Amplification of the CHD1 Gene for Molecular Sexing of Birds using Touchdown-PCR
Hernández-Olicón Aura Patricia1, Palma-Irizarry M2, Santiago-Hernández Juan Carlos1, Lozano-García Carlos1, Carreño-Durán Luis R1*
1Laboratorio de Diagnóstico Molecular, Departamento de Bioquímica, Escuela Nacional de Ciencias Biológicas, Instituto Politécnico Nacional, Mexico City, Mexico
2Subsecretaría de Gestión para la Protección Ambiental, Dirección General de Vida Silvestre, SEMARNAT, Mexico City, Mexico
*Corresponding Author: Carreño Durán Luis Ramón, Laboratorio de Diagnóstico Molecular, Departamento de Bioquímica, Escuela Nacional de Ciencias Biológicas, Instituto Politécnico Nacional, Prolongación de Carpio y, Calle Plan de Ayala s/n, Santo Tomás, Miguel Hidalgo, 11340, Mexico City, Mexico
Received: 13 January 2020; Accepted: 20 January 2020; Published: 23 January 2020
Article Information
Citation: Hernández-Olicón Aura Patricia, Palma-Irizarry M, Santiago-Hernández Juan Carlos, Lozano-GarcÃa Carlos, Carreño-Durán Luis R. Amplification of the CHD1 Gene for Molecular Sexing of birds using Touchdown- PCR. Arch Biochem Mol Biol 11 (2020): 017-026.
View / Download Pdf Share at FacebookAbstract
Birds are a vertebrated group with great ecological importance, it is estimated that 13% of species in the world are at risk of survival. Molecular monitory is an indispensable tool for wild populations conservation. CHD1 gene amplification through PCR has been commonly used for such purpose, however, it is essential to evaluate the primer set CHD1F/CHD1R applicability in a great diversity of species. Individuals from 11 different species of the Galliformes order were sexed, with the use of a touchdown PCR, the amplification products were observed in a 3% agarose gel, male specimens presented a single band corresponding to the CHD1-Z gene, while female specimens presented two bands corresponding to CHD1-Z and CHD1-W genes. The CHD1F/CHD1R primer set, with the use of a touchdown PCR, allowed the differentiation between two highly homologous genes and was efficient in sexing 11 selected species from the Galliformes order belonging to “El Nido” aviary.
Keywords
<p>Birds; Conservation; Sexing; touchdown- PCR; CHD1; Gene</p>
Article Details
Introduction
Birds are one of the most diverse classes of terrestrial vertebrates on the planet [1], which have great ecological importance since they have occupied all kinds of functions in ecosystems such as population control, pollination and seed dispersal, interactions with other animal species that favor survival as a whole and are part of the food chain [2]. Currently, it is estimated that there are approximately 18,000 bird species worldwide [3], however, 13% are at risk of survival, 20% of the world's birdlife is in Mexico and at least 392 species are endangered [4,5]. Due to its importance, the conservation of wild populations has become necessary, thus monitoring is a very useful and indispensable tool that provides information about the context under which the proposed problem is developed [6]. One of the important aspects for these studies is the sexing, which allows knowing the proportion between the number of females and males of a species and thereby estimating the possibility of maintaining the biological continuity through reproduction [7].
Phenotypically there are two types of birds: dimorphic birds, where the male and female have sexual dimorphism, and monomorphic birds, which do not have any character that allows the male to be differentiated from the female [8]. The latter are of special importance in conservation studies due to the difficulty to be sexed by traditional methods [4]. For this purpose molecular biology techniques have been implemented in more recent times due to their greater specificity and higher quality in the results obtained, since these resort to the identification of genes, or specific nucleotide sequences within the DNA of interest that function as genetic markers [7]. In birds, the determination of sex occurs in fertilization by inheritance of the sex chromosomes Z and W, which have evolved from an ancestral autosomal pair where the W chromosome lost most of its genes and acquired a smaller size in comparison with the Z chromosome, which did not change its genetic content. Due to this reason, it is that both have a certain degree of homology. One of the most used genes for the purpose of sexing in birds is CHD1, which is well conserved in a wide variety of species and is found on chromosomes Z and W, designated as CHD1-Z and CHD1-W respectively [7,8], and It is possible to amplify it by the PCR technique.
For this, a series of primers are used that recognize conserved exonic regions of the gene and amplify a less conserved intron that varies in size depending on whether it is the CHD1-Z gene or the CHD1-W gene, thereby observing the size differences in the amplified products of the CHD1 gene on the W and Z chromosomes for sex determination. For this, a series of primers are used that recognize conserved exonic regions of the gene and amplify a less conserved intron that varies in size depending on whether it is the CHD1-Z gene or the CHD1-W gene, thereby observing the size differences in the amplified products of the CHD1 gene on the W and Z chromosomes for sex determination [7,11]. One of the primer sets most used today is CHD1F/CHD1R, which has been implemented in a wide variety of species and it can increase its efficiency by applying the touchdown PCR scheme, which uses a first phase of amplification where the alignment temperature is gradually reduced, starting from certain temperature until reaching an optimal [7].
This protocol also has the potential to largely overcome the problems associated with high annealing temperatures required for some primer–template combinations and is particularly useful for difficult-to-amplify templates, such as those with extensive secondary structures, high % G + C islands in genomes and targets from organisms with more of 60% G + C content [9].Touchdown PCR is a technique to increase the likelyhood of amplifying the product, starts with a very high annealing temperature that decrease every cycle. It is usually applied if your primers tend to bind unspecifically at temperatures close (but lower) to the annealing temperature of the correct positions. That way, the correct product gets a head start.
Materials and Methods
Twenty-one blood samples were taken from 14 different species of birds, 10 of the order Galliformes and 4 of the order Psittaciformes. Sampling was performed by puncture in the brachial or tarsal veins depending on the species, which were provided by the “El Nido” aviary (Table 1) and kept in Microtainer® (BD) tubes at 4 °C. The sex of the sampled birds was provided and recorded during sampling, except for the Chamaepetes unicolor and Pipile natereri species. The DNA was extracted using the Perfect gDNA Blood Mini Kit for Human and Animal Blood (Eppendorf).
The reaction mixture for Touchdown PCR was 25 µL which consisted of Buffer for PCR 1x, 1 unit of recombinant Taq DNA polymerase (Invitrogen ™), 1.5 mM MgCl2, each of the primers in final concentration of 0.2 µM, 0.8 mM dNTP's, 1 µL of DNA, and nuclease-free water. The amplification for the CHD1F/CHD1R primers set of CHD1F 5´-TATCGTCAGTTTCCTTTTCAGGT-3´ and CHD1R 5´-CCTTTTATTGATCCATCAAGCCT-3´ was performed according to the touchdown PCR scheme referenced in previous reports [7] (Table 2).
The PCR products were visualized on a 3% agarose gel, carried out for 90 minutes at 90 V in 1x TBS buffer and subsequently revealed with ethidium bromide.
|
Order |
Specie |
Identification Code |
|
Galliformes |
Gallus gallus |
0A, 0B |
|
Crax blumenbachii |
1A, 1B |
|
|
Penelope purpurascens |
2A, 2B |
|
|
Pauxi pauxi |
3A |
|
|
Lophura nycthemera |
4A |
|
|
Mitu mitu |
5B |
|
|
Chrysolophus pictus pictus |
6A, 6B |
|
|
Lophura nycthemera lineata |
7B |
|
|
Chamaepetes unicolor |
8D, 8D |
|
|
Pipile natereri |
9D, 9D |
|
|
Syrmaticus reevesii |
10A |
|
|
Psittaciformes |
Amazona farinosa |
11D |
|
Amazona autumnalis |
12D |
|
|
Amazona auropalliata |
13D |
|
|
Ara militaris |
14D |
|
|
Anseriformes |
Anas platyrhynchos |
15B |
|
Anser anser |
16A |
|
|
Dendrocygna autumnalis |
17D |
|
|
Aix galericulata |
18A |
|
|
Accipitriformes |
Buteo jamaicensis |
19A* |
|
Parabuteo unicinctus |
20B* |
|
|
Falconiformes |
Caracara plancus |
21D |
|
Strigiformes |
Bubo virginianus |
22A* |
|
Gruiformes |
Grus vipio |
23D |
|
Balearica regulorum |
24B |
|
|
Grus canadensis |
25B* |
|
|
Piciformes |
Ramphastos sulfuratus |
26D |
|
Passeriformes |
Calocitta colliei |
27D |
Table 1: Sampled species in “El Nido” aviary.
|
Step |
Temperature |
Time |
|
Initial denaturation |
94 °C |
4 minutes |
|
Denaturation |
94 °C |
30 seconds |
|
Annealing |
57-50 °C |
45 seconds |
|
Extension |
72 °C |
45 seconds |
|
Repeat for 7 cycles |
||
|
Denaturation |
94 °C |
30 seconds |
|
Annealing |
50 °C |
45 seconds |
|
Extension |
72 °C |
45 seconds |
|
Final extension |
72 °C |
5 minutes |
|
Repeat for 30 cycles |
||
Table 2: Scheme for the CHD1F/CHD1R primers set using the Touchdown PCR technique.
Results
Standardization of the touchdown PCR technique
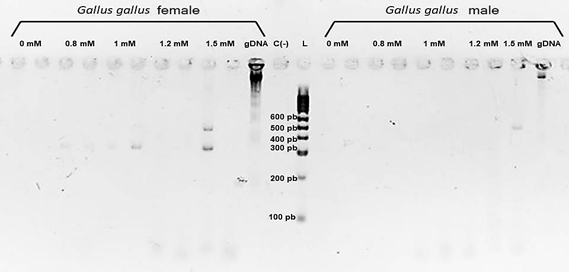
The effect of MgCl2 concentration on the touchdown PCR reaction was evaluated, using female and male Gallus gallus DNA, and it was observed that amplification occurred more efficiently at a final concentration of 1.5 mM MgCl2 in the PCR reaction mixture, additionally the bands obtained using this concentration are observed with much better definition (Figure 1).
Figure 1: MgCl2 concentration curve. A band of 400-500 bp is observed for the male of the species and two bands of 400-500 bp and 300-400 bp for the female, banding patterns are best observed at a concentration of 1.5 mM of MgCl2. L= HyperLadder™ (Bioline), C (-)= Negative control, gDNA= genomic DNA.
The technique was subsequently applied using both Gallus gallus DNA and DNA from the rest of the sampled species; it was observed that under the described reaction conditions. Amplification occurred from a DNA concentration of 8.3 ng/μL.
Results obtained previously showed that amplification does not occur if the touchdown PCR scheme is not used, even if the working conditions are the same.
Results of the application of the touchdown PCR technique in birds of different species
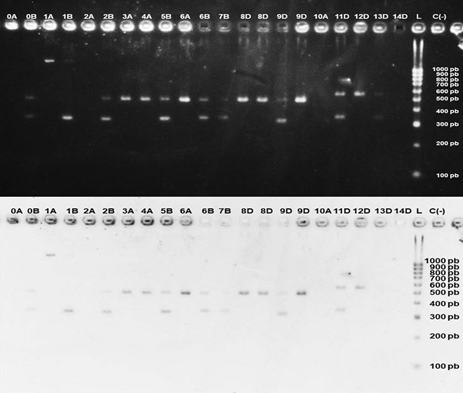
In all the species it was observed that the males presented a single band of 400-500 bp, while the females presented two bands, one of 400-500 bp and another of 300-400 bp, the only exceptions were the Crax blumenbachii species, where the amplicon corresponding to the CHD1-Z gene had a size greater than 1000 bp and the species belonging to the order Psittaciformes, where the same amplicon had a size between 500-600 bp, slightly larger than that of the birds of the order Galliformes.
In some individuals, very faint bands were observed, although they allowed adequate sexing, such as the female of Gallus gallus (0B), the female of Chrysolophus pictus pictus (6B), Lophura nycthemera lineata (7B), and Amazona auropalliata (13D). Additionally, in three of the analyzed samples no amplification was observed, corresponding to the male of Penelope purpurascens (2A), Syrmaticus reevesii (10A), and Ara militaris (Figure 2).
Figure 2: Results obtained for the different bird species sampled from the orders Galliforme and Psittaciforme. (a) Normal image, (b) negative image. Identification codes can be consulted in Table 1. Species sampled in the “El Nido” aviary. L= HyperLadder™ (Bioline), C (-)= Negative control.
In four of the sexed species there was no clear pattern, which were Gallus gallus, Crax blumenbachii, Chrysolophus pictus pictus, and Syrmaticus reevesii (Table 3).
|
Order |
Species |
Identification code |
Phenotypic sexing |
Sex determined by touchdown PCR |
|
Galliformes |
Gallus gallus |
0A |
Male |
Male |
|
0B |
Female |
Female |
||
|
Crax blumenbachii |
1A |
Male |
Male |
|
|
1B |
Female |
Female |
||
|
Penelope purpurascens |
2A |
Male |
ND |
|
|
2B |
Female |
Female |
||
|
Pauxi pauxi |
3A |
Male |
Male |
|
|
Lophura nycthemera |
4A |
Male |
Male |
|
|
Mitu mitu |
5B |
Female |
Female |
|
|
Chrysolophus pictus pictus |
6A |
Male |
Male |
|
|
6B |
Female |
Female |
||
|
Lophura nycthemera lineata |
7B |
Female |
Female |
|
|
Chamaepetes unicolor |
8D |
Male |
Male |
|
|
8D |
Male |
Male |
||
|
Pipile natereri |
9D |
Female |
Female |
|
|
9D |
Male |
Male |
||
|
Syrmaticus reevesii |
10A |
Male |
ND |
|
|
Psittaciformes |
Amazona farinosa |
11D |
Female |
Female |
|
Amazona autumnalis |
12D |
Male |
Male |
|
|
Amazona auropalliata |
13D |
Female |
Female |
|
|
Ara militaris |
14D |
ND |
ND |
Table 3: Correlation between the sex of the birds provided and the sex determined by the touchdown PCR technique.
Discussion
The amplification of the CHD1-Z and CHD1-W genes using the primers set CHD1F/CHD1R with the touchdown PCR technique produced two amplicons of different sizes, which were subsequently related to the sex of the bird in question. Regarding the males, a single band with a size between 400-500 bp corresponding to the CHD1-Z gene was obtained, because the males have a ZZ chromosomal pair and only possess the CHD1-Z gene, females, on the other hand, have a ZW pair and therefore two bands were obtained, one with a size of 400-500 bp corresponding to the CHD1-Z gene and another with a size 300-400 corresponding to the CHD1-W gene. The agarose gel banding patterns for the species Gallus gallus, Crax blumenbachii, Penelope purpurascens, Pauxi pauxi, Lophura nycthemera, Mitu mitu, Chrysolophus pictus pictus, Lophura nycthemera lineata, and Syrmaticus reevesii corresponded to the sex established and provided by the "El Nido" aviary, demonstrating that the present set was specific and accurate for the determination of sex in birds.
In the samples of the species, Chamaepetes unicolor and Pipile natereri, the sex of the birds was unknown by the aviary workers and according to the analysis of the results obtained, the four individuals were male since only one band was observed between the 400 -500 bp. In the species, Gallus gallus, Crax blumenbachii, Chrysolophus pictus pictus, and Syrmaticus reevesii the banding pattern did not occur properly because the amount of DNA extracted by the kit method could have been in excess or in very low concentration, thus the PCR reaction was not carried out properly. It was considered necessary to evaluate the concentration of DNA obtained by the kit before carrying out the amplification in order to determine if any previous dilution was necessary to favor the amplification [7,10,11]. Except for Gallus gallus, all other species in which touchdown PCR was applied with the primers set CHD1F/CHD1R have not been reported in previous work, demonstrating that this set can be applied to other species of birds of the order Galliformes.
In the species of the order Galliformes the males presented a band with a size between 400-500 bp with respect to the molecular size marker, corresponding to the CHD1-Z gene. Whereas the females presented two bands, one of 400-500 bp corresponding also to the CHD1-Z gene and another of 300-400 bp corresponding to the CHD1-W gene, because it is heterozygous and therefore has both variants of the CHD1 gene. On the other hand, in the species of the order Pisttaciformes the same pattern of banding for males and females was observed, except that in the case of the amplicon of the CHD1-Z gene it was slightly larger compared to the species of the order Galliformes, since this was located between 500-600 bp (Error! Reference source not found.). This is because the CHD1 gene, although it is highly conserved in most birds, it presents variations in size between species that are phylogenetically less related to each other.
This situation derives from the evolutionary process in birds, where in some species the genetic information is gradually modified, losing or earning nucleotide sequences, while others have retained it almost entirely [10,12]. It was observed that for the male and female of the Crax blumenbachii species the amplicon corresponding to the CHD1-Z gene was larger than 1000 bp, indicating that the fragment amplified for this species differs in size compared to the rest of the species of the order Galliformes analyzed in the present study, demonstrating that even among phylogenetically related species belonging to the same order there may be variations in the information contained in the CHD1 gene. This banding pattern had not been reported for this particular species, thus it is an important result that should be taken into account for future studies and that increases the relevance of evaluating the applicability of the primers set CHD1F/CHD1R in a greater diversity of species of birds, since as presented in this case, there may be other species with different banding patterns when applying this set and the touchdown PCR technique.
In the cases of the male Penelope purpurascens (2A), Syrmaticus reevesii (10A), and Ara militaris (14D), an adequate amplification of the gene was not observed due to a possible contamination with DNAses or proteases that would affect the sample or the amplification reaction. This situation can occur from the sampling because it was not taken under controlled conditions to avoid microbiological contamination or could have occurred during the extraction process or the reagent mixture for the PCR touchdown reaction. Of these three individuals, only Ara militaris is of unknown sex, so it will be necessary to repeat the procedure from DNA extraction to ensure obtaining a result. This situation is evident in other species such as Gallus gallus, Lophura nycthemera lineata, Pipile natereri and Amazona auropalliata, where there was amplification but poorly visible bands were obtained. The reaction conditions for all samples were the same but the amount of DNA extracted was different depending on the bird and the sex of the individual in question. The sex of 13 individuals was known; all belonging to the order Galliformes, which when undergoing the PCR touchdown technique had a 100% concordance with the sex provided by the workers of the “El Nido” aviary (Error! Reference source not found.) with the exception of the males of Penelope purpurascens and Syrmaticus reevesii.
Whereas, the other 8 individuals of unknown sex; two of the species Chamaepetes unicolor, two of Pipile natereri, one of Amazona farinosa, one of Amazona autumnalis, and one of Amazona auropalliatase were successfully sexed comparing their banding patterns mainly with that of Gallus gallus and subsequently with the rest of the species. Additionally, the species with unknown sex were also monomorphic species, since they did not have any phenotypic characteristic that allowed distinguishing the male from the female; therefore, the results presented for these species are important to verify the efficiency of the primers set CHD1F/CHD1R in the application of the Touchdown PCR technique and that could be implemented for the sexing of a wide diversity of species with the purpose of carrying out adequate monitoring in wild populations of ecological interest and supporting their conservation.
Conclusions
The primers set CHD1F/CHD1R through touchdown PCR allowed the differentiation of two highly homologous genes and was efficient for sexing individuals of the 11 species selected from the order Galliformes from the “El Nido” aviary. This study opens the possibility of being able to implement it in other bird species that are of interest for conservation both in Mexico and in the world.
Acknowledgments
The authors would like to thank “El Nido” aviary for providing the birds blood samples and Salvador Trujillo González for their technical help.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Aritio LB, Atlas temático de zoología, vertebrados. Barcelona, España. Idea Books (1996): 101-106.
- Ali Tabur M, y Ayvaz Y. Ecological Importance of Birds. In: 2nd International Symposium on Sustainable Development, Sarajevo 2010: 560-565.
- Barrowclough GF, Cracraft J, Klicka J y Zink RM. How Many Kinds of Birds Are There and Why Does It Matter?, PLOS One 11 (2016):
- Red List 2017: seabirds starving, songbirds trapped, hope for pelican and kiwis. Available online: https://www.birdlife.org/worldwide/news/red-list-2017-seabirds-starving-songbirds-trapped-hope-pelican-and-kiwis. (accessed on 13 November 2019).
- NOM-059-SEMARNAT-2010. Diario oficial de la federación, Distrito Federal. Available online: https://www.gob.mx/profepa/documentos/norma-oficial-mexicana-nom-059-semarnat-2010.
- Marzluff JM. Worldwide urbanization and its effects on birds. In: Marzluff J.M., Bowman R., Donnelly R. (eds) Avian Ecology and Conservation in an Urbanizing World. Springer, Boston, MA 2001: 19-47.
- James Chun-I Lee, Li-Chin Tsai, Pei-Yi Hwa, Chia-Ling Chan, Alex Huang, Shih-Chien Chin, Lih-Chiann Wang, Jun-Tsong Lin, Adrian Linacre, Hsing-Mei Hsieh. A novel strategy for avian species and gender identification using the CHD gene. Molecular and Cellular Probes 24 (2010): 27-31.
- Fridolfsson AK, Cheng H, Copeland NG, Jenkins NA, Liu HC, Raudsepp T, Woodage T, Chowdhary B, Halverson J, Ellegren H. Evolution of the avian sex chromosomes from an ancestral pair of autosomes. Proc Natl Acad Sci USA 95 (1998): 8147-8152.
- Korbie DJ, & Mattick JS. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nature Protocols 3 (2008): 1452–1456.
- Prum RO, Berv JS, Dornburg A, Field DJ, Townsend JP, Lemmon EM, y Lemmon AR. A comprehensive phylogeny of birds using targeted next-generation DNA sequencing. Nature 526 (2015): 569-573.
- González Suurbach S. Determinación del sexo en aves. Universidad de La Coruña, La Coruña, España. 2014: 62-65
- EJ Oakes. Lekking and the Evolution of Sexual Dimorphism in Birds: Comparative Approaches. The American Naturalist 140 (1992): 665-684.
Article Views: 4456
Journal Statistics




Discover More: Recent Articles
Grant Support Articles
General Biology-Reproduction and Comparative Development,
Lyon Catholic University (UCLy),
Ecole Pratique des Hautes Etudes,
Lyon, France
Editor In Chief