPV Infection Dysregulates Histone H3 Clipping. Journal of Women’s Health and Development 5 (2022): 233-236
DOI: 10.26502/fjwhd.2644-28840093
View / Download Pdf Share at FacebookAbstract
During infection by various pathogens, there is an accumulation of epigenetic alterations that lead to changes in gene expression or viral reactivation. Oncogenic Human Papillomavirus (HPV) dysregulate various epigenetic mechanisms. Impaired histone H3 clipping constitutes an epigenetic mechanism in cervical cancer. However, if the impaired H3 clipping occurred as a primary effect of the HPV infection or if is a consequence of cervical carcinogenesis due to the high number of alterations is unknown. Using human cervical samples with negative pathology to cancer, but positive and negative to oncogenic HPV, we were able to identify that H3 clipping was low in the positive oncogenic HPV cervix compared to the negative oncogenic HPV cervix. These results suggest that low H3 clipping previously observed in cervical cancer may be a primary effect of oncogenic HPV infection.
Keywords
<p>Histone 3; Epigenetics; Histone modification; Post-translational modifications; Histone H3 clipping; Oncogenic HPV</p>
Article Details
Abbreviations:
AT: Adnexal tumor; AUB: Abnormal uterine bleeding; CC: Cervical cancer; FIGO: International Federation of Gynecology and Obstetrics; H3: Histone H3; HPV: Human papillomavirus; IMC: Body mass index; PTM: Post-translational modification; UM: Uterine myomatosis.
1. Introduction
Infectious diseases are caused by pathogenic bacteria, viruses, parasites and fungi [1]. In infectious diseases, pathogens dysregulate Post-Translational Modifications (PTM) of histones. Alteration of PTM generates changes in gene expression or reactivation of latent viruses [2]. The proteolytic cut of histone H3 is an irreversible PTM that has gained relevance in the area of epigenetics [3-12]. Cleavage of H3 is carried out by cellular proteases or by virulence factors of pathogens [3-7, 11, 13-16]. Cleavage of H3 occurs at either the host histone or the pathogen histone. Thus, in the host-pathogen interaction, the dysregulation of the H3 cleavage has an effect on the expression of cellular genes or of the pathogen [13-15, 17]. The most common viral infection of the reproductive tract is caused by Human Papillomaviruses (HPV). There are about 40 types of HPV that are associated with diseases of the genitals [18]. However, oncogenic HPVs are identified in Cervical Cancer (CC) and include HPV-16, HPV-18, HPV-31, HPV-33, HPV-35, HPV-39, HPV-45, HPV-51, HPV-52, HPV-56, HPV-58, HPV-59, HPV-66, HPV-68, HPV-73, HPV-82 [19]. HPV-16 and HPV-18 and are associated with 70% of invasive cervical neoplasms [18]. Various epigenetic changes are identified during HPV infection both in the virus and in the host cell genome, these include DNA methylation, Post-Translational Modifications (PTM) of histones and noncoding RNA [20]. Commonly, the dysregulation of the epigenetic mechanisms associated with oncogenic HPV generates changes in the expression of genes that may contribute to the development of cancer [21]. Proteolytic cleavage of the amino terminal end of H3 has been documented in cancer [22, 23]. Specifically, we reported that the proteolytic cut of H3 is low in CC [22]. However, if the impaired H3 clipping occurred as a primary effect of the HPV infection or if is a consequence of cervical carcinogenesis as a result of the high number of alterations, is unknown. Therefore, we sought to investigate the H3 clipping status in the cervix infected or not infected with oncogenic HPV but negative for CC.
2. Methods
2.1. Patients
Cervical samples from 8 patients diagnosed with Abnormal Uterine Bleeding (AUB) or Uterine Myomatosis (UM) and Adnexal Tumor (AT) were obtained by hysterectomy procedures. The patients diagnosed with AUB were classified according to the International Federation of Gynecology and Obstetrics (FIGO). The information concerning FIGO classification is included in Table 1. Additionally, all cervix samples, were negative to pathological analysis to cervical cancer. Importantly 4 samples (Lane P5 - P8) were positive for oncogenic HPV by PCR (Table 1). In addition, the sample of each patient was divided into two parts; the first one for obtaining DNA and the remaining part was used to obtain total proteins extracts.
|
P1 |
P2 |
P3 |
P4 |
P5 |
P6 |
P7 |
P8 |
|
|
Age (years) |
49 |
36 |
40 |
38 |
51 |
50 |
39 |
47 |
|
IMC |
16.44 |
27.38 |
26.17 |
37.8 |
31.24 |
26.14 |
30.84 |
37.67 |
|
Marital status |
Single |
Married |
Married |
Divorced |
Married |
Divorced |
Married |
Single |
|
Place of residence |
Mexico City |
Mexico City |
Michoacan |
Mexico state |
Mexico City |
Mexico City |
Michoacan |
Mexico City |
|
Employed |
Not |
Not |
Not |
Not |
Not |
Yes |
Not |
Yes |
|
Diagnostic |
AUB L |
AUB L |
AUB L |
AUB L |
AUB L |
AUB L |
AUB L |
UM and AT |
|
Surgery |
TAH |
TAH |
TAH |
TAH |
TAH |
TVH |
TAH |
TAH |
|
HPV genotype |
NEG |
NEG |
NEG |
NEG |
31 |
31, 66 |
18, 31 |
59 |
|
IMC, body mass index; HPV, human papillomaviruses; UM, uterin myomatosis; AT, anexial tumoration; AUB L, abnormal uterine bleeding leiomyoma; TAH, total abdominal hysterectomy; TVH, total vaginal hysterectomy; NEG, negative. |
||||||||
Table 1: Features of patients with positive oncogenic HPV negative HPV and cervix.
2.2. Immunoblot analysis
Immunoblotting assays were performed using total proteins extracts from cervix samples. All samples were obtained with RSB buffer (10 mM Tris-Cl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1% NP40) supplemented with complete protease inhibitor cocktail. Immunoblotting assays were performed as described elsewhere [22]. Briefly, samples were resuspended in 2X Laemmli-buffer with b-mercaptoethanol, heated to 95ºC for 8 min, and loaded onto a 15% SDS-polyacrylamide gel. Proteins were transferred to a PVDF membrane by damp blotting at 40 mA for 3 h. The PVDF membrane was blocked with 5% milk in TBS-T (10X TBS-T: 198 mM Tris, 1400 mM NaCl, 0.01% Tween 20, pH 7.6) for 1 h. Then the PVDF membrane was incubated overnight at 4ºC with the primary anti-H3 antibody (E.960.2, Thermo Fisher Scientific) or HRP conjugated-GAPDH antibody (14C10, Cell Signaling Technology). Anti-H3 antibody was used at a 1:500 dilution and GAPDH (used as loading control) at a 1:1000 dilution. The membrane was subsequently washed three times in TBS-T followed by incubation with HRP-conjugated secondary antibody (Jackson ImmunoResearch) for 1 h at room temperature. Then, the membrane was washed three times in TBS-T. Signals were detected using by chemiluminescence C-Digit (LI-COR).
2.3. Genotyping HPV
DNA extraction from the patient’s tissue was performed following manufactures instructions (Invitrogen Thermo Fisher Scientific). After this, genotyping determination of HPV was carried out through Anyplex II HPV28 detection kit (Seegene) following the manufactures instructions. Briefly, in 20 ml PCR reaction 1x of HPV28 primer mix A (14 oncogenic HPV types: 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68) or B (5 oncogenic HPV types: 26, 53, 69, 73, 82 and 9 non oncogenic types: 6, 11, 40, 42, 43, 44, 54, 61, 70), 1x Anyplex master mix, and approximately 50 ng of DNA were added. The thermocycling conditions was as follow: an initial step at 50 °C for 4 minutes and a denaturation step at 95 °C for 15 minutes. The samples underwent 30 cycles at 95 °C for 30 seconds, 60 °C for 60 seconds and 72 °C for 30 seconds. The PCR reaction were done in a CFX96 Real Time PCR System (Bio-Rad) as recommended by the manufacturer.
3. Results and Discussion
Histone Clipping has been observed in different cell lines [22, 23] including HeLa cells and previous reports showed that H3 clipping is reduced in samples with CC [22]. However, if the impaired H3 clipping occurred as a primary effect of the HPV infection or if is a consequence of cervical carcinogenesis as result of the high number of alterations, is unknown. To address this, we analyzed the H3 clipping in samples of positive oncogenic HPV cervical tissue with negative pathology to cancer. In addition, negative-HPV samples with negative diagnosis to cancer were included as observed in Figure 1. The analyzed samples characteristics are showed in Table 1. Two forms of truncated H3 were detected principally in almost all the samples (Figure 1). However, the presence of truncated H3 forms was reduced in positive oncogenic HPV samples. These results indicate that H3 clipping is decreased in positive oncogenic HPV cervix samples as compared with negative HPV cervix samples. Thus, this data suggest that aberrant H3 clipping is inherently associated with oncogenic HPV infection. Moreover, since low H3 clipping is also observed in samples with CC, it is possible that impaired H3 clipping is preserved during all the process of cervical carcinogenesis. Whether the low H3 clipping is required for the development of CC remains to be demonstrated. Alterations in histone clipping in host cells promoted by infectious microorganisms have been described previously [13-16]. Proteins named adhesion and penetration protein (App) and Meningococcal Serine Protease A (MspA) are secreted by Neisseria meningitidis, a bacterium that can cause meningitis and/or septicemia in children and young adults [24. These proteins, involved in meningococcal pathogenesis, can bind histones of human cells and generate proteolytic clipping of H3 due to their serine endopeptidase activity [16]. H3 clipping also can be generated by the 3C protease of the foot and mouth disease virus (FDMV). The truncated H3 losses 20 amino acids from the N-terminus [14]. Oncogenic viruses can promote aberrant epigenetic changes in the host [25]. HPV oncoprotein specifically can promote PTM [26]. It is known that E6 protein of HPV can bind cellular serine protease thought the PDZ domain [27] and we recently demonstrated that serine and aspartyl proteases are implicated in the H3 clipping [22]. Even though a limitation is the number of samples, the present work is the first to suggest that low H3 clipping may not be a consequence of the gene expression alteration in CC but a primary step associate to the oncogenic HPV infection. If this event is determinant for viral virulence in the progression to CC requires further investigation but if this is the case, it could offer a potential therapeutic preventive target for the development of the disease.
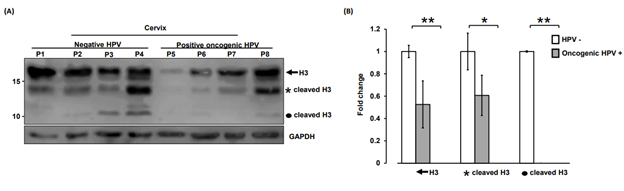
Figure 1: Oncogenic HPV infection dysregulates histone H3 clipping. (A) Immunodetection by western blot showed low levels of the two cleavage forms (asterisk and circle) of histone H3 (H3) in positive oncogenic HPV cervical tissue with negative to pathological analysis to cervical cancer (lane P5 – P8) as compared to negative-HPV cervix samples (lane P1 – P4). Complete H3 (arrow) was identified in all samples (lane P1 – P8). Note the slight decrease of complete h3 in the positive oncogenic HPV cervix samples (lane P5 - P6). GAPDH expression was analyzed in parallel as an internal control. The running position of protein markers is shown on the left. The result is representative of at least three separate experiments. (B) Densitometry profile of complete and processed H3 (asterisk and circle) in cervical tissue samples positive oncogenic HPV vs negative-HPV. The mean and variance of three separate experiments were plotted. The evaluation of the significant difference was determined by the application of Wilconxon statistical test. **P<0.01 for complete H3 (arrow); *P<0.05 for H3 processed (asterisk) and **P<0.01 for H3 processed (circle).
4. Conclusions
Samples positive to oncogenic HPV and negative to pathological analysis of cancer have a decreased in H3 clipping compared with negative HPV cervix samples.
Acknowledgements
We thank Bárcenas I. Jaime from Clínica Integral de la Mujer (Ciudad de México, México).
Funding
The support for this research was provided by Universidad Hipócrates.
Declarations of competing interests
We declared not conflict interests.
References
- HR van Doorn. The epidemiology of emerging infectious diseases and pandemics, Medicine (Abingdon) 49 (2021): 659-662.
- KL Seib, MP Jennings. Epigenetics of infectious diseases, in: Med. Epigenetics, Second Edi, Elsevier (2021): 407-424.
- Y Shin, S Kim, NB Ghate, et al. MMP-9 drives the melanomagenic transcription program through histone H3 tail proteolysis, Oncogene 41 (2022): 560-570.
- P Cheung, S Schaffert, SE Chang, et al. Repression of CTSG, ELANE and PRTN3-mediated histone H3 proteolytic cleavage promotes monocyte-to-macrophage differentiation. Nat Immunol 22 (2021): 711-722.
- JC Rice, BH Weekley, T Kanholm, et al. MMP-2 is a novel histone H3 N-terminal protease necessary for myogenic gene activation, Epigenetics and Chromatin 14 (2021): 1-12.
- K Kim, V Punj, JM Kim, et al. MMP-9 facilitates selective proteolysis of the histone H3 tail at genes necessary for proficient osteoclastogenesis. Genes Dev 30 (2016): 208-219.
- KJ Ferrari, S Amato, R Noberini, et al. Intestinal differentiation involves cleavage of histone H3 N-terminal tails by multiple proteases. Nucleic Acids Res 49 (2021): 791-804.
- E Daura, S Tegelberg, M Yoshihara, et al. Cystatin B-deficiency triggers ectopic histone H3 tail cleavage during neurogenesis. Neurobiol Dis 156 (2021): 105418.
- MAM Ali, JA Garcia-Vilas, CR Cromwell, et al. Matrix metalloproteinase-2 mediates ribosomal RNA transcription by cleaving nucleolar histones. FEBS J 288 (2021): 6736-6751.
- V Paternoster, AV Edhager, P Qvist, et al. Inactivation of the Schizophrenia-associated BRD1 gene in Brain Causes Failure-to-thrive, Seizure Susceptibility and Abnormal Histone H3 Acetylation and N-tail Clipping. Mol. Neurobiol 58 (2021): 4495-4505.
- Y Shin, NB Ghate, B Moon, et al. DNMT and HDAC inhibitors modulate MMP-9-dependent H3 N-terminal tail proteolysis and osteoclastogenesis. Epigenetics and Chromatin 12 (2019): 1-14.
- J Huckriede, F de Vries, M Hultström, et al. Histone H3 Cleavage in Severe COVID-19 ICU Patients. Front Cell Infect Microbiol 11 (2021): 1-7.
- M Tesar, O Marquardt. Foot-and-mouth disease virus protease 3C inhibits cellular transcription and mediates cleavage of histone H3. Virology 174 (1990): 364-374.
- MM Falk, PR Grigera, IE Bergmann, et al. Foot-and-mouth disease virus protease 3C induces specific proteolytic cleavage of host cell histone H3. J Virol 64 (1990): 748-756.
- AVE Capozzo, DJ Burke, JW Fox, et al. Expression of foot and mouth disease virus non-structural polypeptide 3ABC induces histone H3 cleavage in BHK21 cells. Virus Res 90 (2002): 91-99.
- AS Khairalla, SA Omer, J Mahdavi, et al. Nuclear trafficking, histone cleavage and induction of apoptosis by the meningococcal App and MspA autotransporters. Cell Microbiol 17 (2015): 1008-1020.
- AM Herrera-Solorio, SS Vembar, CR MacPherson, et al. Clipped histone H3 is integrated into nucleosomes of DNA replication genes in the human malaria parasite Plasmodium falciparum. EMBO Rep 20 (2019): 1-13.
- A Goodman. HPV testing as a screen for cervical cancer. BMJ 350 (2015): h2372–h2372.
- V Bouvard, R Baan, K Straif, et al. A review of human carcinogens-Part B: biological agents. Lancet Oncol 10 (2009): 321-322.
- MLR Da Silva, BHDR De Albuquerque, TAA De Medeiros Fernandes, et al. The role of hpv-induced epigenetic changes in cervical carcinogenesis (Review). Biomed. Reports 15 (2021): 1-20.
- C Feng, J Dong, W Chang, et al. The Progress of Methylation Regulation in Gene Expression of Cervical Cancer. Int J Genomics 2018 (2018): 1-11.
- J Sandoval-Basilio, N Serafín-Higuera, OD Reyes-Hernandez, et al. Low Proteolytic Clipping of Histone H3 in Cervical Cancer. J Cancer 7 (2016): 1856-1860.
- A Tvardovskiy, K Wrzesinski, S Sidoli, et al. Top-down and Middle-down Protein Analysis Reveals that Intact and Clipped Human Histones Differ in Post-translational Modification Patterns. Mol Cell Proteomics 14 (2015): 3142-3153.
- NG Rouphael, DS Stephens. Neisseria meningitidis: Biology, Microbiology, and Epidemiology. in Methods Mol Biol Humana Press Inc (2012): 1-20.
- J Minarovits, A Demcsák, F Banati, et al. Epigenetic dysregulation in virus-associated neoplasms. in: Adv Exp Med Biol Springer New York LLC (2016): 71–90.
- M Burley, S Roberts, JL Parish. Epigenetic regulation of human papillomavirus transcription in the productive virus life cycle. Semin Immunopathol 42 (2020): 159-171.
- B Stuqui, ALG Conceição, L Termini, et al. The differential role of HTRA1 in HPV-positive and HPV-negative cervical cell line proliferation. BMC Cancer 16 (2016): 840.


 Impact Factor: * 3.4
Impact Factor: * 3.4 Acceptance Rate: 78.89%
Acceptance Rate: 78.89%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 