Prediction of Ovarian Cancer Survival using Machine Learning: A Population-Based Study
Munetoshi Akazawa*, Kazunori Hashimoto
Department of Obstetrics and Gynecology, Tokyo Women's Medical University Adachi Medical Center, Adachi-ku, Kohoku 2-1-10, Tokyo, Japan
*Corresponding author: Munetoshi Akazawa, Department of Obstetrics and Gynecology, Tokyo Women's Medical University Adachi Medical Center, Adachi-ku, Kohoku 2-1-10, Tokyo, Japan.
Received: 20 July 2023; Accepted: 24 July 2023; Published: 04 August 2023
Article Information
Citation: Munetoshi Akazawa, Kazunori Hashimoto. Prediction of Ovarian Cancer Survival using Machine Learning: A PopulationBased Study. Journal of Women’s Health and Development 6 (2023): 68-74.
DOI: 10.26502/fjwhd.2644-288400109
View / Download Pdf Share at FacebookAbstract
Objective: Accurate prediction could lead to risk stratification of patients and can be used as a decision-making tool for adjuvant chemotherapy. This study aimed to predict the prognosis of ovarian cancer using machine learning models.
Materials and Methods: We included patients with epithelial ovarian cancer between 2004 and 2019 extracted from Surveillance, Epidemiology, and End Results (SEER) database. We predicted the 5-year overall survival of patients using 12 clinic-pathological variables. Two machine learning models including gradient boosting machine (XGBoost) and artificial neural network were compared with traditional logistic regression. After 5-fold cross validation, we evaluated the model performance using classification accuracy and area under the curve (AUC) of the receiver operation curve. The importance of the variables in the construction of the prediction models was evaluated.
Results: A total of 18,438 patients were included in the study. Among three prediction models, XGBoost exhibited the best performance, followed by artificial neural network and logistic regression. XGBoost achieved a class accuracy of 0.809 (95%CI: 0.807–0.810) and AUC of 0.808 (95%CI: 0.806–0.809). The class accuracy and AUC were 0.802 (95%CI: 0.794–0.809) and 0.797 (95%CI: 0.784–0.808) in artificial neural network, 0.791 (95%CI: 0.787–0.794) and 0.784 (95%CI: 0.780–0.786) in logistic regression, respectively. In the XGBoost model, the most important variables were “summary stage,” followed by “year of diagnosis” and “M classification”.
Conclusion: Using machine learning, we were able to predict the prognosis of ovarian cancer. The machine learning model showed better prediction performance than the logistic regression models.
Keywords
<p>Machine learning; Ovarian cancer; Prediction; Prognosis; Overall survival</p>
Article Details
Introduction
Ovarian cancer is the second most common gynecologic malignancy, the most common cause of gynecologic cancer death in the United States, and the fourth leading cause of cancer-related deaths in women [1-2]. The majority of ovarian malignancies are of the epithelial type and are diagnosed as late-stage disease. Approximately 75 percent of women have stage III or stage IV disease at diagnosis, and 70–90% of women with advanced stage disease recur within 18 months of diagnosis, which is associated with a poor prognosis [3]. Survival is directly related to disease stage; patients with stage I, II, III, and IV ovarian cancer have median 5-year survival rates of approximately 93%, 70%, 37%, and 25%, respectively [2].The high mortality rate is partly due to its non-typical symptoms, which are difficult to detect in the early stages, and its high invasiveness. Prediction of prognosis is one of the biggest challenges in cancer therapy. Accurate prediction could lead to the risk stratification and triage of patients, which could help guide additional treatment and follow-up strategies. Depending on the risk stratification for the prediction of prognosis, physicians could customize additional treatment in high-risk patients and reduce treatment and follow-up in low-risk patients. Additionally, the accurate prediction of prognosis could be used as an effective tool for decision-making by medical teams and patients. The numerical probabilities of cancer-specific death recurrence could provide patients with high-quality explanations. Historically, pathological characteristics and biomarkers have been studied as the main predictors for the prognosis of ovarian cancers. The role of carbohydrate antigen 125 (CA125) in the prognosis of patients with ovarian cancer has been widely acknowledged [4]. Serum CA125 levels were correlated with survival. Patients who survived for more than 5 years had a lower preoperative CA125 level than patients with a poor prognosis, and there was a higher survival probability in patients with a normal level of preoperative CA125. Similarly, human epididymis protein 4 (HE4) has been studied as a predictor of response to chemotherapy [5]. Regarding pathological characteristics, serous carcinoma, the most common epithelial subtype according to histological classification and high grade are important factors associated with worse prognosis. In addition, previous studies have identified several potential genes or biomarkers for predicting the prognosis of ovarian cancer, but their comprehensiveness and clinical application remain limited. The accuracy and effectiveness of these biomarkers in predicting chemotherapy responses differ among patients with various epidemic and clinical features. Therefore, a multimodal prediction model using clinico-pathological data for ovarian cancer is desired.
Currently, machine learning is considered a possible new predictive technique, and there is a growing interest in its use in prediction models in the medical field. Machine learning, an area of artificial intelligence, can identify patterns in large datasets and construct prediction models that output possible results from the given data. Machine learning models can detect nonlinear correlations in laboratory, demographic, and clinical parameters that cannot be detected by linear methods [6]. Using multiple variables, including numerical and imaginary data, an increasing number of studies have been performed in the medical field to predict individual prognosis in patients. In this study, we attempted to develop prediction models for machine learning algorithms using large amounts of clinicopathological data.
Materials and Methods
Study population and dataset
We included patients with ovarian cancers between January 1, 2004, and December 31, 2019, using the Surveillance, Epidemiology, and End Results (SEER) database (version 8.9.8, National Cancer Institute, USA). SEER is a database of the incidence and survival rates of cancer in the United States, which covers approximately 28% of the population of the United States [7]. The SEER database is national, with information from 18 states, and includes a high proportion of racial/ethnic minorities and foreign-born individuals owing to its targeted sampling strategy. The inclusion criteria were as follows:1) diagnosis of ovarian cancer according to the International Classification of Diseases for Oncology, 3rd Edition (ICD-O-3) or primary site as C56.9 (ovary), the year of diagnosis was between 2004 and 2019, and 3) malignant behavior. The exclusion criteria were as follows:1) ovarian borderline tumor, 2) non-epithelial ovarian cancer, and 3) unknown duration of survival or survival length <5 years in the case of survival. Access to the SEER database did not require ethical approval and was covered by an open access policy.
Variables used for the prediction
A total of 12 parameters were used as variables to predict prognosis:1) year of diagnosis, 2) age, 3) race, 4) pathological grade, 5) pathological type, 6) surgical summary, 7) T classification, 8) N classification, 9) M classification, 10) number of pelvic lymph nodes resected during the surgeries, 11) positive number of lymph nodes, and 12) tumor size. The clinical International Federation of Gynecology and Obstetrics (FIGO) stage was excluded because the TNM classification and clinical stage have a strong correlation, which could lead to multicollinearity disturbing the linear model construction. “Summary stage” was one of the categorical data in SEER database, dived into three classes; Localized, Regional and Distant. The Summary Stage is the most basic way of categorizing how far a cancer has spread from its point of origin, called the General Stage, California Stage, historic stage, and SEER Stage. The histological grades were in accordance with the American Joint Committee on Cancer (AJCC) and FIGO guidelines as follows: G1/well differentiated, G2/ moderately differentiated, G3/poorly differentiated, and G4/undifferentiated. We re-categorized these data into two categories: low-grade (G1) and high-grade (G2, G3, G4). Histological types were divided into serous carcinoma, endometrioid carcinoma, clear cell carcinoma, and mucinous carcinoma. The other types, such as transitional cell carcinoma, mixed cell carcinoma and squamous cell carcinoma, were categorized as “other”, and when the detailed pathologies were unknown, they were categorized as “unknown”. The prediction target was 5-year overall survival (OS), defined as the final outcome of the patients. The time from diagnosis to death or censoring was defined as the survival length. Continuous data, such as age and year of diagnosis, were transformed using z-score normalization. Categorical data were transformed into integer variables as ranked variables, which represented the ordering of the values. Categorical data included TNM classification and surgical stage. The other categorical data, which had no ranks, such as pathology, were transformed using one-hot encoding. Missing data were replaced with the values of the mode in each variable.
Development of machine learning model
We developed three machine learning models: logistic regression, artificial neural network, and gradient boosting machine (XGBoost). Artificial neural networks, frequently named as “deep learning,” are mathematical models mimicking the neurons in the human brain, consisting of multiple layers. Artificial neural networks have demonstrated great success in various predictive tasks, including image, audio, and text data. However, during the last decade, gradient boosting machine (XGBoost) has dominated predictive tasks consisting of tabular data and showed superior performance over artificial neural network [8]. XGBoost is an ensemble of weak traditional models, such as decision trees, and is considered the most powerful machine learning technique, especially for tabular data. XGBoost and artificial neural networks are the two major models among machine learning algorithms and are frequently compared on prediction performance [8].
After randomly assigning the data to the training data (80%) or validation data (20%), we performed 5-fold cross-validation. In cross-validation, the dataset was divided into five subgroups. Four of these were used as the training dataset, and the remaining one was used as the test dataset to estimate the prediction performance. The estimation was calculated five times, and the five results obtained were averaged to obtain a single estimate. The stratified k-cross validation method maintained the rate of survival groups in the training and test sets equal to that of the original dataset. Machine learning was implemented using Python (version 3.7) as a programming language using the scikit-learn (version 1.0) machine learning package and Keras (version 1.2.2). Our study was performed in accordance with the TRIPOD statement [9].
Evaluation technique of prediction performance
Model performance was evaluated using classification accuracy and area under the curve (AUC). Class accuracy was calculated as follows: (class accuracy) = (correctly predicted as survival in the survival groups) + (correctly predicted as death in the dead groups) / total number of cases. The AUC, which is the concordance index (C-index), measures the entire two-dimensional area under the receiver operating characteristic (ROC) curve. We generated 95% confidence intervals and p-values using the empirical bootstrap method with 1000 iterations. ROC curves were created by plotting the true positive rate (TPR) against the false positive rate (FPR) at various threshold settings. Calibration curves were also analyzed. Calibration curves were used to evaluate how calibrated a classifier was, and how the probabilities of predicting each class label differed. Calibration curves were created by plotting the observed frequency of each class against the average predicted probabilities of the models. Calibration curves can be used to measure whether the prediction models are erroneously estimated and over-fitted.
Results
Patients’ demographics
Overall, 18,438 patients with ovarian cancer were included in this study. Among the patients, 7,037 patents (38.1%) were alive and 11,401 patients (61.8%) were dead in 5-year overall survival. The patient selection process is shown in Figure 1 and each clinic-pathological variable in the population is summarized in Table 1. The mean age at diagnosis was 61.6 years (SD, 14.5 years). The mean age in the survival groups were 10 years younger than the dead groups (mean: 54.1 year vs. 65.1 year). Regarding race, Whites were 82.7%, Blacks were 6.5%, and the others comprised 10.7% of the study population. Most of the histological types were serous carcinoma (51.6%), followed by endometrioid carcinoma (9.3%), clear cell carcinoma (6.1%), and mucinous carcinoma (5.1%). The other types, which included transitional cell carcinoma, mixed cell carcinoma, and squamous cell carcinoma, were 5.7%. The detailed pathologies were unknown in 21.9% of cases, most of which were considered as serous carcinoma in a previous study. 8) Most of pathological grades were high-grade (94.1%), and few were low-grade (5.8%). Advanced ovarian cancer was frequent in the viewpoint of TNM classification, with T3 (56.6%) and M1 (30.1%). The median size was 9.9 cm and there were small differences between the survival and dead groups. The median time to event in patients who died of cancer was 23.0 months. The median time to event in the overall population was 114 months.
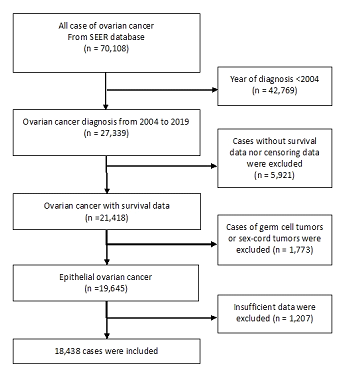
Figure 1: Patient data selection process.
|
N |
% |
|
|
|
|
|
|
All |
|
|
Alive |
Dead |
|
Number of cases |
18,438 |
|
|
7,037 |
11,401 |
|
Mean year at diagnosis |
2009 (3.8) |
2009 (3.1) |
2010 (4.1) |
||
|
Mean age at diagnosis |
61.6 (14.5) |
54.5 (13.3) |
65.1 (13.3) |
||
|
Race |
|||||
|
White |
15,256 |
82.7 |
82.4 |
82.9 |
|
|
Black |
1,192 |
6.5 |
4.6 |
7.6 |
|
|
Others |
1,990 |
10.7 |
12.9 |
9.4 |
|
|
Grade |
|||||
|
Low(G1) |
1082 |
5.8 |
12.4 |
98.2 |
|
|
High(G2, G3, G4) |
17356 |
94.1 |
87.5 |
1.7 |
|
|
Pathology |
|||||
|
Serous |
9,531 |
51.6 |
45.9 |
55.2 |
|
|
Endometrioid |
1,726 |
9.3 |
19.1 |
3.3 |
|
|
Clear cell |
1,134 |
6.1 |
9.2 |
4.2 |
|
|
Mucinous |
938 |
5.1 |
8.1 |
3.2 |
|
|
Others |
1061 |
5.7 |
9.4 |
3.4 |
|
|
Unknown(adenocarcinoma) |
4,048 |
21.9 |
8.1 |
30.5 |
|
|
Surgical summary |
|||||
|
Localized |
2316 |
12.5 |
27.3 |
3.4 |
|
|
Regional |
3641 |
19.7 |
33.7 |
11.1 |
|
|
Distant |
12481 |
67.7 |
38.9 |
85.4 |
|
|
T class |
|||||
|
T1 |
5,742 |
31.1 |
47.6 |
20.9 |
|
|
T2 |
2,257 |
12.2 |
16.1 |
9.8 |
|
|
T3 |
10,439 |
56.6 |
36.2 |
69.2 |
|
|
N class |
|||||
|
N0 |
14584 |
79.1 |
84.7 |
75.6 |
|
|
N1 |
3854 |
20.9 |
15.2 |
24.3 |
|
|
M class |
|||||
|
M0 |
12877 |
69.8 |
89.5 |
57.6 |
|
|
M1 |
5561 |
30.1 |
10.4 |
42.3 |
|
|
Mean tumor size (mm) |
99.0(75.6) |
104.6 (77.9) |
95.5 (73.9) |
||
|
Mean number of examined lymph node |
6.2 (11.2) |
10.6 (12.7) |
3.5 (8.7) |
||
|
Mean number of positive lymph node |
0.66 (2.8) |
0.51 (2.1) |
0.75 (3.2) |
||
|
Vital status |
|||||
|
Alive |
7,037 |
38.1 |
|||
|
Dead |
11,401 |
61.8 |
|
|
|
|
Data are mean (SD) or n(%) |
|||||
Table 1: Baseline characteristics in 5-year overall survival (OS) dataset
Performance of machine learning models
XGBoost showed the best performance, followed by the artificial neural network and logistic regression. XGBoost achieved a class accuracy of 0.809 (95%CI: 0.807–0.810) and AUC of 0.808 (95%CI: 0.806–0.809). The class accuracy and AUC were 0.802 (95%CI: 0.794–0.809) and 0.797 (95%CI: 0.784–0.808) in artificial neural network and 0.791 (95%CI: 0.787–0.794) and 0.784 (95%CI: 0.780–0.786) in logistic regression, respectively. The ROC curve is shown in Figure 2. As shown by the AUC and accuracy, XGBoost showed the best performance compared to the other two models. However, artificial neural network showed a prediction performance similar to that of XGBoost on ROC curves. The calibration curves demonstrated good agreement between XGBoost and logistic regression compared with the artificial neural network shown in Figure 3. Artificial neural network had the possibilities of “overfitting” in the data.
The importance of clinical variables
Because XGBoost is a tree-based classifier, we analyzed the importance of each clinical factor (12 features) in prediction (Figure 4). The importance was calculated through the construction of XGboost models, showing the relative weight of each variables on the prediction. The most important variables were “summary stage,” followed by “year of diagnosis” and “M classification”. The factors that are clinically not weighted, such as size or race, were not important for prediction by machine learning. In contrast, factors that are clinically weighted, such as serous adenocarcinoma or mucinous adenocarcinoma, were not important for prediction by machine learning.
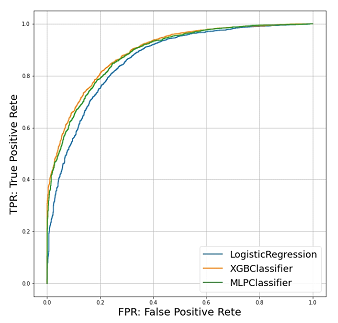
Figure 2: ROC curve of the models targeting 5-year OS.
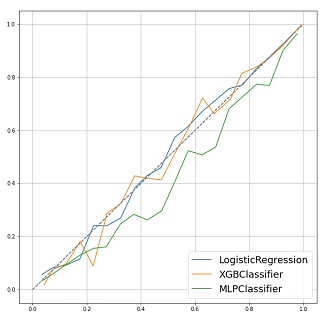
Figure 3: Calibration plots of models targeting 5-year OS
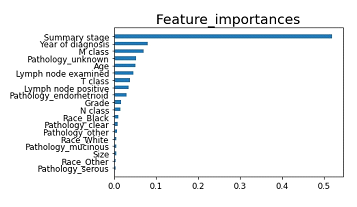
Figure 4: Feature importance of each variables in the prediction by XGboost.
Discussion
To our knowledge, this is one of the largest studies on the application of machine learning in the field of prognosis prediction for ovarian cancers. Although ovarian cancer prognosis largely depends on the response to chemotherapy, analysis of the patients’ background and pathological characteristics could help clarify cancer behavior. Thus, the model using pathological data and patient backgrounds could lead to the accurate prediction of prognosis, accompanied by detailed risk stratification of patients before adjuvant chemotherapy. In this study, we demonstrated the possibility of a machine learning model using large clinicopathological data for survival prediction in patients with ovarian cancer. Currently, the application of machine learning in cancer is a growing field with great potential, and various models have been analyzed for suitable variables and model algorithms. Lee et al. used 64,000 patients with prostate cancer in the SEER study and constructed machine-learning models to predict the prognosis of prostate cancer [10]. Their model achieved an AUC of 0.82 for predicting 10-year cancer-specific mortality in patients with prostate cancer. Bulten et al. reported an artificial intelligence (AI)-based Gleason grading for diagnosing prostate cancer in biopsies. In this study, the largest histopathology-based model competition, joined by 1,290 developers, was held in Kaggle, using 10,616 digitized prostate biopsies; the algorithms achieved AUC of 0.862 and 0.868 with expert uro-pathologists [11]. A new diagnosis and prediction system should be analyzed from a multifaceted aspect of cancer. The construction of accurate models could lead to risk stratification, which could provide cost-effective and personalized medicine. In the past few years, a growing number of studies have been published on prediction models using machine learning models for ovarian cancers (Table 2) [12-19]. However, most studies used a small dataset in a single institute, and machine learning models using large datasets are still difficult to find. In the largest study, Grimley et al. used 25,291 patients in the SEER database, reporting a machine learning model for the prediction of ovarian cancer prognosis [12]. They predicted the 5-year cancer-specific survival using six clinicopathological data, including TNM classes, age, pathology, and grade. The model achieved an AUC (C-index) of 0.739 compared to an AUC of 0.731 in the prediction using only the FIGO stage. In other studies, most used datasets from a single institute, and the size of the dataset was small, consisting of hundreds of cases. Not only tabular data but also image data can be used as variables for the prediction. Paik et al. reported machine learning models using 34 clinical variables for the prediction of 2-year overall survival [13]. The authors included the details of surgical site and the values of CA125 during the chemotherapy as variables. Using the dataset consisting of 1357 patients, their models achieved the AUC of 0.830. A variety of prediction model construction has been attempted in the medical field. The strength of our study is the size of the dataset and its high prediction performance. Considering that the machine learning model shows better performance with larger data, the hundreds of cases were not sufficient for AI studies, and studies using hundreds of thousands of cases were desired. From this viewpoint, our study and one previous study included ten thousands of cases using the national database. Compared with this previous study [12], our study used more variables and up-to-data machine learning algorithms, achieving a better performance (AUC: 0808 in our study vs. 0.739 in the previous study). In addition, we used major prediction models and demonstrated the importance of the variables. The results of our study could be efficient for model construction and the choice of variables in future studies. There is still the possibility of more accurate models in the same dataset, and future studies are desired to analyze suitable variables, algorithms, and preprocessing techniques. The limitations of this study are as follows. First, external validation was lacking because we performed internal validation. The “overfitting” is the biggest challenge in the construction of the machine learning models. In our study, the artificial neural network had the possibility of overfitting, as suggested by the calibration curve. To accurately evaluate the robustness of the prediction models and avoid overfitting, a fresh dataset should be prepared for external validation. Second, the clinical variables were limited. These variables were only used for the data included in the SEER study. Thus, other variables, such as preoperative imaging data, molecular profile, tumor markers, blood examination, and the detailed content of surgery were not used, which could be efficient as strong predictive factors. Third, the cohort distribution was heavily skewed toward advanced-stage disease and included more patients in the dead groups. Since we excluded patients who were alive within 5 years after diagnosis, more deaths occurred. This bias could lead the robustness of the model for the fresh dataset. Currently, the application of machine learning in cancer is a growing field with great potential, which could lead to the risk stratification, promoting optimum allocation of interventions and resources. However, more accurate prediction models are desired for clinical application and suitable variables and algorithms could be found through the trials of model construction. Future studies should analyze more efficient variables and suitable algorithms for the prediction of survival in the patients with ovarian cancers. In this study, we demonstrated the possibility of using machine learning to predict the prognosis in patients with ovarian cancer.
Table 2: Previous reports studying the machine learning model for the prediction of prognosis in the patients with ovarian cancer
Conflict of Interest Statement
All authors declare no conflict of interests.
Acknowledgements
The authors are grateful to Dr. Akimasa Ichinoe for technical assistance. We would like to thank Editage (www.editage.com) for English language editing.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 65 (2015): 87.
- Jayson GC, Kohn EC, Kitchener HC, et al. Ovarian cancer. Lancet 384 (2014): 1376-188.
- Peres LC, Sinha S, Townsend MK, et al. Predictors of survival trajectories among women with epithelial ovarian cancer. Gynecol Oncol 156 (2020): 459-466.
- Zhang M, Cheng S, Jin Y, et al. Roles of CA125 in diagnosis, prediction, and oncogenesis of ovarian cancer. Biochim Biophys Acta Rev Cancer 1875 (2021): 188503.
- Piovano E, Attamante L, Macchi C, et al. The role of HE4 in ovarian cancer follow-up: a review. Int J Gynecol Cancer 24 (2014): 1359-1365.
- Soren SS, Jeffrey CK, Amirhossein A, et al. Development and validation of an ensemble machine learning framework for detection of all-cause advanced hepatic fibrosis: a retrospective cohort study. Lancet Digit Health 4 (2022): e188-99.
- Kemi MD, Alfred R, Julie AS. Practical Guide to Surgical Data Sets: Surveillance, Epidemiology, and End Results (SEER) Database. JAMA Surg 153 (2018): 588-589.
- Ziv RS, Armon A. Tabular Data: Deep Learning is Not All You Need. arXiv 2106 (2021): 03253.
- Gary SC, Johannes BR, Douglas GA, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ 350 (2015): g7594.
- Changhee L, Alexander L, Ahmed A, et al. Application of a novel machine learning framework for predicting non-metastatic prostate cancer-specific mortality in men using the Surveillance, Epidemiology, and End Results (SEER) database. Lancet Digit Health 3 (2021): e158-65.
- Bulten W, Kartasalo K, Chen PHC, et al. Artificial intelligence for diagnosis and Gleason grading of prostate cancer: the PANDA challenge. Nat Med 28 (2022): 154-163.
- Grimley PM, Liu Z, Darcy KM, et al. A prognostic system for epithelial ovarian carcinomas using machine learning. Acta Obstet Gynecol Scand 100 (2021): 1511-1519.
- Paik ES, Lee JW, Park JK, et al. Prediction of survival outcomes in patients with epithelial ovarian cancer using machine learning methods. J Gynecol Oncol 30 (2019): 65.
- Shannon NB, Tan LLY, Tan QX, et al. A machine learning approach to identify predictive molecular markers for cisplatin chemosensitivity following surgical resection in ovarian cancer. Scientific Reports 1252 (2021): 16829.
- Avesani G, Tran HE, Cammarata G, et al. CT-Based Radiomics and Deep Learning for BRCA Mutation and Progression-Free Survival Prediction in Ovarian Cancer Using a Multicentric Dataset. Cancers 14 (2022): 855.
- Laios A, Katsenou A, Tan YS, et al. Feature Selection is Critical for 2-Year Prognosis in Advanced Stage High Grade Serous Ovarian Cancer by Using Machine Learning. Cancer Control 28 (2021): 1225.
- Li HM, Gong J, Li RM, et al. Development of MRI-Based Radiomics Model to Predict the Risk of Recurrence in Patients With Advanced High-Grade Serous Ovarian Carcinoma. AJR Am J Roentgenol 811 (2021): 664-675.
- Feng Y, Wang Z, Cui R, et al. Clinical analysis and artificial intelligence survival prediction of serous ovarian cancer based on preoperative circulating leukocytes. Journal of Ovarian Research 15 (2022): 64.
- Arezzo F, Cormio G, Forgia DL, et al. A machine learning approach applied to gynecological ultrasound to predict progression-free survival in ovarian cancer patients. Arch Gynecol Obstet 306 (2022): 2143-2154.


 Impact Factor: * 3.4
Impact Factor: * 3.4 Acceptance Rate: 78.89%
Acceptance Rate: 78.89%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 