Spontaneous Mutations vs. Prescribed Polymorphisms Review Paper
David Lynn Abel
ProtoBioCybernetics/Protocellular Metabolomics, The Gene Emergence Project, The Origin of Life Science Foundation, Inc USA.
*Corresponding author: David Lynn Abel, ProtoBioCybernetics/Protocellular Metabolomics, The Gene Emergence Project, The Origin of Life Science Foundation, Inc USA.
Received: 22 August 2025; Accepted: 02 September 2025; Published: 15 September 2025.
Article Information
Citation: David Lynn Abel. Spontaneous Mutations vs. Prescribed Polymorphisms Review Paper. Journal of Bioinformatics and Systems Biology. 8 (2025): 68-82.
View / Download Pdf Share at FacebookAbstract
Spontaneous, undirected, pointless “mutations,” whether random or nonrandom, must be differentiated from “prescribed polymorphisms.” Mutations are one-time unique events that occur with no goal or steering toward utility. Purposeless Mutations may be caused by nonrandom physicodynamic factors such as ionizing radiation and carcinogens. But purposeless mutations, nonrandom included, are never “directed” toward any biofunction. This is also true of all natural forces, laws, quantum events, and irreversible nonequilibrium thermodynamics. None of these have ever been observed to pursue or generate nontrivial utility. Prescribed polymorphisms, on the other hand, are both nonrandom and “directed.” Abundant empirical evidence continues to mount that polymorphic programming refinements are quite intentional. The genome not only regularly expands its phase space of polymorphic options, but the genome also actively selects and controls which polymorphisms to employ to meet abrupt and severe environmental challenges. Programming decisions must be active, not passive (not just secondary and after-the-fact of later phenotypic fitness). Programming precedes and prescribes the computation of homeostatic metabolism and the phenotypic fitness of living organisms. Without algorithmic optimization at the genomic level, natural selection would have no fittest organisms to select. Evolution is way too slow to explain so many empirically rapid adaptations. Prescribed Polymorphic Adaptation (PPA) results from efficacious cybernetic commands executed from programming modules called up into upper memory in rapid response to environmental challenges.
Keywords
<p>Nonrandom Mutations; Spontaneous mutations; Directed Polymorphisms; Rapid Adaptation; Genetic Drift; Population Genetics; Genomics; Evolution; Active Selection; Prescriptive Information (PI); Prescribed Polymorphic Adaptation (PPA)</p>
Article Details
Introduction
The capabilities of random mutations to program, process and execute computational haltings of creative new integrated circuits and sophisticated biosystems has long been called into question. The fact that many mutations are now known to be nonrandom has fostered considerable explanatory hope [1-8]. But “nonrandom” is not synonymous with “Directed toward biofunction.” Purely physicodynamic causation can produce nonrandom mutations with no more formal function than random ones. So random vs. nonrandom is not the issue. What matters is elucidating the origin of genomic prescription of biofunction, and the execution of those programming commands to the end of computational halting. Also of interest is the source of the processing machinery (the nanocomputers and extraordinary molecular machinery) needed to execute those programming commands. Both the programming and the processing equipment needed to execute those cybernetic commands would have had to have evolved at the same place and time for either to have had any selectivity. The fundamental question is how all of these halting computations got programmed using representational symbol systems, often superimposed multidimensional codings, and such highly integrated holistic circuitry. How did the symphony of life get so formally orchestrated?
Just being nonrandom alone does not help our model in the slightest. In fact, nonrandomness can preclude efficacious programming choices by severely limiting contingency and eliminating phase space possibilities of active selection. Active selection from among real options at bona fide decision nodes is the essence of programming. All known life is undeniably programmed and cybernetically processed [8-12]. What are the capabilities of the full array of the newly recognized non-random mutations? Are any nonrandom mutations actually “directed” toward new computational success—what this author calls “Prescribed Polymorphic Adaptation (PPA)”? If so, what force or law is doing the directing? Did the same force or law program the genome in the first place? To what degree are genetic variants creatively produced and purposefully controlled? No force or law can program. The program would consist of all the same “choices” (all “1’s,” or all “0’s,” by law!). Programming requires contingency and freedom of active selection from among real options.
Mutations of WHAT?
Have we ever adequately pursued the question, “Mutations of WHAT?” Exactly what is being mutated? How did Prescriptive Information [13-15] and steering mechanisms toward utility arise in an inanimate environment? The question of abiogenic origin of Prescriptive Information (PI) is not very different from the question of origin of algorithmic optimization at the already-existing genomic level. Prescriptive information (PI) is required for both. What caused PI in a prebiotic environment? PI is as abstract, conceptual, nonphysical and formal as the mathematical laws that govern physicodynamics [13-18]. In reality, purposeless mutations are almost always deleterious. This includes many of the supposedly neutral ones. Because of the prevalence of superimposed, multidimensional coding, a neutral mutation in one language can scramble other superimposed languages. The effects of this linguistic corruption in multidimensional languages may not become evident for many generations. Many supposedly neutral mutations are anything but neutral.
Purposeless mutations don’t program. They alter programming, almost always for the worse. What very few cases of seeming benefit are almost always accompanied by additional deleterious effects that outweigh the supposed benefit. Should this be surprising? Only if we are locked into a purely metaphysical Kuhnian paradigm rut that is determined to deny the obvious empirical orchestration of life’s stunning formal cybernetic engineering.
How did genomic programming in protocells and progenotes anticipate such potential first-time stressful encounters, and plan for them? How did various RNA’s and transcription factors get programmed with linguistic-like symbolizations to switch on these reserve programming modules only when needed? We fail to realize that “trial-and-error searches” are just that—active purposeful searching efforts, crude in methodology though they may be. Inanimate nature performs NO “trial-and-error searches.” Not even evolution pursues such a goal. Programming refinement at the genomic level alone increases phenotypic fitness. Even with the new more Lamarckian perspectives arising from epigenetics, increased fitness still stems back to genomic initiation and control—even if it’s only a methyl or acetyl group. Thus, the two questions of PI origin and PI refinement (algorithmic optimization) are very closely related. What force originally programmed the genome in a progenote, and what force further algorithmically optimizes that programming long before environmental selection can even be mentioned?
We should have admitted a century ago that randomness can’t program.
We imagine the capabilities of random mutations to be considerable given extensive genetic drift and the large phase space of possibilities. But neither drift nor a huge phase space of possibilities actually selects for programming fitness. Evolution depends squarely upon selection. For mutations to be heritably beneficial, they must be actively selected from the vast phase space of possibilities at the programming genomic level. Programming choices must be made long before any living organism is processed to differentially survive. This is known as the Genetic Selection (GS) Principle [19]. Fittest programming alone prescribes fittest organisms. Also involved here is the Formalism > Physicality (F > P) Principle [20,21], which posits that formal organization, particularly in the context of life origin, is not a product of physical processes. Formal prescription precedes and directs the physical interactions of life processes. Prescriptive Information, formal orchestration, engineering and genetic algorithms are distinct from and even govern the physical instantiation of biological directives.
Suppose 1% of purposeless mutations were deemed “beneficial.” Could 3.8 million species have been programmed so ingeniously by such extreme inefficiency just since earth’s cooling? Population genetics shows there have not been enough generation times even of prokaryotes since the cooling of earth [22-28]. The model of nothing but mutations plus environmental selection flunks the Universal Plausibility Principle [29-31]. What very few examples we have of beneficial mutations are usually accompanied by far worse secondary effects. Genomic commands are executed computationally as formal processes that arise only from the far side of The Cybernetic Cut [32-34]. The Cybernetic Cut is a great ravine that divides formalism from physicodynamics. The only way this seemingly infinitely deep ravine can be traversed is via the very narrow, one-way Configurable Switch Bridge [32-34] from the far formal side to the near physicodynamic side. Symbolized commands using physical symbol vehicles in a Material Symbol System can alternatively replace physical configurable switches to cross this narrow one-way bridge.
Natural selection is entirely passive, secondary, and after-the-fact of genomically programmed adaptations. The Genetic Selection (GS) Principle [19] distinguishes passive selection of organismic final fitness (undirected natural selection) from active selection for potential function (computationally halting formal selection at decision nodes, logic gates and configurable switch-settings). This dichotomy of selection type also prevails in molecular evolution [11]. Initial active selection must occur largely at the level of nucleoside selection, prior to the realization of any polynucleotide capabilities, let alone holistic integrated biofunction. Many other types of logic gates are employed in three-dimensional genomic programming and control (alternate RNA splicing’s, programmed polymorphisms called up into upper memory from programming modules held in reserve, micro and mini satellites, methylation’s of certain DNA sites, specific acetylations of histones, lncRNA regulation, tandem repeat variability, unique transcription factor binding sites, etc. referenced below). What exactly is computing all of the biofunctional programming success prior to phenotypic realization of fitness? Natural selection is way down the causal line, at the very end.
Sequence variants
“Sequence variant” is generally considered to be a more inclusive term than “polymorphism” (as recommended by the Human Genome Variation Society). In this paper, however, we are trying to distinguish between pointless mutations vs genomically prescribed functional sequence variations. This author uses the term "Prescribed Polymorphic Adaptation (PPA)” to emphasize which sequence variants have been empirically shown to have been purposefully biofunctional. These specifically Prescribed Polymorphisms are genomically executable commands capable of producing far more rapid adaptation than evolution could ever account for. They are usually supplemented by epigenetic regulation. Programming and computation are formal, not physical. Active choices have to be made genomically prior to the realization of phenotypic benefit. What optimized life’s algorithms at the programming level?
The “100,000 Genomes Project Consortium” found that a typical human genome contains three to five million single nucleotide polymorphism (SNP) variants [35]. Wang et al [36] also found 3.1 million unique, non-redundant high-quality sequence variants in chickens, primarily single nucleotide polymorphisms (SNPs). In humans, 99.9% are single nucleotide variants (SNVs) and small indels (short sequences of nucleotides that are inserted or deleted). The consortium also found an average of 2,100 to 2,500 structural variants. The structural variants typically affect 20 million bases [37]. Only 0.1– 0.2% of human DNA differs [37]. Most of the 3-5 million variants involved are inherited. But each of us typically has 70-80 new variants of our own. One small variant can have dramatic effect on our wellbeing. 3% of our DNA consists of short tandem repeats of varying length. This variation can be caused by purposeless mutation (typically resulting in “nucleotide repeat expansion disorders” [very often resulting in neurodegenerative disorders]) [38-47], or by programmed polymorphisms prescribed for very useful adaptive purposes [48-62]. Larger structural variants include deletions and duplications of DNA greater than one kilobase. These are known as “copy number variants” (CNV’s) [63-69]. Large rearrangements and insertions, inversions, translocations can also occur. Epigenetic variations such as DNA methylation and chromatic remodeling from histone protein acetylations do not affect the DNA sequence itself. They are configurable switch settings that help regulate gene function [70-77]. They can cause imprinting disorders, however [78-81]. Variants can occur during embryonic development or from aging, resulting in “mosaicism” (genetically different populations of cells within the same individual) [39,82-84].
Nonrandom Mutations
Numerous papers have provided evidence of nonrandom mutations [1,85-95]. When mutations are shown statistically to be nonrandom, is this the result of purely physicodynamic causation, or are nonrandom mutations somehow actively directed toward algorithmic optimization? If directed, what force or law would be doing the directing toward utility? Physicodynamics has no perception of or appreciation of utility. The laws of motion do not pursue, let alone achieve, computational halting. Pointless mutations have no goal. Physicodynamics has no perception or interest in function. Whether random or nonrandom, programming in the absence of goal “Does not compute!” “Nonrandom” mutation is not synonymous with “directed toward successful adaptation.” Nonrandom mutations can be caused by physicodynamic factors that have nothing to do with biofunction or organismal fitness. How are purposeless mutations able to optimize programming?
Multiple papers have investigated the likelihood of mutations being deleterious [96-112]. Pre-assumed is the dogma, “All adaptive alleles in existence today began as mutations" [96]. Mutation is blindly believed to be the source of Prescriptive Information and efficacious programming commands. The problem with most of these papers is failure to delineate between purposeless mutations vs. prescribed polymorphisms. That failure leads to completely bogus measurement of the percentage of beneficial pointless mutations. We were forced into supporting Physicalism by a beginning purely metaphysical presuppositional imperative. We were enslaved by the starting axiomatic dogma of science that “Mass and energy alone are sufficient. ” Abundant empirical evidence to the contrary is piling up in the biological literature by the month [113-123]. Chance and Necessity are clearly not sufficient [124]. Virtually every activity in any cell is prescribed by formal executable commands leading to formal computational halting. Mass and energy interactions have no mind for “computational success.” Mass/energy interactions cannot even recognize “usefulness” of any kind.
Because of our purely metaphysical presuppositions, we are reluctant to even acknowledge the possibility of intent. But rapidly growing empirical evidence in the fields of genomics and molecular biology proves the reality of genomic intent. Chance and Necessity cannot explain the source of Prescriptive Information and its cybernetic executable commands. This is the essence of genomics. Any cell manifests the intent to keep achieving homeostatic metabolism and to stay alive. The cell is constantly having to outsmart environmental challenges with insightful polymorphic editing. But our protracted denial of the nonphysical, conceptual, formal nature of Prescriptive Information (PI) and its source in nature force us to resort to a crude statistical differentiation between pointless mutations and prescribed polymorphisms. When the frequency of proven beneficial effect rises above 1%, the likelihood of pointless mutation nose-dives. We know fully well that the number of supposedly beneficial pointless mutations is miniscule. We also know that most helpful polymorphisms must be somehow prescribed. But we dare not admit it. We view bench science as superior to philosophy. But then we let pure metaphysics violate Einstein’s minimum-metaphysic principle of science.
When we talk about mutations, we are talking about alterations in programing strings. We are not talking about phenotypes. Programming consists of a recorded string of active selections at the genomic level. Formal choices at decision nodes must be made that function as executable commands. Strings of choices at logic-gate decision nodes must successfully compute. Configurable switch-settings must also be efficacious. Fittest organisms are the organisms with the fittest programming. Halting computations must be prescribed and cybernetically processed before phenotypic fitness can be secondarily selected. No programming and processing—No organism. Fittest programming alone prescribes fittest organisms. Even when pointless mutations seem to have some element of accidental benefit, other adverse effects usually accompany those pointless mutations which are far worse than the benefit. When one studies the defense of the macroevolutionary model, the list of beneficial mutations provided is embarrassingly short, not to mention glaringly contrived. A classic example is the point mutation producing sickle cell anemia. The claimed benefit of this purposeless mutation is malaria resistance. Ask any sickle-cell anemia sufferer if they view their mutation as being “beneficial.” They would far rather have to take anti-malarial drugs like everyone else than to spend a greatly shortened lifetime of ache in hypoxic lactic-acid-build-up agony. This is the number-one example we are given of a “beneficial” purposeless mutation.
Differentiating purposeless mutations from efficacious polymorphisms
Pointless, one-time, unique mutations must never be confused with “prescribed polymorphisms.” This is true whether those pointless mutations are random or nonrandom. “Nonrandom” is not synonymous with “Directed toward biofunction.” The empirical evidence of directed polymorphisms literally doubles each year in the literature. The percentage of “pointless mutations” that are considered to be beneficial simultaneously shrinks as we realize that those “beneficial” pointless mutations were not really spontaneous, purposeless mutations at all, but prescribed polymorphisms. They were executable commands motivated by Prescribed Polymorphic Adaptation (PPA). Statistical measures of the percentage of beneficial purposeless “mutations” is hopelessly skewed by a failure to dichotomize “pointless mutations” from “prescribed polymorphisms.”
Active selections at bona fide decision nodes were necessary to produce computational haltings that were only later realized to produce phenotypic benefit [11]. The programming of Prescriptive Information had to precede phenotypic reality. Reprogramming is required to optimize fitness before natural selection can favor it. For mutations to have programmed all genomes, mutations not only had to be nonrandom, they had to be directed toward biofunction and adaptation from the start. Purposeless mutations and prescribed polymorphisms both introduce new alleles. Both result in the change of nucleotide sequencing. Base substitutions, insertions, deletions, inversions and translocations can all be observed in both pointless mutations and prescribed polymorphisms. It is not always immediately apparent which of the two it is. Yet the difference is absolute. The genomic alteration was either intended and prescribed, or it was not. Practically all beneficial “polymorphisms” are not “purposeless mutations.” Efficacious polymorphisms are genomically prescribed. The genome can actually improve itself with better programming choices. It resembles an AI system of “learning” from previous environmental challenges. This is exactly how virtually all really rapid adaptation is achieved. Evolution would be way too slow to account for bacteria developing resistance to a new antibiotic in only 48 hours, even with prokaryotic generation times of 20 minutes.
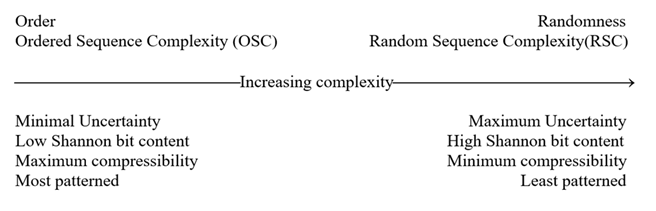
Figure 1: The inverse relationship between order and complexity as demonstrated on a linear vector progression from high order toward greater complexity. Neither Order nor Complexity themselves, or any combination of the two, can produce nontrivial function. To ascertain where on this vector “usefulness”/utility/function/algorithmic optimization could be found requires adding the additional dimension of Choice Causation. Physicodynamic Causation is blind to, and cannot produce nontrivial functionality. For greater understanding of the Universal Determinism Dichotomy (UDD), see Abel, D.L.; Trevors, J.T. Three subsets of sequence complexity and their relevance to biopolymeric information. Theoretical Biology and Medical Modeling 2005, 2, 29-45 www.DavidAbel.us (Last accessed 9/2025) or doi:10.1186/1742-4682-2-29.
This manuscript started out with the title “The capabilities of mutations.” What should be pursued is “the capabilities of Genomically Prescribed Polymorphisms.” Big difference! The capabilities of spontaneous purposeless mutations, not surprisingly, is negligible. Functional polymorphisms are not just random genetic drift that happens to be secondarily beneficial phenotypically. Suppose nonrandom mutations were militated by laws or constraints as their source of nonrandomness. “Necessity” would kill the vast contingency potential of genetic drift. Necessity would eliminate most of the phase space of possibilities. Laws are compression algorithms. Laws describe hidden order. Order shrinks possibility complexity (Figure 1). The opportunity of purposeless mutations to contribute to evolution would be severely compromised by laws or constraints on possibility space. The best of conceptual complexity requires a large possibility space from which to make active selections. We appeal to genetic drift as the source of a large possibility space. This is exactly what Necessity (law) would have eliminated. The usefulness of law-militated nonrandom mutations would be greatly reduced. So, we need to understand that from an evolutionary viewpoint, nonrandom mutations are no friend. They are the enemy of the evolution model because law causation would limit genetic drift. We must dichotomize “purposeless mutations,” whether random or nonrandom, from all of the other forms of “polymorphisms.” The only way nonrandom mutations would be regularly beneficial would be if their nonrandomness was directed toward biofunction. This would require Choice Causation at the genomic programming decision-node level. Chance or Necessity knows nothing of issuing executable commands. Only the Prescriptive Information (PI) of genomes could cause such steering toward usefulness.
The Three fundamental categories of reality
Chance or Necessity is a false dichotomy. There are three fundamental ontological categories, not two: Chance, Necessity and Choice [125]. Physicodynamic Causation must be distinguished from Choice Causation, even within the natural sciences. The Universal Determinism Dichotomy (UDD) [125,126] discounts “chance” as a real cause of any effect. Chance is merely a descriptive term. The science of Biology demands acknowledgement of the UDD. There is no escaping it. Homeostatic metabolism and life are formally orchestrated at bona fide “decision nodes,” not mere “bifurcation points.” Life is not only instructed, it is steered and directed by executable commands that are then processed by nanocomputers and very sophisticated molecular machines. Mere genetic drift and large phase spaces alone do not constitute active selection for utility. Quantum Mechanics and irreversible nonequilibrium thermodynamics manifest zero preference for function. We are not talking about passive, secondary, natural selection of the fittest organisms. That is far down the road. We are talking about the superior programming and computation that produces fittest phenotypic organisms.
Any programmer will affirm that programming refinement is not achieved by pointless “fork-in-the-road coin flips.” Real active selections must be made at bona fide “decision nodes” from among real options. This is the essence of “programming.” Efficacious commands must be issued that arise from Choice Causation alone, not Physicodynamic Causation. Disallow Choice Causation, and we disallow life. Everything about life depends upon efficacious choices at bona fide decisions nodes. Programming is impossible without purposeful choices in pursuit of computational success. Mutations are “bugs” that corrupt programming. They are not a succession of ingenious programming choices that we observe in any genome. These recorded command choices produce Prescribed Polymorphisms that alone produce rapid adaptation. The religion of blind belief in “purposeless mutations’ all-sufficiency in programming genomes” is more childish and archaic than Ptolemaic astronomy. Purposeless accidents do not construct Sydney Opera Houses. Typographical errors do not write PhD theses. Raw mass/energy interactions did not produce our cell phones. Chance and Necessity can explain none of these formal achievements. Yet they pale in comparison to the orchestration of homeostatic metabolism of any prokaryote. Nothing is more formal and choice-instructed than life itself. Life arises only out of efficacious executed commands. And those commands can be executed only by machinery that itself had to be manufactured with efficacious executed commands. Physicodynamically-caused mutations can have just as random effects as random mutations. The alteration lacks purpose. Programming “bugs” corrupt successful computations; they do not produce them. This is the reason why pointless mutations are almost never beneficial. Evolution has no goal. As long as we insist on limiting our thought to macroevolution, we will never be able to elucidate the mechanism of rapid adaptation though Genomically Prescribed Polymorphisms (GPP). Prescribed polymorphisms can arise only from preprogrammed genomic intent and goal. These executable commands are called up into upper memory from pre-programmed modules triggered by environmental challenges. Configurable switches are turned on by environmental challenges.
The mountain of empirical evidence for “Genomically Prescribed Polymorphisms’
First, the distribution of polymorphisms of any kind is statistically less common specifically in critical exons [1,56,60,127]. It’s as though nature knows better than to “mess” with “what already seems to work best.” Why would this be the case if all polymorphisms were pointless and unregulated? If mutations are unrelated to utility, the mutation rate should be constant across the board. Helpful and nonhelpful mutations should occur equally in all exons if they are 1) the result of random typographical copying errors or 2) the nonrandom result of physicodynamic causation that is blind to “usefulness.” Mere mass/energy interactions are oblivious to formal function. Nonrandom purely physicodynamic causation could care less what might be helpful. Our traditional concept of mutation has no causative “good vs. bad” connotation. So, the question remains, “Why is the distribution of mutations statistically less common specifically in critical exons [1,56,60,127]?
Says Monroe et al [1]
“Mutations occur less often in functionally constrained regions of the genome—mutation frequency is reduced by half inside gene bodies and by two-thirds in essential genes.”
“Epigenomic and physical features explain over 90% of variance in the genome-wide pattern of mutation bias surrounding genes.
“Observed mutation frequencies around genes in turn accurately predict patterns of genetic polymorphisms in natural Arabidopsis accessions (r = 0.96).”
“Genes subject to stronger purifying selection have a lower mutation rate.”
This suggests a causal link between sequence alterations and biological pragmatism. The reduced rate of polymorphisms is clearly not pointless. It is being directed from the perspective of utility. In critical exons, the genome is commanding itself, “Don’t alter proven efficacious programming with purposeless mutations.” The genome then obeys its own command. “Don’t mess with a good thing!” There is purpose in not mutating.
Prescribed Polymorphic Adaptation (PPA) is the real key to life’s adaptation success in rapidly changing environments. Prerecorded modules are called up into upper memory as needed to meet sudden challenges. Genomically prescribed adaptive polymorphisms generate more fit organisms for the environment to then only secondarily favor. According to Albert [61], most common polymorphisms are potential regulatory polymorphisms located in noncoding regions, including promoter/upstream and downstream intron regions, that may affect transcription [128]. Polymorphisms located within introns and untranslated regions transcribed as RNA affect transcription, RNA splicing, stability or translation [129]. Such polymorphisms can also be located in intergenic regions of unknown function [62]. Albert proposes four classes of genetic polymorphisms [61]. Notice that functional polymorphisms are freely acknowledged. That many polymorphisms are functional rather than purposeless is empirically based and well-observed:
- • Class 0: Function not determined. Either (A) no function is known, or (B) theoretical function is predicted but has not been experimentally demonstrated.
- • Class 1: Functional in vitro. The functional effect of the polymorphism on a target DNA element or regulatory mechanism has been demonstrated using in vitro assays (e.g., gel shift, reporter assay, ligand binding); however, the function of the polymorphism on endogenous gene expression or in vivo is unknown.
- • Class 2: Functional in vivo. In addition to class 1 requirements, (A) function effect of the polymorphism on the endogenous gene has been tested in model cellular systems (e.g., human transformed cell lines, human B lymph-oblasts, primary cell cultures) using methods such as relative allelic expression and chromatin immunoprecipitation, and (B) in vitro function is correlated with a functional change in human tissue.
- • Class 3: Functional phenocopy. In addition to class 1 requirements, (A) function has been demonstrated in vivo using model organisms such as knockin mice, and (B) function is correlated with a functional change in human tissue.
Other factors such as specific transcription factors and lncRNA regulation are involved in determining which polymorphisms are functional. But even epigenetic configurable-switch-settings are genomically controlled. These controls do not arise from “purposeless mutations.” Such sophisticated functionality does not just spontaneously generate. It is genomically prescribed. All sorts of factors such as micro and mini satellites can be involved in adaptation. Alternate splicing, transcription factor binding at specific DNA sites, and the specific unit and number variations of tandem repeats are no accidents. Purposeless mutations introduce only constraints describable with probability bounds. But prescribed polymorphisms and other regulatory factors define formal controls. Much of the evidence of prescribed polymorphic functionality comes in the form of research dealing with the loss of that functionality [48-57,130,131]. The loss of functionality is often caused by purposeless mutations. But research emphasizes and reinforces the existence of prescriptive commands in the wild type that produce very sophisticated benefits prior to their mutated loss. What exactly generated those executable commands in the first place?
Programming modules are called up into upper memory when needed.
“Prescribed Polymorphic Adaptation (PPA) uses prior programming modules held in reserve and called up real-time to meet previously-anticipated changing environmental challenges. “That’s impossible,” we say. “Evolution cannot anticipate need.” And right we are. The problem is, that is exactly what genomic programming does, as empirically demonstrated in large numbers of papers summarized by geneticist Jeffrey P. Tomkins [76,132-137]. What is negated is our purely metaphysical starting axiom that disallows any formalisms in nature. Not only is life filled with formalisms at every turn, but even the laws of physics are abstract, conceptual mathematical formalisms. Robert Endres expands on Wigner’s [17] and Hamming’s [16] “unreasonable effectiveness of mathematics in the natural sciences” with his “unreasonable likelihood of being: origin of life, terraforming, and AI” [18]. There is nothing unreasonable at all with the formalisms of reality. The only thing unreasonable is their exclusion form scientific investigation because of bankrupt naturalistic philosophy. Directed polymorphisms such as tandem repeat unit and number variation, micro and mini satellites, highly tailored transcription factor binding sites that control the DNA that produced them, programmable epigenetic configurable switch-settings, promotors and widely dispersed enhancers are all used by these reserve programming modules when called up into upper memory in life’s nanocomputers and mind-boggling molecular machinery. Down the line of genomic controls from DNA, lncRNA alternate splicing and its regulatory function become central [138-141]. Life becomes about RNA and riboprotein machinery executing cybernetic choice commands:
- highly coordinated alternate splicing [142-148].
- changing the number of tandem repeats to provide immediate adaptation [133,149-155].
- formal organization, structure, development, growth, reproduction, and integration of hundreds of disparate subcellular formal functions [156-159].
“Beneficial pointless mutations” are almost always Prescribed Polymorphisms
The paucity of beneficial purposeless mutations is severe [160]. Even in those extremely rare instances of supposed beneficial purposeless mutations, how was it determined that they were not actually prescribed polymorphisms? If accompanied by even more deleterious effects than the supposed benefit, we can be reasonably sure that the polymorphism really was a “purposeless mutation.” It is not likely to have been commanded by Prescribed Polymorphic Adaptation (PPA). But when adaptive new function is produced by a polymorphism and its accompanying regulation is obviously ingenious, genomic programming is highly likely. Failure to recognize this severely skews the percentage of purposeless mutations given credit for being beneficial.
Conclusions
Prescribed Polymorphic Adaptation (PPA) is the only reason bacteria can develop resistance to new antibiotics in way too few generation times for such adaptation to be attributable to evolution. The Prescriptive Information was already there waiting to be called up when needed to adapt. In these cases, mutation as we have always conceived of mutation is not applicable. The alterations in nucleic acid sequences were ultimately prescribed in the same fashion as the genome was programmed in the first place, prior to any mutations. Active selections at bona fide decision nodes alone account for rapid Prescribed Polymorphic Adaptation (PPA). Such polymorphisms are directed, not pointless mutations. As with many other undeniable formal controls of subcellular metabolism attest, a third fundamental category of reality exists in addition to Chance and Necessity: Choice Causation, not just Physicodynamic Causation. No Choice Causation—No programming or cybernetic processing by sophisticated molecular machines—No life. “Chance and Necessity” is a grossly inadequate view of scientifically observable, searchable and discoverable reality. We just presuppose genomic programming and its regulation without any justification from naturalistic presuppositions. We just take initial Prescriptive Information (PI) [13,15] for granted, as though it “spontaneously generated” out of thin air. Worse yet, we try to attribute it to nothing but typographical errors. We only want to talk about PI’s alteration, never about its origin. No question is more germane to abiogenesis research than PI’s origin [161]. The fittest phenotypes are the fittest cybernetically executed programs. The fittest phenotypes are integrated symphonies of halted computations. Computations are formal, not physical. Programming choices are formal. They have to be made prior to the realization of any phenotypic function. Not even evolution selects for function. Evolution selects only for fittest already-programmed, already-cybernetically processed, already-living organisms.
How did inanimate nature assign meaning to symbols that represent coded instructions? Biosemiotic coding is vast. More than 200 biological codes have now been discovered [162]. Coding is fundamental to most of life’s processes. Life is inherently semiotic [163-165]. Life uses and interprets signs and symbols extensively. Barbieri’s Code Biology uses standard scientific methods to treat life as a form of information processing [166]. The vast findings of medical genomics are fast becoming a total embarrassment to old-time disingenuous evolutionary biologist arguments. No serious geneticist or molecular biologist can any longer sustain the gospel of pointless mutations. This is why interest has been so great in the reality of nonrandom mutations. But “nonrandom” is not synonymous with “directed.” What we empirically observe in every area of the cell is formal orchestration of a symphony. We observe control, not mere constraint. We observe highly integrated circuitry, metabolic intent and goal. If directed polymorphisms rather than pointless mutations are finally to be acknowledged, the question only becomes “What directs those efficacious controlled polymorphisms?” The religion of physicalism must be retired if genomics and molecular biology are to be included in scientific endeavor. A new naturalism is in order that includes nature’s formalisms.
References
- Monroe, J.G.S., T.; Carbonell-Bejerano, P.; Becker, C.; Lensink, M.; Exposito-Alonso, M.; Klein, M.; Hildebrandt, J.; Neumann, M.; Kliebenstein, D.; et al. . Mutation bias reflects natural selection in Arabidopsis thaliana. . Nature 2022, 602, 101-105, , doi:10.1038/s41586-021-04269-6.
- Yao, X.F.; Liu, Y.; Li, Z.; Jiang, G.Q.; Wang, W.; Lu, H.; Li, H.; Lu, Z.; Liu, C.M. Non-Random Distribution of EMS-Induced Mutations Reveals Preference for Open Chromatin and Expressed Genes in Rice. Adv Sci (Weinh) 2025, e10034, doi:10.1002/advs.202510034.
- Retnakumar, R.J.; Chettri, P.; Lamtha, S.C.; Sivakumar, K.C.; Dutta, P.; Sen, P.; Biswas, S.; Agarwal, N.; Nath, A.N.; Devi, T.B.; et al. Genome-wide accumulations of non-random adaptive point mutations drive westward evolution of Helicobacter pylori. BMC microbiology 2025, 25, 229, doi:10.1186/s12866-025-03944-2.
- Pessino, S.; Nestares, G.; Bianchi, M.B.; Katzaroff, I.; Amato, L.; Bocchini, M.; Marconi, G.; Albertini, E.; Ochogavia, A.C. Diploid aposporous sunflower forms triploid BIII progeny displaying increased apospory levels and non-random genetic mutations. Sci Rep 2025, 15, 4808, doi:10.1038/s41598-025-89105-x.
- Matsushita, T.; Kano-Sueoka, T. Non-random Codon Usage of Synonymous and Non-synonymous Mutations in the Human HLA-A Gene. J Mol Evol 2023, 91, 169-191, doi:10.1007/s00239-023-10093-5.
- Levine, A.J. Non-Random Selection of Cancer-Causing Mutations in Tissue-Specific Stem Cells Cause Cancer. J Clin Oncol Res 2020, 8.
- Alade, A.A.; Buxo-Martinez, C.J.; Mossey, P.A.; Gowans, L.J.J.; Eshete, M.A.; Adeyemo, W.L.; Naicker, T.; Awotoye, W.A.; Adeleke, C.; Busch, T.; et al. Non-random distribution of deleterious mutations in the DNA and protein-binding domains of IRF6 are associated with Van Der Woude syndrome. Mol Genet Genomic Med 2020, 8, e1355, doi:10.1002/mgg3.1355.
- Abel, D.L. The Common Denominator of All Known Lifeforms. Journal of Bioinformatics and Systems Biology 2025, 8, 29-35 doi:10.26502/jbsb.5107099.
- Abel, D.L. Life is Programmed Computation. Journal of Bioinformatics and Systems Biology 2025, 8, 1-16, doi:10.26502/jbsb.5107097
- Abel, D.L. “Assembly Theory” in life-origin models: A critical review. Biosystems 2025, 247, 105378, doi:10.1016/j.biosystems.2024.105378.
- Abel, D.L. Selection in molecular evolution. Stud Hist Philos Sci 2024, 107, 54-63, doi:10.1016/j.shpsa.2024.07.004.
- Abel, D.L. What is Life? Archives of Microbiology and Immunology 2024, 8, 428-443, doi:10.26502/ami.936500189.
- Abel, D.L. The biosemiosis of prescriptive information Semiotica 2009, 2009, 1-19 (Available at www.DavidAbel.us Last accessed 18/2025), doi:10.1515/semi.2009.026.
- D'Onofrio, D.J.; Abel, D.L.; Johnson, D.E. Dichotomy in the definition of prescriptive information suggests both prescribed data and prescribed algorithms: biosemiotics applications in genomic systems (Last accessed 9/2024). Theor Biol Med Model 2012, 9, 8, doi:10.1186/1742-4682-9-8.
- Abel, D.L. Prescriptive Information (PI) [Scirus SciTopic Page] (www.DavidAbel.us Last accessed 8/2025). Available online: www.DavidAbel.us (accessed on
- Hamming, R.W. The unreasonable effectiveness of mathematics. The American Mathematical Monthly 1980, 87, 81-90.
- Wigner, E.P. The unreasonable effectiveness of mathematics in the natural sciences. Comm. Pure Appl. 1960, 13.
- Endres, R.G. The unreasonable likelihood of being: origin of life, terraforming, and AI arXiv:2507.18545 2025, v1 [q-bio.PE] doi:https://arxiv.org/abs/2507.18545.
- Abel, D.L. The Genetic Selection (GS) Principle In The First Gene: The Birth of Programming, Messaging and Formal Control, Abel, D.L., Ed.; LongView Press--Academic: New York, N.Y., 2011; pp. 161-188 (http://DavidAbel.us Last accessed in Nov 2024).
- Abel, D.L. The Formalism > Physicality (F > P) Principle In The First Gene: The Birth of Programming, Messaging and Formal Control, Abel, D.L., Ed.; LongView Press Academic: New York, N.Y., 2011; pp. 325-351 (http://DavidAbel.us (last accessed 311/2024).
- Abel, D.L. Formalism > Physicality (F > P) Principle SciTopic Paper Available online: http://DavidAbel.us (accessed on 4/1/2025).
- Sanford, J.C. Genetic Entropy; Publisher: FMS Publications: 2014.
- Sanford, J.C. Biological Information and Genetic Theory. In Biological Information: New Perspectives, Marks II, R.J., Behe, M.J., Dembski, W.A., Gordon, B.L., Sanford, J.C., Eds.; World Scientific Cornell Conference Proceedings, 2013; pp. 203-209.
- Nelson, C.W.; Sanford, J.C. Computational evolution experiments reveal a net loss of genetic information despite selection. In Biological Information: New Perspectives, Marks II, R.J., Behe, M.J., Dembski, W.A., Gordon, B.L., Sanford, J.C., Eds.; World Scientific Cornell University Proceedings, 2013; pp. 338-368.
- Marks II, R.J.; Behe, M.J.; Dembski, W.A.; Gordon, B.L.; Sanford, J.C., (Eds.) Biological Information: New Perspectives. World Scientific: Cornell University Proceedings, 2013.
- Sanford, J.C.; Baumgardner, J.R.; Brewer, W.H. Selection Threshold Severely Constrains Capture of Beneficial Mutations. In Biological Information — New Perspectives, Marks II, R.J., Behe, M.J., Dembski, W.A., Gordon, B.L., Sanford, J.C., Eds.; World Scientific: Cornell University Proceedings, 2012.
- Montañez, G.; Marks II, R.J.; Fernandez, J.; Sanford, J.C. Multiple Overlapping Genetic Codes Profoundly Reduce the Probability of Beneficial Mutation. In Biological Information: New Perspectives, Marks II, R.J., Behe, M.J., Dembski, W.A., Gordon, B.L., Sanford, J.C., Eds.; World Scientific: Cornell Conference Proceedings, 2011; pp. 139-167.
- Loewe, L.; Hill, W.G. The population genetics of mutations: good, bad and indifferent. Philos Trans R Soc Lond B Biol Sci 2010, 365, 1153-1167, doi:10.1098/rstb.2009.0317.
- Abel, D.L. The Universal Plausibility Metric and Principle In The First Gene: The Birth of Programming, Messaging and Formal Control, Abel, D.L., Ed.; LongView Press--Academic: New York, N.Y., 2011; pp. 305-324 (http://DavidAbel.us (last accessed in Jan 2025)
- Abel, D.L. The Universal Plausibility Metric (UPM) & Principle (UPP) [Scirus SciTopic Page] (www.DavidAbel.us Last accessed 5/2025). Available online: http://DavidAbel.us (accessed on 5/2025).
- Abel, D.L. The Universal Plausibility Metric (UPM) & Principle (UPP). Theoretical Biology and Medical Modelling 2009, 6, 27, doi:10.1186/1742-4682-6-27.
- Abel, D.L. The ‘Cybernetic Cut’: Progressing from Description to Prescription in Systems Theory The Open Cybernetics and Systemics Journal 2008, 2, 252-262 (Also available at www.DavidAbel.us Last accessed 259/2025), doi:10.2174/1874110X00802010252.
- Abel, D.L. The Cybernetic Cut and Configurable Switch (CS) Bridge In The First Gene: The Birth of Programming, Messaging and Formal Control, Abel, D.L., Ed.; LongView Press--Academic, Biol. Res. Div. 55-74 New York, N.Y., 2011; pp. 55-74 (http://DavidAbel.us Last accessed in 58/2025).
- Abel, D.L. The Cybernetic Cut: Progressing from Description to Prescription in Systems Theory [Scirus SciTopic Page] (www.DavidAbel.us Last accessed 8/2025). Available online: www.DavidAbel.us (accessed on
- Trost, B.; Loureiro, L.O.; Scherer, S.W. Discovery of genomic variation across a generation. Hum Mol Genet 2021, 30, R174-r186, doi:10.1093/hmg/ddab209.
- Wang, J.; He, X.; Ruan, J.; Dai, M.; Chen, J.; Zhang, Y.; Hu, Y.; Ye, C.; Li, S.; Cong, L.; et al. ChickVD: a sequence variation database for the chicken genome. Nucleic Acids Research 2005, 33, D438-D441, doi:10.1093/nar/gki092.
- Core Concepts, G.P. What's a Little Variation? Available online: (accessed on August, 2025).
- Moon, J. Tandem repeat disorders: from diagnosis to emerging therapeutic strategies. Encephalitis 2025, 5, 27-35, doi:10.47936/encephalitis.2024.00122.
- Doss, R.M.; Lopez-Ignacio, S.; Dischler, A.; Hiatt, L.; Dashnow, H.; Breuss, M.W.; Dias, C.M. Mosaicism in Short Tandem Repeat Disorders: A Clinical Perspective. Genes (Basel) 2025, 16, doi:10.3390/genes16020216.
- Wu, Y.; Song, T.; Xu, Q. R-LOOPs on Short Tandem Repeat Expansion Disorders in Neurodegenerative Diseases. Molecular neurobiology 2023, 60, 7185-7195, doi:10.1007/s12035-023-03531-4.
- Paucar, M.; Laffita-Mesa, J.; Niemela, V.; Malmgren, H.; Nennesmo, I.; Lagerstedt-Robinson, K.; Nordenskjold, M.; Svenningsson, P. Genetic screening for Huntington disease phenocopies in Sweden: A tertiary center case series focused on short tandem repeat (STR) disorders. J Neurol Sci 2023, 451, 120707, doi:10.1016/j.jns.2023.120707.
- Panoyan, M.A.; Wendt, F.R. The role of tandem repeat expansions in brain disorders. Emerg Top Life Sci 2023, 7, 249-263, doi:10.1042/ETLS20230022.
- Annear, D.J.; Kooy, R.F. Unravelling the link between neurodevelopmental disorders and short tandem CGG-repeat expansions. Emerg Top Life Sci 2023, 7, 265-275, doi:10.1042/ETLS20230021.
- Stevanovski, I.; Chintalaphani, S.R.; Gamaarachchi, H.; Ferguson, J.M.; Pineda, S.S.; Scriba, C.K.; Tchan, M.; Fung, V.; Ng, K.; Cortese, A.; et al. Comprehensive genetic diagnosis of tandem repeat expansion disorders with programmable targeted nanopore sequencing. Sci Adv 2022, 8, eabm5386, doi:10.1126/sciadv.abm5386.
- Chintalaphani, S.R.; Pineda, S.S.; Deveson, I.W.; Kumar, K.R. An update on the neurological short tandem repeat expansion disorders and the emergence of long-read sequencing diagnostics. Acta Neuropathol Commun 2021, 9, 98, doi:10.1186/s40478-021-01201-x.
- Hannan, A.J. Expanding genes, repeating themes and therapeutic schemes: The neurobiology of tandem repeat disorders. Neurobiol Dis 2020, 144, 105053, doi:10.1016/j.nbd.2020.105053.
- Ryan, C.P. Tandem repeat disorders. Evol Med Public Health 2019, 2019, 17, doi:10.1093/emph/eoz005.
- Zheng, S.; Wang, Y.; Li, G.; Qin, S.; Dong, Z.; Yang, X.; Xu, X.; Fang, G.; Li, M.; Zhan, S. Functional polymorphism of CYCLE underlies the diapause variation in moths. Science 2025, 388, eado2129, doi:10.1126/science.ado2129.
- Yang, H.; Chu, M.; Naominggaowa; Zhang, X.; Shan, M.; Lu, X.; Pan, Z.; He, J. Tissue-specific expression, functional analysis, and polymorphism of the KRT2 gene in sheep horn. Genomics 2025, 117, 110990, doi:10.1016/j.ygeno.2025.110990.
- Refaat, S.; Al-Rashidi, H.E.; El Azeem, R.A.A.; Nouh, W.E.; Hamed, S.; Attia, Z.R. The functional TNF-alpha(-308)G > a single-nucleotide polymorphism (rs1800629): association with the predictive indices of breast cancer carcinogenesis. Breast Cancer Res Treat 2025, 210, 57-70, doi:10.1007/s10549-024-07536-y.
- Goncalves, S.M.; Leite, L.; de Vasconcelos, P.; Pereira, I.; Dewi, I.M.W.; Mercier, T.; Aerts, R.; Ligeiro, D.; Mendes, F.; Freitas, F.; et al. A Functional Polymorphism in IL-36beta Modulates Macrophage Antifungal Effector Functions and Increases Susceptibility to Invasive Pulmonary Aspergillosis. J Infect Dis 2025, doi:10.1093/infdis/jiaf301.
- Ganai, I.; Goswami, A.M.; Sultana, N.; Sultana, S.; Laha, A.; Biswas, H.; Moitra, S.; Podder, S. Functional insights into PTGS2 rs689466 polymorphism associated to asthma in West Bengal, India. Gene 2025, 962, 149592, doi:10.1016/j.gene.2025.149592.
- Dirisipam, K.; Madduru, D.; Jahan, P.; Gujrati, D. TGF-beta1 promoter functional gene polymorphism -509 C/T in the maternal susceptibility to recurrent pregnancy loss in South Indian women. Hum Immunol 2025, 86, 111182, doi:10.1016/j.humimm.2024.111182.
- Carvalho, P.P.; Souza, M.; Medina, L.; Munoz, M.; Schriefer, A. Proteophosphoglycan functional motifs display genetic polymorphism in a natural population of Leishmania (Viannia) braziliensis. Acta Trop 2025, 265, 107606, doi:10.1016/j.actatropica.2025.107606.
- Wang, C.; Chen, Y.; Zhao, J.; Feng, X.; Ma, R.; Wang, H.; Xue, L.; Tian, J.; Yang, L.; Gu, Y.; et al. Association of SPP1 and NCAPG genes with milk production traits in Chinese Holstein cows: polymorphism and functional validation analysis. Front Vet Sci 2024, 11, 1435128, doi:10.3389/fvets.2024.1435128.
- Fraszczak, M.; Liu, J.; Mielczarek, M.; Dobosz, P.; Szyda, J. EXPLORING THE DISTRIBUTION OF SINGLE NUCLEOTIDE POLYMORPHISMS ACROSS HUMAN EXONS AND INTRONS. bioRxiv 2024, 2024.2003.2023.586436, doi:10.1101/2024.03.23.586436.
- Ahmadifard, A.; Maroofi, N.; Maleki Tehrani, M.; Dabestani, T.; Sadat Mousavi Maleki, M.; Bayrami, S.; Banan, M. Genome editing in K562 cells suggests a functional role for the XmnI Gg polymorphism: a widely used genetic marker in beta-thalassemia and sickle cell disease patients. Cell Mol Biol (Noisy-le-grand) 2024, 70, 230-236, doi:10.14715/cmb/2024.70.7.33.
- Justen, H.; Hasselmann, T.; Illera, J.C.; Delmore, K.E.; Serrano, D.; Flinks, H.; Senzaki, M.; Kawamura, K.; Helm, B.; Liedvogel, M. Population-specific association of Clock gene polymorphism with annual cycle timing in stonechats. Scientific Reports 2022, 12, 7947, doi:10.1038/s41598-022-11158-z.
- Alonso-Blanco, C.; Andrade, J.; Becker, C.; Bemm, F.; Bergelson, J.; Borgwardt, K.M.; Cao, J.; Chae, E.; Dezwaan, T.M.; Ding, W. 1,135 Genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 2016, 166, doi:10.1016/j.cell.2016.05.063.
- Kurmangaliyev, Y.Z.; Sutormin, R.A.; Naumenko, S.A.; Bazykin, G.A.; Gelfand, M.S. Functional implications of splicing polymorphisms in the human genome. Human Molecular Genetics 2013, 22, 3449-3459, doi:10.1093/hmg/ddt200.
- Albert, P.R. What is a functional genetic polymorphism? Defining classes of functionality. Journal of Psychiatry and Neuroscience 2011, 36, 363-365, doi:10.1503/jpn.110137.
- Liao, P.-Y.; Lee, K.H. From SNPs to functional polymorphism: The insight into biotechnology applications. Biochemical Engineering Journal 2010, 49, 149-158.
- Scaglione, D.; Ciacciulli, A.; Gattolin, S.; Caruso, M.; Marroni, F.; Casas, G.L.; Jurman, I.; Licciardello, G.; Catara, A.F.; Rossini, L.; et al. Deep resequencing unveils novel SNPs, InDels, and large structural variants for the clonal fingerprinting of sweet orange [Citrus sinensis (L.) Osbeck]. Plant Genome 2025, 18, e20544, doi:10.1002/tpg2.20544.
- Landi, M.; Carluccio, A.V.; Shah, T.; Niazi, A.; Stavolone, L.; Falquet, L.; Gisel, A.; Bongcam-Rudloff, E. Genome-wide comparison reveals large structural variants in cassava landraces. BMC genomics 2025, 26, 362, doi:10.1186/s12864-025-11523-y.
- Jana, U.; Rodriguez, O.L.; Lees, W.; Engelbrecht, E.; Vanwinkle, Z.; Peres, A.; Gibson, W.S.; Shields, K.; Schultze, S.; Dorgham, A.; et al. The human immunoglobulin heavy chain constant gene locus is enriched for large complex structural variants and coding polymorphisms that vary in frequency among human populations. bioRxiv 2025, doi:10.1101/2025.02.12.634878.
- Battlay, P.; Craig, S.; Putra, A.R.; Monro, K.; De Silva, N.P.; Wilson, J.; Bieker, V.C.; Kabir, S.; Shamaya, N.; van Boheemen, L.; et al. Rapid Parallel Adaptation in Distinct Invasions of Ambrosia Artemisiifolia Is Driven by Large-Effect Structural Variants. Mol Biol Evol 2025, 42, doi:10.1093/molbev/msae270.
- Ryan, M.; McDonough, J.A.; Ward, M.E.; Cookson, M.R.; Skarnes, W.C.; Merkle, F.T. Large structural variants in KOLF2.1J are unlikely to compromise neurological disease modeling. Cell Stem Cell 2024, 31, 290-291, doi:10.1016/j.stem.2024.02.006.
- Tsai, H.H.; Kao, H.J.; Kuo, M.W.; Lin, C.H.; Chang, C.M.; Chen, Y.Y.; Chen, H.H.; Kwok, P.Y.; Yu, A.L.; Yu, J. Whole genomic analysis reveals atypical non-homologous off-target large structural variants induced by CRISPR-Cas9-mediated genome editing. Nat Commun 2023, 14, 5183, doi:10.1038/s41467-023-40901-x.
- Rice, E.S.; Alberdi, A.; Alfieri, J.; Athrey, G.; Balacco, J.R.; Bardou, P.; Blackmon, H.; Charles, M.; Cheng, H.H.; Fedrigo, O.; et al. A pangenome graph reference of 30 chicken genomes allows genotyping of large and complex structural variants. BMC Biol 2023, 21, 267, doi:10.1186/s12915-023-01758-0.
- Cantu Gutierrez, M.E.; Hill, M.C.; Largoza, G.E.; Gillespie, W.B., 3rd; Martin, J.F.; Wythe, J.D. Mapping the transcriptional and epigenetic landscape of organotypic endothelial diversity in the developing and adult mouse. Nat Cardiovasc Res 2025, 4, 473-495, doi:10.1038/s44161-025-00618-0.
- Alva, V.; Jain, P.; Khan, K.; Zameer, F.; Kumar, P.P.; Prashanth Kv, H.; Gopal, S.; Niranjan, V.; Sahu, B.; H, R.; et al. Helicobacter pylori associated MicroRNA and transcriptional networks in gastric pathogenesis: Exploring the epigenetic dialogue landscape. Microb Pathog 2025, 207, 107864, doi:10.1016/j.micpath.2025.107864.
- Tall, A.R.; Fidler, T.P. An epigenetic switch in macrophages promotes fibrosis in the failing heart. Nat Cardiovasc Res 2024, 3, 254-255, doi:10.1038/s44161-024-00439-7.
- Smith, J.P.; Paxton, R.; Medrano, S.; Sheffield, N.C.; Sequeira-Lopez, M.L.S.; Gomez, R.A. Inhibition of Renin Expression Is Regulated by an Epigenetic Switch From an Active to a Poised State. Hypertension 2024, 81, 1869-1882, doi:10.1161/HYPERTENSIONAHA.124.22886.
- Mitesser, V.; Simantov, K.; Dzikowski, R. Time to switch gears: how long noncoding RNAs function as epigenetic regulators in Apicomplexan parasites. Curr Opin Microbiol 2024, 79, 102484, doi:10.1016/j.mib.2024.102484.
- Jakobsen, S.T.; Jensen, R.A.M.; Madsen, M.S.; Ravnsborg, T.; Vaagenso, C.S.; Siersbaek, M.S.; Einarsson, H.; Andersson, R.; Jensen, O.N.; Siersbaek, R. MYC activity at enhancers drives prognostic transcriptional programs through an epigenetic switch. Nat Genet 2024, 56, 663-674, doi:10.1038/s41588-024-01676-z.
- Tomkins, J.P. Epigenetic Mechanisms: Adaptive Master Regulators of the Genome. Acts and Facts 2023, 52, 14=17.
- Jin, L.; Chen, Y.; Muzaffar, S.; Li, C.; Mier-Aguilar, C.A.; Khan, J.; Kashyap, M.P.; Liu, S.; Srivastava, R.; Deshane, J.S.; et al. Epigenetic switch reshapes epithelial progenitor cell signatures and drives inflammatory pathogenesis in hidradenitis suppurativa. Proc Natl Acad Sci U S A 2023, 120, e2315096120, doi:10.1073/pnas.2315096120.
- Kagami, M.; Hara-Isono, K.; Sasaki, A.; Amita, M. Association between imprinting disorders and assisted reproductive technologies. Epigenomics 2025, 17, 397-410, doi:10.1080/17501911.2025.2471269.
- Ivannikova, E.M.; Degtyarevskaya, T.Y.; Tarasova, N.N.; Tinyatov, E.A.; Magomedova, A.N.; Magomedova, K.A. [Sleep disorders in imprinting disorders]. Zh Nevrol Psikhiatr Im S S Korsakova 2025, 125, 75-80, doi:10.17116/jnevro202512505275.
- Ye, M.; Reyes Palomares, A.; Iwarsson, E.; Oberg, A.S.; Rodriguez-Wallberg, K.A. Imprinting disorders in children conceived with assisted reproductive technology in Sweden. Fertil Steril 2024, 122, 706-714, doi:10.1016/j.fertnstert.2024.05.168.
- Baekgaard, C.H.; Lester, E.B.; Moller-Larsen, S.; Lauridsen, M.F.; Larsen, M.J. NanoImprint: A DNA methylation tool for clinical interpretation and diagnosis of common imprinting disorders using nanopore long-read sequencing. Ann Hum Genet 2024, 88, 392-398, doi:10.1111/ahg.12556.
- Linthorst, J.; Sistermans, E.A. Noninvasive Prenatal Testing: Mosaic Ratio Score as a Predictor for Confined Placental Mosaicism. Clin Chem 2025, doi:10.1093/clinchem/hvaf095.
- Huang, Y.; Li, F.; Li, X.; Wang, H. Prenatal diagnosis and genetic counseling of a case with trisomy 20 mosaicism and mixed-type maternal UPD20. Pract Lab Med 2025, 46, e00495, doi:10.1016/j.plabm.2025.e00495.
- Comel, M.; Lamairia, M.; Boute, O.; Cenni, C.; Bergougnoux, A.; Cossee, M.; Koenig, M.; Mansard, L.; Vincent, M.C. Maternal Mosaicism Challenges in Non-Invasive Prenatal Diagnosis. Prenat Diagn 2025, doi:10.1002/pd.6868.
- Parkhomchuk, D.; Amstislavskiy, V.; Soldatov, A.; Ogryzko, V. Use of high throughput sequencing to observe genome dynamics at a single cell level. Proceedings of the National Academy of Sciences 2009, 106, 20830-20835, doi:doi:10.1073/pnas.0906681106.
- Brundin, L.Z. Evolution by orderly stepwise subordination and largely nonrandom mutations. Systematic Biology 1986, 35, 602-607.
- Sun, J.; Hwang, P.; Sakkas, E.D.; Zhou, Y.; Perez, L.; Dave, I.; Kwon, J.B.; McMahon, A.E.; Wichman, M.; Raval, M.; et al. GNN Codon Adjacency Tunes Protein Translation. Int J Mol Sci 2024, 25, doi:10.3390/ijms25115914.
- Scala, M.; Traverso, M.; Capra, V.; Vari, M.S.; Severino, M.; Grossi, S.; Zara, F.; Striano, P.; Minetti, C. Pelizaeus-Merzbacher Disease due to PLP1 Frameshift Mutation in a Female with Nonrandom Skewed X-Chromosome Inactivation. Neuropediatrics 2019, 50, 268-270, doi:10.1055/s-0039-1688954.
- Werner, B.; Sottoriva, A. Variation of mutational burden in healthy human tissues suggests non-random strand segregation and allows measuring somatic mutation rates. PLoS Comput Biol 2018, 14, e1006233, doi:10.1371/journal.pcbi.1006233.
- Yang, X.; Hoshino, A.; Taga, T.; Kunitsu, T.; Ikeda, Y.; Yasumi, T.; Yoshida, K.; Wada, T.; Miyake, K.; Kubota, T.; et al. A female patient with incomplete hemophagocytic lymphohistiocytosis caused by a heterozygous XIAP mutation associated with non-random X-chromosome inactivation skewed towards the wild-type XIAP allele. J Clin Immunol 2015, 35, 244-248, doi:10.1007/s10875-015-0144-6.
- Martincorena, I.; Luscombe, N.M. Non-random mutation: the evolution of targeted hypermutation and hypomutation. Bioessays 2013, 35, 123-130, doi:10.1002/bies.201200150.
- Livnat, A. Interaction-based evolution: how natural selection and nonrandom mutation work together. Biol Direct 2013, 8, 24, doi:10.1186/1745-6150-8-24.
- Boonyawat, B.; Dhanraj, S.; Al Abbas, F.; Zlateska, B.; Grunenbaum, E.; Roifman, C.M.; Steele, L.; Meyn, S.; Blanchette, V.; Scherer, S.W.; et al. Combined de-novo mutation and non-random X-chromosome inactivation causing Wiskott-Aldrich syndrome in a female with thrombocytopenia. J Clin Immunol 2013, 33, 1150-1155, doi:10.1007/s10875-013-9927-9.
- Martincorena, I.; Seshasayee, A.S.; Luscombe, N.M. Evidence of non-random mutation rates suggests an evolutionary risk management strategy. Nature 2012, 485, 95-98, doi:10.1038/nature10995.
- Filisetti, D.; Ostermann, G.; von Bredow, M.; Strom, T.; Filler, G.; Ehrich, J.; Pannetier, S.; Garnier, J.M.; Rowe, P.; Francis, F.; et al. Non-random distribution of mutations in the PHEX gene, and under-detected missense mutations at non-conserved residues. Eur J Hum Genet 1999, 7, 615-619, doi:10.1038/sj.ejhg.5200341.
- Bao, K.; Melde, R.H.; Sharp, N.P. Are mutations usually deleterious? A perspective on the fitness effects of mutation accumulation. Evol Ecol 2022, 36, 753-766, doi:10.1007/s10682-022-10187-4.
- Wagner, A. Evolvability-enhancing mutations in the fitness landscapes of an RNA and a protein. Nature Communications 2023, 14, 3624, doi:10.1038/s41467-023-39321-8.
- Valdmanis Paul N., V., Dominique J. and Rouleau, Guy A. The Proportion of Mutations Predicted To Have a Deleterious Effect Differs Between Gain and Loss of Function Genes in Neurodegenerative Disease 2008, doi:https://onlinelibrary.wiley.com/doi/pdf/10.1002/humu.20939.
- Eyre-Walker, A.; Keightley, P.D. The distribution of fitness effects of new mutations. Nature Reviews Genetics 2007, 8, 610-618, doi:10.1038/nrg2146.
- Wang, Y.; Yang, Y.; Han, Z.; Li, J.; Luo, J.; Yang, H.; Kuang, J.; Wu, D.; Wang, S.; Tso, S.; et al. Efficient purging of deleterious mutations contributes to the survival of a rare conifer. Hortic Res 2024, 11, uhae108, doi:10.1093/hr/uhae108.
- Verma, S.; Menon, R.; Sowdhamini, R. Structural insights into the role of deleterious mutations at the dimeric interface of Toll-like receptor interferon-beta related adaptor protein. Proteins 2024, 92, 1242-1258, doi:10.1002/prot.26707.
- Shafie, A.; Ashour, A.A.; Anjum, F.; Shamsi, A.; Hassan, M.I. Elucidating the Impact of Deleterious Mutations on IGHG1 and Their Association with Huntington's Disease. J Pers Med 2024, 14, doi:10.3390/jpm14040380.
- Matheson, J.; Masel, J. Background Selection From Unlinked Sites Causes Nonindependent Evolution of Deleterious Mutations. Genome Biol Evol 2024, 16, doi:10.1093/gbe/evae050.
- Lavanchy, E.; Cumer, T.; Topaloudis, A.; Ducrest, A.L.; Simon, C.; Roulin, A.; Goudet, J. Too big to purge: persistence of deleterious Mutations in Island populations of the European Barn Owl (Tyto alba). Heredity (Edinb) 2024, 133, 437-449, doi:10.1038/s41437-024-00728-8.
- Hu, C.; Liu, G.; Zhang, Z.; Pan, Q.; Zhang, X.; Liu, W.; Li, Z.; Li, M.; Zhu, P.; Ji, T.; et al. Genetic linkage disequilibrium of deleterious mutations in threatened mammals. EMBO Rep 2024, 25, 5620-5634, doi:10.1038/s44319-024-00307-2.
- Hasselgren, M.; Dussex, N.; von Seth, J.; Angerbjorn, A.; Dalen, L.; Noren, K. Strongly deleterious mutations influence reproductive output and longevity in an endangered population. Nat Commun 2024, 15, 8378, doi:10.1038/s41467-024-52741-4.
- Faraggi, E.; Jernigan, R.L.; Kloczkowski, A. Rapid discrimination between deleterious and benign missense mutations in the CAGI 6 experiment. Hum Genomics 2024, 18, 89, doi:10.1186/s40246-024-00655-z.
- Chen, G.; Shi, G.; Dai, Y.; Zhao, R.; Wu, Q. Graph-Based Pan-Genome Reveals the Pattern of Deleterious Mutations during the Domestication of Saccharomyces cerevisiae. J Fungi (Basel) 2024, 10, doi:10.3390/jof10080575.
- Bao, K.; Strayer, B.R.; Braker, N.P.; Chan, A.A.; Sharp, N.P. Mutations in yeast are deleterious on average regardless of the degree of adaptation to the testing environment. Proc Biol Sci 2024, 291, 20240064, doi:10.1098/rspb.2024.0064.
- Badve, S.B.; Kim, E.; Sibia, U.S.; Borrego, O.T.; Vara, S.; Damron, A.; Riker, A.I. Male Breast Cancer With Dual BRCA2 and BRIP1 Deleterious Gene Mutations. Ochsner J 2024, 24, 157-161, doi:10.31486/toj.23.0119.
- Aubier, T.G.; Galipaud, M. Senescence evolution under the catastrophic accumulation of deleterious mutations. Evol Lett 2024, 8, 212-221, doi:10.1093/evlett/qrad050.
- Matheson, J.; Masel, J. Background selection from unlinked sites causes non-independent evolution of deleterious mutations. bioRxiv 2023, 2022.2001.2011.475913, doi:10.1101/2022.01.11.475913.
- Wu, X.; Bellagio, T.; Peng, Y.; Czech, L.; Lin, M.; Lang, P.; Epstein, R.; Abdelaziz, M.; Alexander, J.; Caton-Darby, M.; et al. Rapid adaptation and extinction across climates in synchronized outdoor evolution experiments of Arabidopsis thaliana. bioRxiv 2025, doi:10.1101/2025.05.28.654549.
- Wang, J.L.; Zhang, W.D.; Yang, X.D.; Zhao, P.G.; Wang, X.Y.; Zhao, S.Y.; Chen, L.Y. Chromosome-level genome assembly of Pontederia cordata L. provides insights into its rapid adaptation and variation of flower colours. DNA Res 2025, 32, doi:10.1093/dnares/dsaf002.
- Shahmohamadloo, R.S.; Gabidulin, A.R.; Andrews, E.R.; Rudman, S.M. Microbiome evolution plays a secondary role in host rapid adaptation. bioRxiv 2025, doi:10.1101/2025.06.27.661976.
- Peng, Q.; Qin, J.; Xu, H.; Kao, G.; Yang, F.; Sun, Z.; Zhang, X.; Slamti, L.; Guo, S.; Song, F. Rapid adaptation of Bacillus thuringiensis to alkaline environments via the L-lactate metabolism pathway regulated by the CRP/FNR family regulator LtmR. Pestic Biochem Physiol 2025, 208, 106255, doi:10.1016/j.pestbp.2024.106255.
- Metheringham, C.L.; Plumb, W.J.; Flynn, W.R.M.; Stocks, J.J.; Kelly, L.J.; Nemesio Gorriz, M.; Grieve, S.W.D.; Moat, J.; Lines, E.R.; Buggs, R.J.A.; et al. Rapid polygenic adaptation in a wild population of ash trees under a novel fungal epidemic. Science 2025, 388, 1422-1425, doi:10.1126/science.adp2990.
- Leiva, C.; Torda, G.; Zhou, C.; Pan, Y.; Harris, J.; Xiang, X.; Tan, S.; Tian, W.; Hume, B.; Miller, D.J.; et al. Rapid Evolution in Action: Environmental Filtering Supports Coral Adaptation to a Hot, Acidic, and Deoxygenated Extreme Habitat. Glob Chang Biol 2025, 31, e70103, doi:10.1111/gcb.70103.
- Kwakye, A.; Reid, K.; Wund, M.A.; Heins, D.C.; Bell, M.A.; Veeramah, K.R. Rare "Jackpot" Individuals Drive Rapid Adaptation in Threespine Stickleback. bioRxiv 2025, doi:10.1101/2025.03.25.642177.
- Grieshop, M.P.; Behr, A.A.; Bowden, S.; Lin, J.D.; Molari, M.; Reynolds, G.Z.; Brooks, E.F.; Doyle, B.; Rodriguez-Nava, G.; Salinas, J.L.; et al. Replicative selfish genetic elements are driving rapid pathogenic adaptation of Enterococcus faecium. bioRxiv 2025, doi:10.1101/2025.03.16.643550.
- Coomber, A.; Rasmussen, D.A.; Ristaino, J.B. Experimental Evolution of Phytophthora infestans on Tomato Reveals Rapid Genotypic and Phenotypic Adaptation and Dynamic RXLR Genome Variation. Phytopathology 2025, 115, 998-1007, doi:10.1094/PHYTO-12-24-0401-R.
- Cassin-Sackett, L.; Tsuchiya, M.T.N.; Dikow, R.B. Rapid adaptation to a globally introduced virulent pathogen in a keystone species. PNAS Nexus 2025, 4, pgaf199, doi:10.1093/pnasnexus/pgaf199.
- Brazier, T.; Merot, C. Genomic architecture of rapid adaptation illustrated by biological invasions. Nat Ecol Evol 2025, 9, 1317-1318, doi:10.1038/s41559-025-02774-9.
- Trevors, J.T.; Abel, D.L. Chance and necessity do not explain the origin of life Cell Biol Int 2004, 28, 729-739 (Also available at www.DavidAbel.us Last accessed 711/2024), doi:https://doi.org/10.1016/j.cellbi.2004.06.006.
- Abel, D.L. The three fundamental categories of reality In The First Gene: The Birth of Programming, Messaging and Formal Control, Abel, D.L., Ed.; LongView Press-Academic: Biolog. Res. Div.: New York, N.Y., 2011; pp. 19-54 (http://DavidAbel.us Last accessed in 9/2025).
- Abel, D.L. The Universal Determinism Dichotomy (UDD): Physicodynanamic Determinism (PD) vs Choice Determinism (CD). Scopus Sci Topic Paper 2005, doi:https://www.davidabel.us/papers/The-Universal-Determinism-Dichotomy-UDD4.pdf.
- Sane, M.; Parveen, S.; Agashe, D. Mutation bias alters the distribution of fitness effects of mutations. PLoS Biol 2025, 23, e3003282, doi:10.1371/journal.pbio.3003282.
- Chorley, B.N.; Wang, X.; Campbell, M.R.; Pittman, G.S.; Noureddine, M.A.; Bell, D.A. Discovery and verification of functional single nucleotide polymorphisms in regulatory genomic regions: current and developing technologies. Mutation Research/Reviews in Mutation Research 2008, 659, 147-157.
- Sadee, W.; Wang, D.; Papp, A.; Pinsonneault, J.; Smith, R.; Moyer, R.; Johnson, A. Pharmacogenomics of the RNA world: structural RNA polymorphisms in drug therapy. Clinical Pharmacology & Therapeutics 2011, 89, 355-365.
- Zhu, H.; Wu, Z.; Wang, J.; Zhang, E.; Zhang, S.; Yang, Y.; Li, W.; Shi, H.; Yang, G.; Lv, L.; et al. DLG2 rs11607886 polymorphism associated with schizophrenia and precuneus functional changes. Schizophr Res 2025, 280, 50-58, doi:10.1016/j.schres.2025.04.004.
- Liang, J.; Xu, M.; Wang, X.; Li, H.; Luo, X.; Christoforou, A.; Wang, Q.M. Effect of COMT Val158Met Polymorphism on Stroke Functional Outcome and Recovery. Neurorehabil Neural Repair 2025, 39, 612-623, doi:10.1177/15459683251340926.
- Tomkins, J.P. Long Non-Coding RNAs: The Unsung Heroes of the Genome. Acts and Facts 2025, 54.
- Tomkins, J.P. Genomic Tandem Repeats: Where Repetition is Purposely Adaptive. Acts and Facts 2025, 54, 14-17.
- Tomkins, J.P. Small Heritable RNAs Pack a Big Adaptive Punch. . Acts & Facts 2024, 53, 12–15.
- Tomkins, J.P. RNA Editing: Adaptive Genome Modification on the Fly. Acts and Facts 2024, 53, 14-17.
- Tomkins, J.P. RNA Editing: Biocomplexity Hits a New High. Available online: http://www.icr.org/article/8649/ (accessed on
- Tomkins, J.P. Three-Dimensional DNA Code Defies Evolution. Available online: http://www.icr.org/article/8691/ (accessed on
- Walter, N.G. Are Non-Protein Coding RNAs Junk or Treasure? . BioEssays 2023, 46, e2300201.
- Mattick, J.S.e.a. Long Non-Coding RNAs: Definitions, Functions, Challenges and Recommendations. Nature Reviews Molecular Cell Biology 2023, 24, 430–447.
- Ye, W.e.a. Comprehensive Analysis of Hub mRNA, lncRNA and miRNA, and Associated ceRNA Networks Implicated in Grass Carp (Ctenopharyngodon Idella) Growth Traits. . Genomics 2021, 113, 4004–4014.
- Statello, L.e.a. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nature Reviews Molecular Cell Biology 2021, 22, 96–118.
- Hang, J.; Wan, R.; Yan, C.; Shi, Y. Structural basis of pre-mRNA splicing. Science 2015, doi:10.1126/science.aac8159.
- Norris, A.D.; Calarco, J.A. Emerging roles of alternative pre-mRNA splicing regulation in neuronal development and function. Frontiers in Neuroscience 2012, 6, doi:10.3389/fnins.2012.00122.
- Siddaway, R.; Milos, S.; Vadivel, A.K.A.; Dobson, T.H.W.; Swaminathan, J.; Ryall, S.; Pajovic, S.; Patel, P.G.; Nazarian, J.; Becher, O.; et al. Splicing is an alternate oncogenic pathway activation mechanism in glioma. Nat Commun 2022, 13, 588, doi:10.1038/s41467-022-28253-4.
- Matalkah, F.; Jeong, B.; Sheridan, M.; Horstick, E.; Ramamurthy, V.; Stoilov, P. The Musashi proteins direct post-transcriptional control of protein expression and alternate exon splicing in vertebrate photoreceptors. Commun Biol 2022, 5, 1011, doi:10.1038/s42003-022-03990-w.
- Le, B.T.; Paul, S.; Jastrzebska, K.; Langer, H.; Caruthers, M.H.; Veedu, R.N. Thiomorpholino oligonucleotides as a robust class of next generation platforms for alternate mRNA splicing. Proc Natl Acad Sci U S A 2022, 119, e2207956119, doi:10.1073/pnas.2207956119.
- Karlebach, G.; Aronow, B.; Baylin, S.B.; Butler, D.; Foox, J.; Levy, S.; Meydan, C.; Mozsary, C.; Saravia-Butler, A.M.; Taylor, D.M.; et al. Betacoronavirus-specific alternate splicing. Genomics 2022, 114, 110270, doi:10.1016/j.ygeno.2022.110270.
- Mvubu, N.E.; Pillay, B.; Pillay, M. Infection of pulmonary epithelial cells by clinical strains of M. tuberculosis induces alternate splicing events. Gene 2020, 750, 144755, doi:10.1016/j.gene.2020.144755.
- Reinar, W.B.e.a. Length Variation in Short Tandem Repeats Affects Gene Expression in Natural Populations of Arabidopsis thaliana. Plant Cell 2021, 33, 2221–2234.
- Reinar, W.B.; Lalun, V.O.; Reitan, T.; Jakobsen, K.S.; Butenko, M.A. Length variation in short tandem repeats affects gene expression in natural populations of Arabidopsis thaliana. The Plant Cell 2021, 33, 2221-2234, doi:10.1093/plcell/koab107.
- Sawaya, S.e.a. Microsatellite tandem repeats are abundant in human promoters and are associated with regulatory elements. . PLoS ONE 2013, 8.
- Gemayel, R.; Cho, J.; Boeynaems, S.; Verstrepen, K.J. Beyond junk-variable tandem repeats as facilitators of rapid evolution of regulatory and coding sequences. Genes (Basel) 2012, 3, 461-480, doi:10.3390/genes3030461.
- Ma, L.; Jensen, J.S.; Mancuso, M.; Hamasuna, R.; Jia, Q.; McGowin, C.L.; Martin, D.H. Variability of Trinucleotide Tandem Repeats in the MgPa Operon and Its Repetitive Chromosomal Elements in Mycoplasma genitalium. J Med Microbiol 2011, doi:jmm.0.030858-0 [pii] 10.1099/jmm.0.030858-0.
- Gemayel, Ρ.e.a. Variable Tandem Repeats Accelerate Evolution of Coding and Regulatory Sequences. Annual Reviews of Genetics 2010, 44, 445–477.
- Ma, L.; Martin, D.H. Single-nucleotide polymorphisms in the rRNA operon and variable numbers of tandem repeats in the lipoprotein gene among Mycoplasma genitalium strains from clinical specimens. J Clin Microbiol 2004, 42, 4876-4878, doi:42/10/4876 [pii] 10.1128/JCM.42.10.4876-4878.2004.
- , N. Dynamic Self-organization in an Open Reaction Network: A Principle for the Emergence of Life. ChemRxiv. Cambridge: Cambridge Open Engage 2022, Open Engage; 2022; This content is a preprint and has not been peer-reviewed.
- Levit, G.S.; Hossfeld, U.A. Self-Organization Meets Evolution: Ernst Haeckel and Abiogenesis. In Self-Organization as a New Paradigm in Evolutionary Biology: From Theory to Applied Cases in the Tree of Life, Dambricourt Malassé, A., Ed.; Springer International Publishing: Cham, 2022; pp. 11-32.
- Pereto, J. Crystals and the debates on the nature, recognition and origin of life: Comment on "Mineral self-organization on a lifeless planet" by J.M. Garcia-Ruiz et al. Phys Life Rev 2020, 34-35, 86-88, doi:10.1016/j.plrev.2020.05.001.
- McCusker, D. Cellular self-organization: generating order from the abyss. Molecular Biology of the Cell 2020, 31, 143-148, doi:10.1091/mbc.E19-04-0207.
- Crombie, T.A.; Rajaei, M.; Saxena, A.S.; Johnson, L.M.; Saber, S.; Tanny, R.E.; Ponciano, J.M.; Andersen, E.C.; Zhou, J.; Baer, C.F. Direct inference of the distribution of fitness effects of spontaneous mutations from recombinant inbred Caenorhabditis elegans mutation accumulation lines. Genetics 2024, 228, doi:10.1093/genetics/iyae136.
- Abel, D.L. Primordial Prescription: The Most Plaguing Problem of Life Origin Science LongView Press Academic: New York, N. Y., 2015.
- Society, T.C.B. Code Bilogy. Available online: https://www.codebiology.org/#:~:text=The-study-of-the-biological,the-standard-methods-of-science. (accessed on 9/9/2025)
- Barbieri, M. Overview of the third special issue in code biology. Biosystems 2021, 210, 104553, doi:https://doi.org/10.1016/j.biosystems.2021.104553.
- Barbieri, M. A general model on the origin of biological codes. Biosystems 2019, 181, 11-19, doi:10.1016/j.biosystems.2019.04.010.
- Barbieri, M. What is code biology? Biosystems 2018, 164, 1-10, doi:https://doi.org/10.1016/j.biosystems.2017.10.005.
- Barbieri, M. Overview of the fourth special issue in code biology. Biosystems 2024, 235, 105074, doi:10.1016/j.biosystems.2023.105074.


 Impact Factor: * 4.2
Impact Factor: * 4.2 Acceptance Rate: 77.66%
Acceptance Rate: 77.66%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 