Nanozymedb: A Manually Curated Database to Understand and Match Kinetics of Nanozymes with Natural Enzymes
Shreya Sharma, Kartik Singh, Ankit Kalra, Shilpa Sharma*
Department of Biological Sciences and Engineering, Netaji Subhas University of Technology, Sector-3, Dwarka, New Delhi, 110078, India
*Corresponding author: Shilpa Sharma, Department of Biological Sciences and Engineering, Netaji Subhas University of Technology, Sector-3, Dwarka, New Delhi, 110078, India.
Received: 09 May 2023; Accepted: 21 December 2023; Published: 26 December 2023
Article Information
Citation: Shreya Sharma, Kartik Singh, Ankit Kalra, Shilpa Sharma. Nanozymedb: A Manually Curated Database to Understand and Match Kinetics of Nanozymes with Natural Enzymes. Journal of Nanotechnology Researc. 5 (2023): 22-27.
DOI: 10.26502/jnr.2688-85210039
View / Download Pdf Share at FacebookAbstract
Nanoparticle-based catalysts, also known as Nanozymes, have played a crucial role in growing success of nanotechnology owing to their intrinsic enzyme-like characteristics. They have gained significant interest among the research community in the past decade due to effective mode of action, higher stability, lowcost industrial production, wide spectrum of physical and chemical properties and tunable catalytic activities compared to both natural and artificial enzymes. Currently, the knowledge of these nanozymes is restricted either to individual research papers available across platforms such as Google Scholar and PubMed, or literature reviews incorporating a subset of few such papers. Hitherto, there exists no platform or repository summarizing an exhaustive list of nanozymes and their kinetics. In an attempt to benefact the scientific community, we present NanozymeDB, a comprehensive data framework serving as a single platform for addressing and collating catalytic and kinetic parameters of nanozymes.
Keywords
<p>Nanozyme; Nanomaterials; Catalytic; Kinetic parameters; Database; Web portal</p>
Article Details
Introduction
Enzymes are powerful biocatalysts capable of facilitating majority of biological reactions that occur in living systems [1]. The intrinsic shortcomings of natural enzymes including ease of denaturation, high cost related to preparation and purification, challenges in recycling/reusing, sensitivity to environmental conditions, low operational stability, etc. restrict their practical applications in biomedicine, environmental protection, biosensing, food processing, and many more [2]. To subjugate these limitations, scientists have been staunched to the exploration of artificial enzymes as low-cost and stable alternatives to natural enzymes. Since, the year 2007, after the discovery of iron oxide Fe3O4 nanoparticles reported to mimic peroxidase activity, several studies on nanomaterial based artificial enzymes have been continually surfacing [3]. Nanozymes or nanoparticle (particles of size ranging from 1-100 nm) based catalysts obey the classical definition of catalysis and inhabit the conspicuous growing part of success of nanotechnologies [4,5]. They have enticed substantial interest over the past decade attributable to their apparent precedence to not just natural enzymes but also many artificial enzymes in use, including, high stability, easy large scale production, low cost, wider physical and chemical operational window, and high and tunable catalytic activities [6]. In order to claim them enzyme mimics, the nanoparticles in use must possess inherent catalytic properties displaying similar catalytic mechanism as the natural enzymes [7]. The eccentric physiochemical attributes of nano-sized materials confer nanozymes with several possibilities for chemical functionalization and structural modifications, providing supercilious tactics for calibrating their catalytic activity [8]. Profiting from quick advancement of catalysis science, biotechnology, nanotechnology, and computational design, critical progress has been attained in mirroring novel enzymatic activities with high-performance nanomaterials, regulating the nanozyme activities, elucidating the catalytic mechanisms, and broadening prospective applications [9,10]. Several research laboratories around the globe are actively working on nanozymes, highlighting significance and influence of the field [11].
Hithero, the most well-known nanozymes are nanozymes with activities of oxidases and peroxidases [12,13]. Others, more intricate, for example activities of superoxide dismutase [14], laccase [15], phosphatase [16], catalase [17], and hydrolase are significantly less introduced. A few nanomaterials have multi-enzymatic activity [18]. The synergist action of nanomaterials is dependent upon Michaelis-Menten energy and is calibrated by the Michaelis-Menten steady (Km, mM) and the reactant rate constant (Kcat, s−1). Nanozymes have applications in malignant growth theranostics [19], environmental protection [20], cytoprotection [21], biosensing [22], and other applications, and of significant consideration is the capacity to direct the reactant movement of nanomaterials by changing their composition, shape, size, gem framework, and surface science. Given the steadily developing intricacy of nanozyme frameworks, information driven approaches begin to penetrate the field of nanozyme discovery empowering sane speculation detailing and in silico evaluation [23]. Unlike regular enzymes that ordinarily show high synergist action and substrate selectivity, nanozymes’ functional application are limited by low activity, and poor specificity [1]. To make nanozymes better alternatives to natural enzymes, much exertion has been exhausted to move along the presentation of nanozymes, including managing their substrate selectivity, working on their reactant action, and fostering their multienzyme mimetic action [24,25]. Up to this point, iron-based and carbon-based nanozymes have been the most contemplated and recreated in light of their great biocompatibility and extraordinary potential for biomedical applications. With the standard synergist energy, nanozymes could be quantitively looked at as far as the catalytic proficiency (kcat/Km), substrate explicitness (Km), catalytic rate constant (kcat), and maximal response rate (Vmax) for explicit substrates. Attributable to the mind-boggling reliance of physicochemical properties and synergist attributes, nanozyme-based discovery applications are seriously restricted by mediocre repeatability and unwavering quality. With the expeditious escalation in nanozymes data, nanozymes and their related catalytic properties, it becomes essential to forge a platform with structured data to interrogate salvage pertinent data. Present knowledge of nanozymes is restricted to individual papers that can be searched through Google Scholar, PubMed or reviews where a subset of studies has been summarized. However, there exists absence of repository that collates the existing nanozymes and their kinetics related information. A recent study by Razlivina and co-workers explored collation of a database of exiting inorganic nanozymes as reported in the scientific literature majorly focusing on their peroxidase activity [26]. In the present study, we report NanozymeDB (www.nanozymedb.com or https://nanozymedb.vercel.app/home), an open-source database, a benefaction to the scientific community actively researching the area of nanozymes. This is an extensive data framework that fills in as a common portal for addressing and comprehending nanozyme catalytic and kinetic parameters, all in a single platform. Our database is not limited to collation of information of inorganic nanozymes with peroxidases activity alone as done by Razlivina et al., [26]. We have included all the currently existing nanozymes and their catalytic activity that is subject to Michaelis-Menten kinetics. The web module developed amalgamates several web-based tools for searching/browsing facilities expedite users in the extraction and analysis of data.
Methods
1 Data mining and curation
The data was manually curated using google scholar and PubMed to obtain research and review articles using keywords, “nanozymes,” “nanoparticles used as enzymes,” “bioinspired nanozymes.” The papers were sorted since 2007 when the first nanozyme (ferromagnetic nanoparticles with peroxidase-like activity) was reported [27]. Nanozymes are classified mainly into ‘oxidase family,’ ‘peroxidase family,’ ‘catalase,’ and ‘superoxide dismutase.’ Other nanozymes, not belonging to any above categories have also been identified and their classification has been denoted as ‘others.’
2 Data processing and enrichment
Filtering of raw data collected from different sources was carried out. To avoid redundancy, all duplicate data was removed. DOI of articles, author information, article type, journal, publication date, abstract, title was extracted and recorded in a tabular format.
3 Database and web browser
The approach is based on NoSQL database namely MongoDB due to its advantages like developer friendliness, fast queries, flexible data models, and horizontal scaling. Following the processing and assembling of data in a csv file, the data was uploaded on the MongoDB server. The imported csv file header columns were used as keys in MongoDB and Python script was employed to insert values to these keys following which different key-value pairs were generated. Next, to deploy the database in the form of website, first, different application programming interface (API) routes were created for the database search portal. Node.js was further used for developing backend of website environment. The search inputs given by the user are stored in a cookie and the cookie values are used to compare and fetch the results utilizing the search tags stored in the database. The search tags are created with every new entry in the database and are modifiable. The frontend of the website was developed using EJS, HTML, CSS, and Javascript.
Result and Discussions
The NanozymeDB database proffers thorough information to study the novel concept of biomimetic optimization of nanozymes. The compatibility of the web application is excellent with several web browsers tested including Microsoft Edge, Internet Explorer, Chrome, Firefox, etc. The processing of data collected from different sources resulted in >600 entries. The integrated data fields included Nanozyme/Monomaterial (structural), enzyme like activity, Substrate, Temperature, pH, Km, Vmax, kcat:
1. Nanozyme/monomaterial (structural)
The properties of nanomaterials qualify them as propitious candidates for enzyme mimetics. The nanomaterials with specific nanostructures catalyze the biochemical reactions of specific substrates. Different nanomaterials mimic the activity of different enzymes. The reported nanozymes collected from different sources included (A) Carbon based nanozymes, (B) Metal-oxide/Sulphide based nanozymes, (C) Metal based nanozymes.
2. Enzyme like activity
The catalytic mechanisms of the nanozymes with unique catalytic activities may help in better understanding of the nanozymes [28]. The catalytic mechanisms of the reported nanozymes have been found to mainly exhibit excellent oxidase-like (enzyme that catalyzes oxidation-reduction reactions), peroxidase-like (enzyme that catalyzes oxidation-reduction reactions employing free radical transformation), catalase-like (enzyme that catalyzes decomposition of hydrogen peroxide to water and oxygen), and superoxide dismutase-like (catalyzes the dismutation of superoxide radicals into hydrogen peroxide and oxygen) properties.
3. Substrate
Nanozymes can catalyze substrates of natural enzymes under physiological conditions with similar catalytic mechanism and kinetics [25].
4. Temperature
It is important to know the temperature range (optimum temperature) in which maximal rate of reaction (optimum activity) is achieved by the nanozyme.
5. pH
Changes in pH affect the catalytic activity of nanozymes. The shape and charge of the substrate are also influenced by pH changes.
6. km
The apparent steady state kinetic parameters determined by the Michaelis-Menten model of enzyme kinetics, like the Michaelis constant km (substrate concentration that allows the enzyme to achieve half Vmax) will aid in prediction of nanozymes’ affinity to the substrate thus permitting determination of best substrate for the nanozyme [8].
7. Vmax
The reactivity of the nanozyme upon saturation with the substrate will aid in the estimation of nanozyme concentration [8].
8. kcat
Curation of kcat values of nanozyme to the knowledge pool aids in determination of the number of substrate molecules trasnformed into products per unit time.
The relevant DOI records of the articles used to extract the information were linked with corresponding clickable hyperlinks. An interactive web interface NanzoymeDB was created using the NoSQL database and NodeJS. The NanozymeDB proffers a variety of user-friendly web interfaces for extraction of data utilizing wide range of options:
1. Search
Access and effortless retrieval of data has been made possible to the user utilizing the simple search option. The user can proffer query against any field of database using the keywords like nanomaterial/monomaterial, nanozyme, substrate, Temperature, pH, Km, Vmax, kcat. Once the input is provided by the user, the server delivers all the relevant data linked with the keywords in a tabular format (Figure 1).
2. Download excel sheet of search results
The web interface also allows the user to download the data compilation of the searched results in excel format so that it is easy for the users to understand the data thoroughly and extract actionable insights.
3. Contribution to the database
This option allows the user to contribute to the unique resource collating and comparing the catalytic activity and kinetics of the nanozymes. The user is directed to a contribution form wherein the new entries added by default go unapproved and are forwarded to the editor. The editor is then responsible for verifying the data. The display of new entries on the database is dependent on the approval/disapproval by the editor (Figure 2).
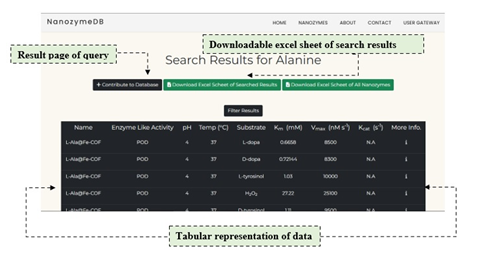
Figure 1: Search option allows users to input relevant data with keywords to deliver outputs in a tabular format.
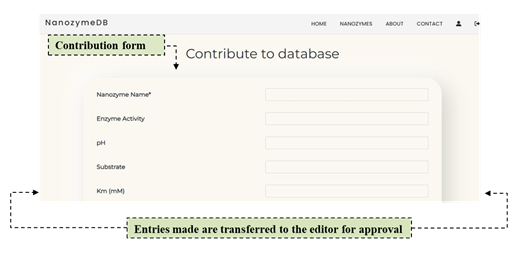
Figure 2: ‘Contribute to database’ feature allows the user to contribute to the unique resource collating
Availability
The web portal is amenable and compatible with all the latest gadgets. All probable users of the database including researchers, clinicians, students, advisors, practitioners, etc. can access the interface without any prior login credentials requirement. NanozymeDB can consequently be accessed freely at nanozymedb.com or https://nanozymedb.vercel.app/home
Discussion
In the present scenario, where the field of nanozymes is now clasping attention, there is an alacritous requirement of a web platform that could transform the discourse of attending to the nanozymes and learning more about them. In the absence of such a platform, researchers would have to surf various websites at a particular time for reasoning and to study about different parameters relevant to the nanozyme. The immersing data accessible by most web services is in the form of cumbersome texts which take time to fathom. The tabular presentation of data makes it advantageous to look at terrifically significant parameters of the nanozymes clearly and in a hassle free manner. NanozymeDB simplifies the process of data assortment and recovery. The staggering measure of information spread across the web in different vaults is scattered. NanozymeDB makes it helpful for the client, as it accumulates and presents the data from this large number of stores immediately, under a solitary umbrella. This will save time and make the course of data recovery by means of dispersed written works and weblinks less baffling. Various aspects of the nanozyme like optimum temperature, pH, substrate, structure, etc. can be mined by the user.
Utility of database
Prospective applications of NanozymeDB are:
- To the best of authors’ knowledge, presently no database subsists to the need of scientific community working actively in the domain of nanozymes including the design of nanozymes and use of nanozymes for several applications. We proffer a structured platform to the scientific community that allows comparison of catalytic activity and kinetics of nanozymes with that of natural enzymes and resource for initiating new studies.
- The tabular presentation of the data permits the user in better analysis thus allowing the researcher to join all the dots utilizing the platform.
- The platform provides a holistic way to reveal the catalytic principles of nanozymes. The compilation of the data at a single stop will intrigue graduate students and professional researchers across academia and industry working to integrate key catalytic modules of natural enzymes into the structure of nanozyme thus making the process hassle free and saving of time to excavate the enormous information pool dissipated on the internet.
- Our web interface proffers opportunity to look for >600 unique entries of nanozyme summarizing their kinetic parameters knowledge.
- The platform will be ceaselessly updated with sufficient literature accumulation ensuring high level and up to date quality of information from present research trends in arena of nanozymes.
Conclusion
The NanozymeDB database delivers thorough information on nanozymes and their catalytic and kinetic properties. The database has been manually curated using publicly available abstracts/articles/reviews since 2007 when first nanozyme was reported. The rapid advancement of nanotechnology has accorded to the emergence and refinement of novel class of artificial enzymes known as nanozymes that have exhibited promising applications especially in medicine and industry. Nanomaterials with enzyme like properties are extensively employed in agricultural science, environmental remediation, sensors, food processing, imaging, and therapy. Design of efficient nanozymes would require consideration of biomimicry through the amalgamation of nanoscience, materials, biology, and chemistry. The recent progress in nanozyme research has reached a state where a specifically focused repository on nanozymes and their comparable catalytic properties is a requisite to steer to avenue of research. The NanozymeDB will serve as a vital resource tool spanning, >600 nanozymes, and their catalytic and kinetic parameters explored through various angles.
References
- Liang M, Yan X. Nanozymes: From New Concepts, Mechanisms, and Standards to Applications. Accounts of Chemical Research 52 (2019): 2190-2200.
- Huang Y, Ren J, Qu X. Nanozymes: Classification, Catalytic Mechanisms, Activity Regulation, and Applications. Chemical Reviews 119 (2019): 4357-4412.
- Gao L, Yan X. Nanozymes: Biomedical Applications of Enzymatic Fe3O4 Nanoparticles from In Vitro to In Vivo. Advances in Experimental Medicine and Biology 1174 (2019): 291-312.
- Zandieh M, Liu J. Nanozyme Catalytic Turnover and Self-Limited Reactions. ACS Nano 15 (2021): 15645-15655.
- Robert A, Meunier B. How to Define a Nanozyme. ACS Nano (2022).
- Yang W, et al. Nanozymes: Activity origin, catalytic mechanism, and biological application. Coordination Chemistry Reviews 448 (2021): 214170.
- Yan X. Nanozymology Connecting biology and nanotechnology (2020).
- Ballesteros CAS, et al. Recent trends in nanozymes design: From materials and structures to environmental applications. Materials Chemistry Frontiers 5 (2021): 7419-7451.
- Li S, et al. Nanozyme-Enabled Analytical Chemistry. Analytical Chemistry 94 (2022): 312-323.
- Zandieh M, Liu J. Surface Science of Nanozymes and Defining a Nanozyme Unit. Langmuir 38 (2022): 3617-3622.
- Abedanzadeh S, Moosavi-movahedi Z, Sheibani N. Nanozymes?: Supramolecular perspective. Biochemical Engineering Journal 183 (2022): 108463.
- Attar F, et al. Nanozymes with intrinsic peroxidase-like activities’, Journal of Molecular Liquids 278 (2019): 130-144.
- Liu Q, et al. A Review on Metal- and Metal Oxide-Based Nanozymes: Properties, Mechanisms, and Applications, Nano-Micro Letters. Springer Singapore (2021).
- Singh N, et al. A Cerium Vanadate Nanozyme with Specific Superoxide Dismutase Activity Regulates Mitochondrial Function and ATP Synthesis in Neuronal Cells. Angewandte Chemie - International Edition 60 (2021): 3121-3130.
- Ren X, et al. One-pot synthesis of active copper-containing carbon dots with laccase-like activities’, Nanoscale 7 (2015): 19641-19646.
- Liu H, Liu J. Self-limited Phosphatase-mimicking CeO2 Nanozymes. ChemNanoMat, 6 (2020): 947-952.
- Jiao M, et al. Solving the H2O2 by-product problem using a catalase-mimicking nanozyme cascade to enhance glycolic acid oxidase. Chemical Engineering Journal 388 (2020): 124249.
- Zhang X, et al. Co2V2O7 Particles with Intrinsic Multienzyme Mimetic Activities as an Effective Bioplatform for Ultrasensitive Fluorometric and Colorimetric Biosensing. ACS Applied Bio Materials 3 (2020): 1469-1480.
- Peng Y, et al. Size- and shape-dependent peroxidase-like catalytic activity of MnFe2O4 Nanoparticles and their applications in highly efficient colorimetric detection of target cancer cells. Dalton Transactions 44 (2015): 12871-12877.
- Feng YB, et al. High-efficiency catalytic degradation of phenol based on the peroxidase-like activity of cupric oxide nanoparticles’, International Journal of Environmental Science and Technology 12 (2015): 653-660.
- Xu ZQ, et al. Highly Photoluminescent Nitrogen-Doped Carbon Nanodots and Their Protective Effects against Oxidative Stress on Cells. ACS Applied Materials and Interfaces 7 (2015): 28346–28352.
- Niu X, et al. Metal-organic framework based nanozymes: promising materials for biochemical analysis. Chemical Communications 56 (2020): 11338-11353.
- Korschelt K, Tahir MN, Tremel W. A Step into the Future: Applications of Nanoparticle Enzyme Mimics. Chemistry (Weinheim an der Bergstrasse, Germany) 24 (2018): 9703-9713.
- Wang X, Hu Y, Wei H. Nanozymes in bionanotechnology: From sensing to therapeutics and beyond. Inorganic Chemistry Frontiers 3 (2016): 41-60.
- Wang P, et al. Nanozymes: A New Disease Imaging Strategy. Frontiers in Bioengineering and Biotechnology 8 (2020): 1–10.
- Razlivina J, et al. DiZyme: Open-Access Expandable Resource for Quantitative Prediction of Nanozyme Catalytic Activity. Small 18 (2022): 1-10.
- Gao L, et al. Intrinsic peroxidase-like activity of ferromagnetic nanoparticles. Nature Nanotechnology 2 (2007): 577-583.
- Wu Y, Darland DC, Zhao JX. Nanozymes-hitting the biosensing “target”’, Sensors, 21 (2021).

 Impact Factor: * 2.9
Impact Factor: * 2.9 Acceptance Rate: 78.36%
Acceptance Rate: 78.36%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 