Structure and Dynamics of the Penicillin-Binding Protein 3 from Staphylococcus Epidermidis Native and in Complex with Cefotaxime and Vaborbactam
Martin Schwinzer*, 1, Hévila Brognaro1, Holger Rohde2 Christian Betzel*, 1
1University of Hamburg, Institute for Biochemistry and Molecular Biology, Laboratory for Structural Biology of Infection and Inflammation, c/o DESY, 22603, Hamburg, Germany
2University Medical Center Hamburg-Eppendorf, Institute for Medical Microbiology, Virology and Hygiene, Martinistrasse 52, 20246 Hamburg, Germany
*Corresponding author: Martin Schwinzer, University of Hamburg, Institute for Biochemistry and Molecular Biology, Laboratory for Structural Biology of Infection and Inflammation, c/o DESY, 22603, Hamburg, Germany. Christian Betzel, University of Hamburg, Institute for Biochemistry and Molecular Biology, Laboratory for Structural Biology of Infection and Inflammation, c/o DESY, 22603, Hamburg, Germany.
Received: 11 October 2023; Accepted: 20 October 2023; Published: 29 November 2023
Article Information
Citation: Martin Schwinzer, Hévila Brognaro, Holger Rohde Christian Betzel. Structure and Dynamics of the Penicillin-Binding Protein 3 from Staphylococcus Epidermidis Native and in Complex with Cefotaxime and Vaborbactam. International Journal of Applied Biology and Pharmaceutical Technology. 14 (2023): 60-75.
View / Download Pdf Share at FacebookAbstract
Staphylococcus epidermidis (Se) is a highly abundant gram-positive bacterium predominantly found on human skin. It poses significant threat to immunocompromised patients due to its ability to form biofilms on medical devices. In this study, we determined and refined the first structure of a penicillin-binding protein (PBP) from SePBP3 to a resolution of 2.5 Å. The apo form analysis revealed a shift in the head sub-domain (HSD) relative to the homologous structure in Staphylococcus aureus (Sa). The discovery led us to conduct an analysis of SePBP3’s flexibility applying also X-ray solution scattering. Additional molecular dynamics simulations revealed a rigid transpeptidase domain paired with a flexible pedestal domain, displaying an open and closed interface between the N-terminal anchor domain and the HSD. Furthermore, we solved and refined the structure of SePBP3 in complex with the β-lactam antibiotic cefotaxime and the boron-based antibiotic vaborbactam to 2.51 and 2.3 Å resolution, respectively. Both ligands demonstrated high binding affinity, as confirmed by ITC measurements. Since Staphylococcus epidermidis is a potential major contributor to nosocomial infections, the new structural insights into a highly affine PBP capable of binding various classes of antibiotics provide valuable information for future drug design investigations.
Keywords
<p>Staphylococcus epidermidis, penicillin-binding protein, antimicrobial resistance, boron-based anitibiotic, X-ray structure</p>
Article Details
1. Introduction
Staphylococcus epidermidis is classified within the coagulase-negative staphylococci (CoNS) subset and, under standard physiological conditions, functions as a commensal organism ubiquitously present within the human cutaneous microbiota [1, 2]. However, it has evolved as an opportunistic pathogen and is now unfortunate the etiological agent for implant-associated infections. Its inherent capacity for biofilm formation on medical prosthetics, including catheters and surgical apparatus, poses significant risks to patients with compromised immune systems, such as those diagnosed with HIV or those receiving immunosuppressive regimens post-transplantation [3-10]. Nosocomial infections, coupled with the burgeoning resistance to antibiotic regimens, often necessitate the surgical excision of medical implants, including artificial cardiac valves, cerebrospinal fluid shunts, intravascular catheters, and joint prosthetics [11, 12]. Alarmingly, a preponderance of these hospital-acquired Staphylococcus epidermidis isolates has manifested multidrug-resistance, further exacerbating the challenges associated with therapeutic interventions [13-15]. Peptidoglycan, ubiquitously present in bacterial cell walls, is an indispensable structural component synthesized by penicillin-binding proteins (PBPs). Its paramount function is to confer cellular stability, safeguarding the bacterial cell against external osmotic pressures [16-18]. Exclusively prevalent in prokaryotic cells, PBPs, localized within the periplasmic space, play a pivotal role in bacterial cell wall biosynthesis, making them an optimal target for antimicrobial agents [19-20]. PBPs can be taxonomically classified based on molecular weight into high-molecular weight (HMW) and low-molecular weight (LMW) categories. Notably, HMW PBPs are multidomain enzymes tasked with peptidoglycan crosslinking. These enzymes can be further stratified into Class A and Class B. HMW Class B PBPs are characterized by the presence of a transpeptidase domain and an N-terminal pedestal, which is postulated to mediate interactions with auxiliary proteins. The process of transpeptidation, integral to bacterial cell wall integrity, facilitates the crosslinking of the peptide stem, composed of D-alanine residues, from an emerging peptidoglycan chain to an extant chain integrated within the cell wall matrix [16, 21,22]. In both gram-positive and gram-negative bacteria, the transpeptidation domains are characterized by highly conserved motifs. Motif 1, denoted as [S]XX[K], incorporates a catalytic and indispensable serine residue, succeeded by two variable amino acids and culminating in a lysine residue. This motif is oriented towards the catalytic cavity and anchored on a α-helix. The second motif, [S/Y]X[N/C], is situated on a loop interlinking two α-helices, while motif 3, defined as [K/H][T/S]GT, is located on a β-sheet. Both motifs 2 and 3 straddle motif 1 [21]. PBPs are inherently multidomain enzymes. For instance, PBP3 from Staphylococcus aureus (SaPBP3) comprises three distinct domains (PDBID: 3vsk). The N-terminal domain, colloquially termed the pedestal, integrates a membrane anchor, a linker domain, and a functionally enigmatic head sub-domain. In contrast, the C-terminus is typified by the transpeptidase domain [23]. The N-terminal anchor domain is also referred to as the disordered "sugar tong" domain, postulated to play a role in PBP polymerization [24,25]. While dimerization of PBPs has been evidenced in E. coli, no such dimeric form of SaPBP3 has been observed either by mass spectrometry or analytical ultracentrifugation analyses [23-26]. Previous studies discussed the role of SaPBP3 in Staphylococcus aureus including its role regarding the cell division. Staphylococcus aureus cells with inactivated SaPBP3 showed abnormal cell morphology and septa defects while treated with sub-minimal inhibitory concentration (MIC) of antibiotics. SaPBP3 is not essential for the cell under normal conditions, but a loss of this protein activated inhibition of the remaining PBPs. It is hypothesized that SaPBP3 protects other PBPs by its high affinity against β-lactam antibiotics [27]. Methicillin resistance in bacteria is normally explained by the MecA gene encoding PBP2a with low affinity towards β-lactam antibiotics but clinical Staphylococcus epidermidis strains lacking the MecA gene with mutated SePBP3 showed an increased methicillin resistance [28-29]. β-lactam antibiotics are suicide inhibitors of PBPs and divided into five classes, the narrow spectrum penicillins, broad spectrum penicillins, cephalosporins, monobactams and carbapenems [30]. They all are mimicking the peptide stem of the substrate and thereby able to bind covalently at the catalytic serine of the transpeptidase domain [31]. Nosocomial strains are able to escape the treatment of β-lactams antibiotics by expressing β-lactamases which are able to hydrolyze the β-lactam ring or by expressing low affine PBPs [28,32,33]. An additional and by now proven approach of new antibiotic are boron-atom containing antibiotics, which are able to covalently bind to the active site serine of serine proteases by mimicking the tetrahedral transition state of β-lactam acylation and diacylation [34]. Vaborbactam, Taniborbactam for instance are known boron-atom containing β-lactamase inhibitors and a novel study reveals also inhibition of β-lactamases by Bortozomib and Ixazomib, both know as proteasome inhibitors, in application of anti-cancer treatment [35,36]. In this study we presented the structure of SePBP3 from Staphylococcus epidermidis and explore the question of the role of SePBP3 as a potential decoy receptor by demonstrating the affinities towards the β-lactam antibiotic cefotaxime and the boron-based antibiotic vaborbactam. Furthermore, we revealed the flexibility of the different domains in relation to the potential protein-protein-interface between the anchor domain and the head-subdomain (HSD).
2. Material and Methods
2.1 Molecular cloning, protein expression and purification
The truncated Δ47Penicillin-binding protein 3 gene was molecularly cloned into a pET28a plasmid, a process facilitated by BioCat GmbH. For protein expression, this recombinant plasmid was introduced into chemically competent E. coli BL21 DE3 cells. These transformed cells were cultured in LB medium supplemented with 50 µg/ml kanamycin at 37°C. Upon reaching an optical density (OD600) of 0.6, the culture temperature was adjusted to 30°C, followed by the induction of protein expression via the addition of 1 mM IPTG. Expression was allowed to continue for an additional 3 hours post-induction. Post-expression, cells were harvested by centrifugation at 4,000 x g, then resuspended and washed in phosphate-buffered saline (PBS). Cell lysis was achieved by sonicating the pellet in a lysate/equilibration buffer containing 50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, and set at pH 8.0. The resultant cell debris was separated by centrifuging at 20,000 x g, and the clarified supernatant was then loaded onto a Ni-NTA affinity chromatography column for purification. After a washing step with 20 mM imidazole, the protein of interest was eluted using 250 mM imidazole. The eluted protein solution was dialyesed overnight in a buffer comprising 20 mM Tris-HCl, 100 mM NaCl, and adjusted to pH 8.0. Following, the protein solution was further purified using size exclusion chromatography. Peak fractions corresponding to the desired protein were pooled and concentrated using a 50 kDa Amicon concentrator, achieving a final concentration of 20 mg/ml.
2.2 Protein crystallization
For crystallization experiments a monodisperse solution of SePBP3 at a concentration of 20 mg/ml was prepared; Dynamic light scattering was applied prior to crystallization experiments to analyse the dispersity in solution. X-ray suitable crystals were obtained by the hanging drop vapor diffusion technique at 20°C. 1 µl of the protein solution was mixed with an equal volume of crystallization agent solution composed of 2.2 M AmSO4 at pH 8. This combination was set against a 1 ml reservoir liquor with identical composition. Crystals were observed within 3 days. Prior to data collection, these crystals were cryo-protected using a solution of 2.2 M AmSO4, 12% glycerol, and pH 8. For co-crystallization experiments, the aforementioned protocol was followed with a slight modification: 0.1 µl of an inhibitor solution (100 mM concentration, solubilized in 5% DMSO) was added to the drop containing the protein solution.
2.3 Diffraction data collection and structure determination
X-ray diffraction data were collected applying synchrotron radiation collected at 100 K at the beam line P13 (PETRA IIII, DESY) [46]. This data were subsequently processed with the XDS program package [47]. Data collection and refinement parameters are summarized in Table 1. The phase problem was solved by molecular replacement, using SaPBP3 from Staphylococcus aureus (PDID: 3vsk) and using the Phaser-MR's one-component interface program embedded within the Phenix software suite (version 1.19.2-4158) [48,49]. The structure refinement was performed with the Phenix.refine tool [48][50], while the structure modelling was performed with WinCoot 0.9.8.7.1 [51].
Table 1: X-ray data collections and refinements
|
Data collection |
|||
|
SePBP3 |
SePBP3 - Cefotaxime |
SePBP3 - Vaborbactam |
|
|
X-ray source |
P13, Petra III, DESY |
P13, Petra III, DESY |
P13, Petra III, DESY |
|
Detector |
EIGER 16M |
EIGER 16M |
EIGER 16M |
|
Space group |
P41212 |
P41212 |
P41212 |
|
Cell dimensions |
|||
|
a,=b, c (Å) |
83.67, 305.16 |
83.21, 305.28 |
83.15, 306.14 |
|
Wavelength (Å) |
0.97625 |
1.033 |
0.82655 |
|
Resolution (Å) |
80.69- 9.01 (5.88-2.5) |
49.24 - 9.02 (2.512-2.51) |
80.24 - 9.20 (2.37-2.3) |
|
Total reflections |
71280 (7833) |
70884 (7997) |
89288 (8349) |
|
Total unique reflections |
46353 (2318) |
38258 (11304) |
48019 (3160) |
|
Wilson B-factor (Å2) |
56.24 |
90.32 |
36.29 |
|
Rmeas |
0.014 (0.464) |
0.026 (0.227) |
0.030 (0.283) |
|
CC1/2 |
0.999 (0.996) |
0.999 (0.956) |
0.9642 (0.872) |
|
I/σI |
16.8 (3.1) |
20.5 (3.0) |
25.2 (4.8) |
|
Completeness |
99.7 (95.2) |
0.999 (0.950) |
98.1 (99.3) |
|
Refinement |
|||
|
Reflection used |
35596 |
36231 |
63242 |
|
Rwork |
0.22 |
0.23 |
0.2 |
|
Rfree |
0.26 |
0.25 |
0.23 |
|
No. atoms |
|||
|
Protein |
4870 |
4944 |
4870 |
|
Ligand/ion |
- |
26 |
20 |
|
Water |
60 |
5 |
338 |
|
Average B-factor (Å2) |
|||
|
Protein |
71.73 |
110.21 |
50.9 |
|
Ligand/ion |
- |
81.05 |
35.97 |
|
Water |
66.32 |
119.58 |
46.34 |
|
R.m.s. deviations |
|||
|
Bond lenghts (Å) |
0.0082 |
0.018 |
0.0079 |
|
Bond angles (°) |
1.04 |
1.641 |
0.995 |
|
Ramachandran |
|||
|
favored (%) |
93.02 |
84.34 |
94.29 |
|
allowed (%) |
5.55 |
12.34 |
3.62 |
|
outliers (%) |
1.43 |
3.32 |
2.09 |
|
PDBID |
8C5B |
8C5W |
8C5O |
2.4 SAXS data collection and analyses
SAXS data were collected at the beamline P12 (PETRA III, DESY) at a temperature of 293.15 K employing the in-batch measurement approach. A protein concentration of 5 mg/ml was applied first, with systematically performing serial dilutions to achieve concentrations as low as 1 mg/ml, using the dialysis buffer as the diluent. The acquired data from these experiments were subsequently processed and visualized using the BioXTAS RAW software, complemented by further analysis with ATSAS GNOM [52,53].
2.5 ITC measurements and analyses
The protein utilized for isothermal titration calorimetry (ITC) experiments was prepared following previously detailed protocols and was subsequently concentrated to a final concentration of 40 µM. ITC measurements were performed at a temperature of 293.15 K using the Malvern PEAQ-ITC instrument. The ligand intended for titration was prepared at a concentration 10 times higher than the protein concentration, ensuring comprehensive saturation during titration. The experiment commenced with an initial mock injection of 0.4 µl, serving as a system equilibration step. This was succeeded by 13 systematic injections, each dispensing a volume of 3 µl over 6 seconds, with a 150-second interlude between injections to guarantee equilibration and full heat dissipation. A consistent stirring speed of 750 rpm was maintained throughout the procedure to facilitate rapid mixing and even heat distribution. The data accrued from these experiments were processed utilizing NITPIC (version: 1.2.7). Further thermodynamic analysis and data fitting were executed with SEDPHAT (version 15.2b), and data representation was accomplished via the Gussi software package (version: 1.3.0). Each interaction between the protein and a unique ligand was assayed individually to ensure distinct binding profiles [54-56].
2.6 Molecular dynamics simulation
The crystallographic structure of SePBP3 served as the starting point for our molecular dynamic simulation studies. Prior to simulation, the protein structure underwent rigorous preparation using the protein preparation workflow embedded within the Schroedinger's Maestro suite [57]. As an initial step, missing hydrogen atoms were systematically added to the protein structure to ensure its chemical completeness. To refine the hydrogen-bonding network, we employed PROPKA, optimizing H-bond assignments based on predicted pKa values and the protein's local environment. Subsequent restrained minimization was performed utilizing the OPLS4 force field, with any crystallographic waters beyond the first solvation shell being pruned [58]. For the molecular dynamics (MD) simulation, the solvated system was constructed using the TIP4P water model, encapsulated in an orthorhombic periodic boundary box [59]. To ensure no direct interaction with its periodic images, a minimum distance of 20 Å was maintained between the protein and the box edges. Moreover, to closely mimic physiological conditions, a salt concentration of 150 mM NaCl was introduced. The MD simulations were executed under the NPT ensemble (N: constant particle number; P: constant pressure at 1.01325 bar; T: constant temperature at 300 K). The integration was carried out over a 100 ns trajectory with configurations saved every 20 ps, yielding a total of 250 frames for subsequent analysis [60].
3. Results and Discussion
3.1 Transpeptidase and Pedestal - Linker, Anchor and Head-Sub-domain
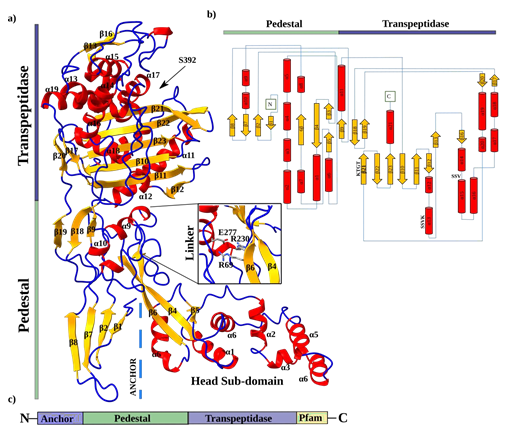
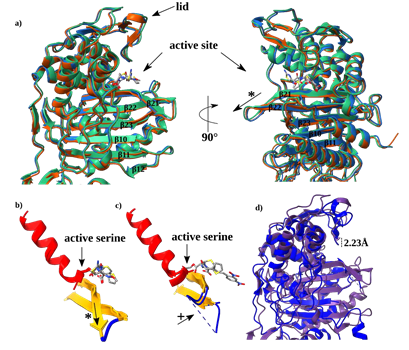
SePBP3 exhibits structural homology close to the canonical domains found in class B PBPs, as previously described by Sauvage [22]. The structure can be divided into two prominent domains: a transpeptidase domain and a pedestal domain consisting of three sub-domains. These sub-domains include the N-terminal anchor domain, a head sub-domain, and a linker domain (Figure 1a). The topology plot provides a 2-dimensional representation of these domains and highlights particular the interdomain linkers (Figure 1b). The transpeptidase domain consists of ten β-sheets (β12-17, β20-23) and twelve α-helices (α8-10), exhibiting the characteristic structure of the DD-alanine transpeptidase / transpeptidase-like body. At the core of this domain, a central structural ladder is formed by five β-sheets (β10, β11, β21, β22, β23). Several α-helices surrounding this central ß-structure. Importantly, all three highly conserved fingerprint motifs, namely the active-site serine (SSVK), SSN motif, and KTGT motif, are present in close hydrogen-bonding proximity distance to each other. The active-site serine is located at the end of helix 13, the SSN motif is located between α-helices 14 and 15, and the KTGT motif is part of β-sheet 21. It is postulated that the active serine (S392) is deprotonated by the nearby located base of the lysine (K395) of the first motif. Subsequently, the active form of the serine is able to perform a nucleophilic attack of the carbonyl carbon of the peptide stem of the peptidoglycan [21]. Within the linker domain, there are three β-sheets (β9, β18, β19) and three α-helices (α8-10). Notably, Goffin and Ghuysen identified a motif unique to class B Penicillin-binding proteins located within this linker region [21]. This motif is postulated to serve as a stabilization factor and is composed of two arginine residues and one glutamine residue. Despite their dispersed positions within the protein sequence, these amino acids (R69, R230, E277) are in a 3-dimensional arrangement (Figure 1a). In SePBP3, the N-terminal domain, which is typically connected to the membrane anchor, consists of four antiparallel β-sheets (β1, β2, β7, β8). This particular arrangement is also referred to a disordered sugar tong domain; however in SePBP3, this region does not display a particular disorder but rather exhibits lower electron density [23-25]. This observation suggests the possibility of this domain being inherently flexible. Yoshida et al. reported the observation of dimerization between two molecules within the crystal lattice specifically at this particular domain. This data suggest that the domain might serve as a dimerization domain, however, a dimeric form of the protein could not be detected in solution [23]. SePBP3 at this domain forms none contact between two molecules in the crystal lattice as well as none in solution.
Figure 1: Cartoon and topology plot of SePBP3. a) The secondary structure is indicated in different colours. Loops are depicted in blue, sheets in yellow and helices in red. The membrane anchor is indicated by dashed lines. The linker domain is highlighted as it contains a 3-dimensional motif. b) The topology plot is depicted in the same colour scheme and highlights the interdomain connections. c) A simplified cartoon of SePBP3 includes the position of the membrane anchor.
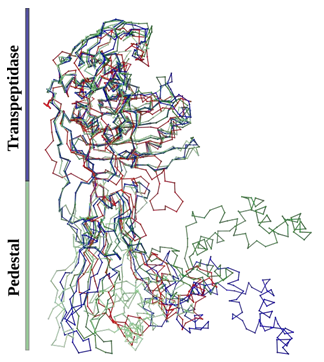
Consistent with the characteristics of PBPs, the transpeptidase domain displays a highly conserved backbone. This conservation is not limited to HMW and LMW PBPs; β-lactamases also manifest this structural feature. Conversely, pedestal domains exhibit significant variability, rendering them non-alignable. Figure 2 underscores that, even among proteins of the same class, the pedestals of distinct class B PBPs diverge in terms of position and amino acid length. A superposition of the domains and corresponding RMSD values (considering the entire protein versus the transpeptidase domain exclusively) highlight this observation. For SePBP3 and SaPBP3, the Cα RMSD value is rather high, 7.6 Å when considering the entire protein structure, and it narrows to 0.35 Å for just the transpeptidase domain. In the case of SePBP3 and PaPBP3, the RMSD values are 3.6 Å and 1.5 Å for the entire protein and the transpeptidase domain, respectively. When comparing SePBP3 and SpPBP2B, the corresponding RMSD values are 6.8 Å for the entire protein and 0.8 Å for the transpeptidase domain. The sequence homology varies notably among SePBP3 and its counterparts: SaPBP3, PaPBP3, and SpPBP2B. SaPBP3 have 85% homology, PaPBP3 22%, and SpPBP2B 35%. These percentages are consistent with the RMSD values observed for the entire protein. A sequence alignment, emphasizing both the low-affinity residues and the highly conserved motifs, is illustrated in Figure 3.
Figure 2: Comparative Cα backbone analysis of penicillin-binding protein structures: Conserved transpeptidase and linker domains with varied N-terminal anchor and head sub-domains. Superimposed crystal structures of penicillin-binding proteins from Staphylococcus epidermidis (SePBP3; blue), Staphylococcus aureus (SaPBP3/3vsk, dark-green), Pseudomonas aeruginosa (PaPBP3/7auh, green), and Streptococcus pneumoniae (SpPBP2B/2wad, red).
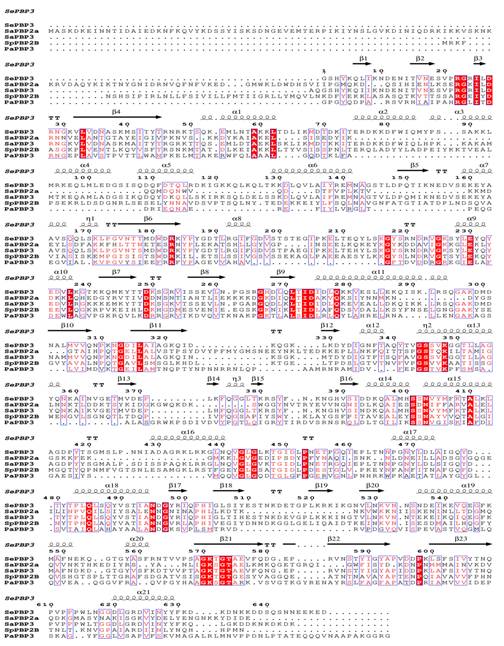
Figure 3: Sequence alignment of SePBP3 from Staphylococcus epidermidis, SaPBP2a and SaPBP3 from Staphylococcus aureus, SpPBP2B from Streptococcus pneumoniae and PaPBP3 from Pseudomonas aeruginosa including secondary structure of SePBP3. Identical amino acids are highlighted in red. Secondary structure elements of SePBP3 are placed on top of the sequences. Figure was created by ESPript 3.0 [38].
3.2 Rigid Transpeptidase Domain and Flexible Pedestal
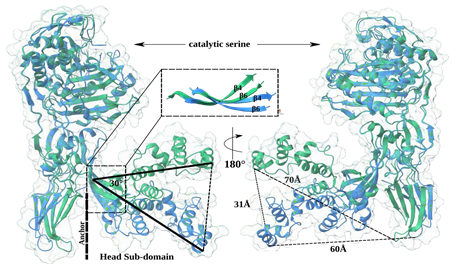
The structural flexibility of PBPs has emerged as a significant topic of interest due to discussions to identify potential interaction partners. Particular the interplay between shape, elongation, division, and sporulation (SEDS) proteins and class B PBPs has been found to play a crucial role in facilitating the entire process of peptidoglycan crosslinking. In this process, the membrane anchor and the N-terminal anchor domain of PBPs interact with membrane proteins such as RodA, which functions as a glycosyltransferase. Furthermore, the region between the head sub-domain and the N-terminal anchor domain is proposed to serve as an interface for proteins associated with RodA, such as MreC [18]. Additional variations exist, like the low-affinity PBP2a possesses a unique allosteric centre that is located between the pedestal domains and has been shown to be involved in β-lactam antibiotic resistance [28]. The precise role of the HDS in class B PBPs remains uncertain. Besides the potential protein-protein interaction function it is suggested that the head sub-domain functions as a steric hindrance, preventing the transpeptidase from approaching too closely to the cell membrane [39]. The pedestal region has been observed to exhibit different conformations, like for PBP5 from Enterococcus faecium, which has been described in closed and open forms [40]. We made a similar observation by comparing the crystal structures of SePBP3 and SaPBP3 (PDBID: 3vsk). Both structures share a similar structural arrangement in their head sub-domains, which comprises seven α-helices (α1-7) and three β-sheets (β4-6). While the domains alone align well (RMSD = 0.81 Å [37]), the presence of bends in β-sheets 4 and 6 induces a displacement of the entire domain. The HSD undergoes a shift of approximate 30° and shows a distance of approximate 30 Å between the two forms. Furthermore, the distance from the top of the HSD to the N-terminal anchor domain increases by 10 Å from SePBP3 to SaPBP3. In spite of, in this analysis, a closed conformation was not observed, two distinct conformational states were identified, namely an extended and a compressed state (Figure 4). Interestingly, AlphaFold predicts confidently (90 > pLDDT > 70) a closed conformation for both proteins of this interface area (ENTRYID: SePBP3: Q5HNZ7; SaPBP3: A0A389VKU4). Not only the HSD, but the entire pedestal appears to be flexible, as evidenced by the lack of a clearly interpretable electron density for the N-terminal anchor domain. SaPBP3 displays enhanced electron density at this position, likely due to crystal contacts situated at the same location, which might restrict flexibility at this domain. Generally, the N-terminal anchor domain in PBPs often exhibits weak electron density, especially when the crystal packing space permits movement, like for EfPBP4 from Enterococcus faecalis [40].
Figure 4: Superimposed structures of Penicillin-Binding Protein 3 (PBP3) from Staphylococcus epidermidis (SePBP3; blue) and Staphylococcus aureus (SaPBP3, green). The head sub-domain (HSD) exhibits two distinct states, an extended and a compressed conformation towards the N-terminal anchor domain. SaPBP3, with a separation of 70 Å, shows a greater distance than that observed for SePBP3, with 60 Å. The distance between the two ends of the head subdomain is 31 Å and the angle between them is approximate 30°. The origin of the curved conformation is located in β-sheets 4 and 6. No well-interpretable electron density was found in the N-terminal anchor domain of SePBP3, suggesting relative high mobility of this region.
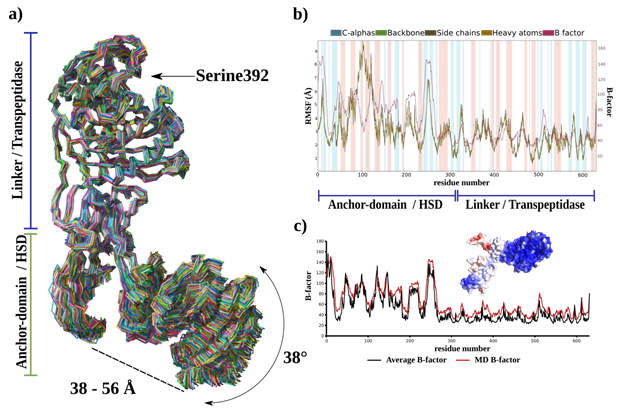
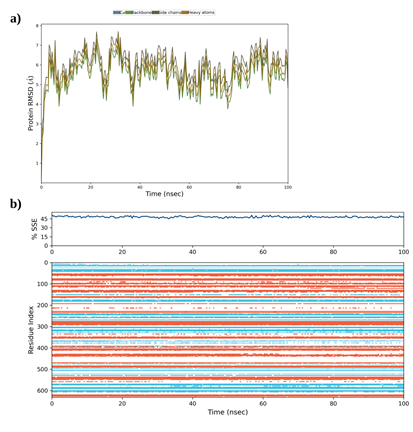
- a) Superimposition of 250 frames extracted over a 100 ns simulation duration. Delineations of flexible and rigid regions, span between the N-terminal anchor domain and HSD and angular mobility of the HSD are annotated. b) Root Mean Square Fluctuation (RMSF) and B-factor plots characterizing the fluctuations of Cα, backbone, side chains, and heavy atoms for each residue. Secondary structure elements are highlighted with different colours: α-helices are highlighted in blue and β-sheets in pink. Regions of flexibility and rigidity are annotated in accordance with their corresponding domains. c) Comparative B-factor plot derived from both diffraction data (average B-factor) and the MD simulation. The surface representation of SePBP3 is color-coded to emphasize variations in B-factor values, highlighting structural dynamics.
The dynamics of SePBP3 using a 20 ns molecular dynamics simulation, was further explored, applying the Schroedinger's Maestro suite [57]. As illustrated in Figure 2 for various PBPs, the alignment of the α carbons (Cα) of SePBP3 across all simulation frames shows a similar phenomenon: a stable transpeptidase body accompanied by a robust linker region. The N-terminal anchor domain demonstrates only modest shifts, since it is situated in a crystal packing region that permits such movements, which could explain the observed weak electron density in this region for SePBP3. Within the crystal lattice, the head sub-domain is close to neighbouring proteins, constraining its freedom of movement. Nonetheless, after 100 ns of simulation time, SePBP3 exhibits a notable displacement of 38° for this domain. The distance between the N-terminal anchor domain and the HSD fluctuates from 38 to 56 Å, yielding both closed and open conformations (Figure 5a).
Moreover, the linker domain displays stability, possibly owing to the postulated 3-dimensional motif that may act as a stabilizing entity. The calculated Root Mean Square Fluctuation (RMSF) indicates a flexible N-terminal and a rigid C-terminal transpeptidase (Figure 5b), while the B-factor values obtained from the simulation calculations closely mirror those obtained from the diffraction data refinement, which is underscoring the congruence between our simulation data and empirical X-ray data (Figure 5c). The RMSD values oscillate within a range of 5 to 7 Å, suggesting intrinsic flexibility of the protein structure. Concurrently, the preservation of secondary structure elements throughout the 100 ns simulation duration ensures that elevated RMSD values are not attributed to the loss of these structural components (Supplementary Figure 1). These observations are also in line with the previously observed domain shift of PBP5 from Enterococcus faecium discussed to be resulting of varied crystal packing [40]. Thereby the interface between the N-terminal anchor domain and HSD can adopt both 'open' or 'closed' states. This duality might allow and modulate potential interactions with binding partners, while ensuring the transpeptidase body retains its rigidity.
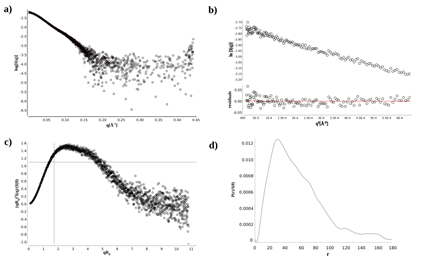
We conducted also small-angle X-ray scattering (SAXS) experiments to further validate the flexibility of SePBP3 in solution. The dimensionless Kratky plot analysis confirmed a non-globular folding of SePBP3, as evidenced by the peak for globular proteins being shifted to the right with higher qRg values, exceeding the threshold of 1.73 (Figure 6c), fitting to the overall shape of class B PBPs [22]. Furthermore, the SAXS data indicate an elongated conformation of SePBP3 in solution, evident from the extended tail observed in the distance distribution function curve [P(r)], which challenges the largest particle dimension (Dmax) determination (Figure 6d), making it unfeasible to precisely validate the shift of the head sub-domain (HSD) across different conformational states (table 2) [41].
Table 2: Guinier anylsis and P(r) function analysis of SAXS measurements
|
Guinier analysis |
P(r) function analysis |
||
|
Rg (Å) |
43.41 ± 0.23 |
44.62 ± 0.37 |
Rg (Å) |
|
I(0) (arb.) |
0.06 ± 1.47e-4 |
0.06 ± 2.12e-4 |
I(0) (arb.) |
|
q-range (1/Å) |
0.0032-0.02555 |
0.0098-0.4443 |
q-range (1/Å) |
|
qRg range |
0.139-1.109 |
unclear |
Dmax (Å) |
|
R2 |
0.982 |
0.968 |
R2 |
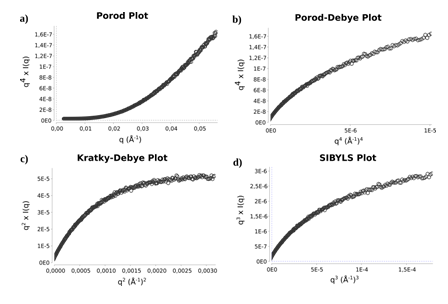
Subsequently, we applied the Porod-Debye law to assess the compactness and flexibility. According to the law the scattering intensity of a compact particle decays as q-4, and for some small range of q a plot of q4I(q) vs. q4 will achieve a plateau, explained by the scattering particle with a sharp homogenous electron density contrast between the particle and the solvent (Figure 7b). On the other hand, proteins that have considerable flexibility show a slower slope than q−4 and therefore do not reach a plateau in the Porod-Debye region, instead they show a plateau on the Kratky-Debye plot (q2 I(q) versus q2), as observed for the SePBP3 SAXS data analysis (Figure 7c), hence validating and confirming the flexibility of the protein [42,43].
The complementary observations suggest that SePBP3 is a protein with inter-domain flexibility. The transpeptidase domain demonstrates a rigid body behaviour, as it is structurally conserved and aligns well with other class B PBPs. Similarly, the linker domain exhibits similar traits with structurally conserved motifs such as R69, R230, and E277 in SePBP3. In contrast, the pedestal region of different class B PBPs crystal structures often exhibits weak electron density at the N-terminal anchor domain. Furthermore, the HDS displays variable spatial orientations towards the N-terminal anchor domain, which enables an open and closed state of the interface at the region.
3.3 Open active site facilitates susceptibility of the active serine to vaborbactam and cefotaxime
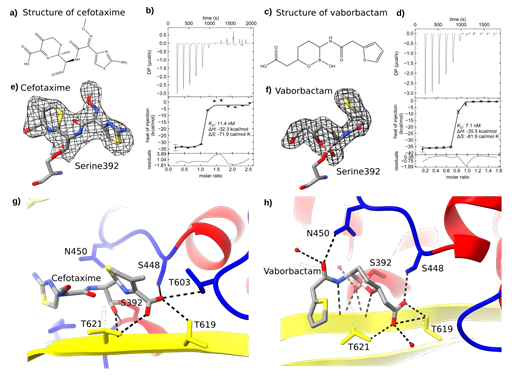
We investigated the SePBP3 complex structures of β-lactam and boron-based antibiotics utilizing co-crystallization assays. The antibiotics were introduced at a concentration of 100 mM, applying 5% DMSO as solvent. Notably, the crystallization time extended significantly, ranging from initial 3 days to 3-4 weeks. Despite this extension, the protein crystallized with same cell constants and crystal symmetry as observed for the native structure. The SePBP3- cefotaxime complex diffracted to 2.51 Å resolution, while the SePBP3-vaborbactam complex diffracted to 2.3 Å (Table 1). An intriguing observation was that the electron density map for cefotaxime did not correlate with a closed β-lactam ring configuration but suggested an open conformation. This observation suggests that SePBP3 catalyzed the antibiotic during the co-crystallization process. (Figure 8a, c, e and f)
Elucidating the putative role of SaPBP3 as a decoy receptor led us to gauge the affinity of both ligands to it. Recent publications reveal that SaPBP3 plays a vital role in cell survival and the missing activity of SaPBP3 applying sub-MIC levels induces an alteration in cellular morphology [27]. Intriguingly, SePBP3 exhibited a substantial affinity towards both cefotaxime, a third-generation cephalosporin antibiotic and vaborbactam, a non-suicidal β-lactamase inhibitor. The approximate dissociation constant (KD) for these interactions was measured at a proximate value of 10 (nM) (Figure 8b and d). This notably low KD intimates a robust affinity of SePBP3 for not only canonical β-lactam inhibitors, but also for boron-based inhibitors. All three highly conserved motifs participate in the binding process, as they covalently link the catalytic serine 329 (motif 1/SSNK), also engaged in hydrogen bonds with Serine 446 (motif 2/SSN), and Threonine 619 and 621 (motif 3/KTGT) (Figure 8g and h).
Figure 8: Binding Characterization of Cefotaxime and Vaborbactam with SePBP3. Panel (a) illustrates the open-form structure of cefotaxime, catalyzed by SePBP3 during co-crystallization, while panel (c) depicts the structure of vaborbactam, which is not catalysed. Both ligands bind with high affinity: (b) cefotaxime with a KD- value of 11.4 nM and (d) vaborbactam with a KD- value of 7.1 nM. The ligands are embedded into the electron density at sigma levels of e) 2.0 for cefotaxime and f) 1.0 for vaborbactam in the omit map, respectively. Dashed lines represent the hydrogen bonds for (g) cefotaxime and for (h) vaborbactam.
To characterize the relative high affinity of SePBP3 towards cefotaxime and vaborbactam we analysed the active site cavity in detail. The binding of cefotaxime and vaborbactam is not inducing any conformational changes. This is normally not the case, as binding of compounds in active site regions are supported by the flexibility of distinct loops and sheets. For instance, there are discussions regarding the flexibility of the β sheets and the corresponding interconnection regional loops. These connection regions are found to be flexible in NgPBP2 (PDBID: 6xqv) from Neisseria gonorrhoeae and in mutants of SpPBP2B (PDBID: 2wad) from Streptococcus pneumonia, as no traceable electron density was observed, whereas in drug-sensitive strains those regions exhibit suitable electron density [44, 45]. In SePBP3 the interconnection region is rather short but well defined, the main difference is the longer β21 sheet, which bends away from the active Serine 392 (Figure 9a). Its position relative to the Serine 392 is located much lower than in SaPBP2a (PDBID: 1vqq) [28]. This shielding position of the β sheet in PBP2a is the main reason for the relative low affinity, as the serine must bend up to bind a ligand. In SePBP3 the position of β21 sheet maintained at the same location, also the active serine is constantly in an exposed position (Figure 9 b and c), explaining the susceptibility of the active serine to vaborbactam and cefotaxime. The overall structure of SePBP3 has a relative wide-open active site cavity, contributing to SePBP3s substrate binding profile. For instance, the lid region of the cavity remains unaffected during ligand binding. When compared to SaPBP2a, the lid is positioned approximate 2 Å higher, influencing the accessibility of the cavity (Figure 9 d). SePBP3 not only displays a rigid active site but also maintains a open state of the active cavity, indicating it as a highly affine PBP.
Figure 9: Comparative structural analysis of SePBP3 and SaPBP2a. a) Superposition of SePBP3 in its apo form (blue) and complexed with cefotaxime (orange) and vaborbactam (green). A structural superimposition of b) the SePBP3 apo structure and the complexes with cefotaxime and vaborbactam and c) SaPBP2a in apo form and in complex with penicillin G. d) A superimposition of the transpeptidase domains between SePBP3 (blue) and SaPBP2a (violet) reveals an open lid, exhibiting a deviation of 2.23 Å, highlighting the open cavity of SePBP3. The asterix (*) symbol highlights the bending of β-sheet 21 and the plus (+) symbol the flexible loop region.
Figure 9: Comparative structural analysis of SePBP3 and SaPBP2a. a) Superposition of SePBP3 in its apo form (blue) and complexed with cefotaxime (orange) and vaborbactam (green). A structural superimposition of b) the SePBP3 apo structure and the complexes with cefotaxime and vaborbactam and c) SaPBP2a in apo form and in complex with penicillin G. d) A superimposition of the transpeptidase domains between SePBP3 (blue) and SaPBP2a (violet) reveals an open lid, exhibiting a deviation of 2.23 Å, highlighting the open cavity of SePBP3. The asterix (*) symbol highlights the bending of β-sheet 21 and the plus (+) symbol the flexible loop region.
4. Conclusion
We described the structure of the penicillin-binding protein PBP3 from Staphylococcus epidermidis and compared it in detail with known homologous structures. The data presented provide new insights for and about the structure and dynamics of SePBP3 from Staphylococcus epidermidis, supporting future drug discovery investigations. Our data and results show that SePBP3 exhibits a rigid and in part extended active site cavity, a structural configuration suggestive of heightened susceptibility to antibiotics. The complementary structural and biochemical analyses elucidate the notable affinity of SePBP3 for both the canonical β-lactam cefotaxime and the boron-based antibiotic vaborbactam. Also, the observed binding affinities are in the nanomolar range, emphasizing that SePBP3's has superior capability to bind inhibitors more effectively compared to other known PBPs. Furthermore, our study and data obtained highlight the inherent flexibility of SePBP3 and precisely mapped the location of the inter-domain flexibility within the protein structure. While the exact functional significance of the pedestal domain in class B PBPs, particularly within the head-sub domain, remains till now enigmatic, our data highlight a pivotal aspect: the rigidity of the transpeptidase domain remains unaltered by the mobility of the pedestal domain. This stability is potentially conferred by the rigid linker region, which provides anchorage and stabilizes the transpeptidase domain. Considering these structural attributes, the mobile pedestal may provide the interface for potential interacting partners, whereas the transpeptidase domain retains its autonomy for crosslinking reactions.
Acknowledgments
The authors acknowledge financial support obtained from the Cluster of Excellence 'Advanced Imaging of Matter' of the Deutsche Forschungsgemeinschaft (DFG) - EXC 2056 - project ID 3907159 and support obtained by BMBF (Federal Ministry of Eduction and Research, Germany) via projects 05K19GU4 and 05K20GUB.
Conflict of Interest
The authors declare no conflict of interest.
References
- Becker K, Heilmann C and Peters G. Coagulase-negative staphylococci. Clin. Microbiol. Rev 27 (2014): 870-926.
- Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat. Rev. Microbiol 16 (2018): 143-155.
- Otto M. Staphylococcus epidermidis - the ‘accidental’ pathogen,” Nat Rev Microbiol 7 (2009): 555-567.
- Olson ME, Todd DA, Schaeffer CR, Paharik AE, Van Dyke MJ, Büttner H, et al. Staphylococcus epidermidis agr quorum-sensing system: Signal identification, cross talk, and importance in colonization. J Bacteriol 196 (2014): 3482-3493.
- Büttner H, Mack D, Rohde H. Structural basis of Staphylococcus epidermidis biofilm formation: Mechanisms and molecular interactions. Front. Cell. Infect. Microbiol 5 (2015): 1-15, 2015.
- Mack D, Rohde H, Harris LG, Davies AP, Horstkotte MA, Knobloch JKM. Biofilm formation in medical device-related infection. Int. J. Artif. Organs 29 (2006): 343-359.
- Darouiche RO. Treatment of infections associated with surgical implants. N. Engl J Med 350 (2004): 1422-1429.
- Goldmann DA, Pier GB. Pathogenesis of infections related to intravascular catheterization. Clin. Microbiol. Rev 6 (1993): 176.
- Geipel U, Herrmann M. The infected implant: bacteriology. Unfallchirurg 108 (2005): 961-975.
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science (80-. ) 284 (1999): 1318-1322.
- Rupp ME, Archer GL. Coagulase-negative staphylococci: pathogens associated with medical progress. Clin. Infect Dis 19 (1994): 231-245.
- Rohde H, Burdelski C, Bartscht K, Hussain M, Buck F, Horstkotte MA, et al. Mack. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol 55 (2005): 1883-1895.
- Lee JYH, Monk IR, da Silva AG, Seemann T, Chua KYL, Kearns A, et al. Howden. Global spread of three multidrug-resistant lineages of Staphylococcus epidermidis. Nat. Microbiol 3 (2018): 1175-1185.
- Li M, Wang X, Gao Q, Lu Y. Molecular characterization of Staphylococcus epidermidis strains isolated from a teaching hospital in Shanghai, China. J. Med. Microbiol 58 (2009): 456-461.
- Du X, Larsen J, Li M, Walter A, Slavetinsky C, Both A, et al. Peschel. Staphylococcus epidermidis clones express Staphylococcus aureus-type wall teichoic acid to shift from a commensal to pathogen lifestyle Nat Microbiol 6 (2021): 757-768.
- Höltje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev 62 (1998): 181-203, 1998.
- Zapun A, Vernet T, Pinho MG. The different shapes of cocci. FEMS Microbiol Rev 32 (2018): 345-360.
- Sjodt M, Rohs PDA, Gilman MSA, Erlandson SC, Zheng S, Green AG, et al. Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex. Nat. Microbiol 5 (2020): 813-820.
- Ghuysen JM, Frere JM, Leyh-Bouille M, Nguyen-Distèche M, Coyette J. Active- site-serine D-alanyl-D-alanine-cleaving-peptidase-catalysed acyl-transfer reactions. Procedures for studying the penicillin-binding proteins of bacterial plasma membranes. Biochem. J 235 (1986): 159.
- Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev 36 (1972): 407.
- Goffin C, Ghuysen J-M. Multimodular Penicillin-Binding Proteins: An Enigmatic Family of Orthologs and Paralogs. Microbiol. Mol. Biol. Rev 62 (1998): 1079.
- Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev 32 (2008): 234-258.
- Yoshida H, Kawai F, Obayashi E, Akashi S, Roper DI, Tame JRHT, et al. Crystal structures of penicillin-binding protein 3 (PBP3) from methicillin-resistant Staphylococcus aureus in the apo and cefotaxime-bound forms. J. Mol. Biol 423 (2012): 351-364.
- Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48 (2020): D265, 2020.
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. The Pfam protein families database. Nucleic Acids Res 40 (2012).
- Zijderveld CAL, Aarsman MEG, Den Blaauwen T, Nanninga Penicillin- binding protein 1B of Escherichia coli exists in dimeric forms. J. Bacteriol 173 (1991): 5740-5746.
- Pinho MG, De Lencastre H, Tomasz A. Cloning, Characterization, and Inactivation of the Gene pbpC, Encoding Penicillin-Binding Protein 3 of Staphylococcus aureus. J. Bacteriol 182 (2000): 1074.
- Lim D, Strynadka NCJ. Structural basis for the β lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat. Struct. Biol 9 (2002): 870-876.
- Petinaki E, Dimitracopoulos G, Spiliopoulou I. Decreased affinity of PBP3 to methicillin in a clinical isolate of Staphylococcus epidermidis with borderline resistance to methicillin and free of the mecA gene. Microb. Drug Resist 7 (2001): 297-300.
- Bush K, Bradford PA. β-Lactams and β-Lactamase Inhibitors: An Overview,” Cold Spring Harb. Perspect. Med 6 (2016).
- Tipper DJ, Strominger JL. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proc. Natl. Acad. Sci USA 54 (1965): 1133.
- Bush K, Bradford PA. Interplay between β-lactamases and new β-lactamase inhibitors. Nat. Rev. Microbiol 175 (2019): 295-306.
- Munita JM, Arias CA. Mechanisms of Antibiotic Resistance. Microbiol. Spectr 4 (2015): 464-472.
- Fu H, Fang H, Sun J, Wang H, Liu A, Sun J, et al. Boronic Acid-based Enzyme Inhibitors: A Review of Recent Progress. Curr. Med. Chem 21 (2014): 3271-3280.
- Hecker SJ, Reddy KR, Totrov M, Hirst GC, Lomovskaya O, Griffith DC, et al. Discovery of a cyclic boronic acid β-lactamase inhibitor (RPX7009) with utility vs class A serine carbapenemases. J. Med. Chem 58 (2015): 3682-3692.
- Perbandt M, Werner N, Prester A, Rohde H, Aepfelbacher M, Hinrichs W, et al. Structural basis to repurpose boron-based proteasome inhibitors Bortezomib and Ixazomib as β-lactamase inhibitors. Sci. Reports 12 (2022): 1-12.
- Holm L, Laiho A, Törönen P, Salgado M. DALI shines a light on remote homologs: One hundred discoveries. Protein Sci 32 (2023): 1-18.
- Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42 (2014): W320-W324.
- Macheboeuf P, Contreras-Martel C, Job V, Dideberg O, Dessen A. Penicillin binding proteins: Key players in bacterial cell cycle and drug resistance processes.FEMS Microbiol. Rev 30 (2006): 673-691.
- Moon TM, D'Andréa ÉD, Lee CW, Soares A, Jakoncic J, Desbonnet C, et al. The structures of penicillin-binding protein 4 (PBP4) and PBP5 from Enterococci provide structural insights into -lactam resistance. J. Biol. Chem 293 (2018): 18574-18585.
- Receveur-Brechot V, Durand D. How random are intrinsically disordered proteins? A small angle scattering perspective. Curr Protein Pept Sci (2012): 55-75.
- Rambo RP, Tainer JA. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers 95 (2011): 559-71.
- Burger VM, Arenas DJ, Stultz CM. A Structure-free Method for Quantifying Conformational Flexibility in proteins. Sci Rep 30 (2016): 29040.
- Fenton BA, Tomberg J, Sciandra CA, Nicholas RA, Davies C, Zhou P. Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the β3-β4 loop to favor a low-affinity drug-binding state. J. Biol. Chem 279 (2021): 101188.
- Contreras-Martel C, Dahout-Gonzalez C, Martins ADS, Kotnik M, Dessen A. PBP Active Site Flexibility as the Key Mechanism for β-Lactam Resistance in Pneumococci. J. Mol. Biol 387 (2009): 899-909, 2009.
- Cianci M, Bourenkov G, Pompidor G, Karpics I, Kallio J, Bento I, et al. P13, the EMBL macromolecular crystallography beamline at the low-emittance PETRA III ring for high- and low-energy phasing with variable beam focusing. J. Synchrotron Radiat 24 (2017): 323-332.
- Kabsch W. XDS,” Acta Crystallogr. Sect. D Biol. Crystallogr 66 (2010): 125.
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr 40 (2007): 658-674.
- Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll Ti, et al. Adams. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. Sect. D, Struct. Biol 75 (2019): 861- 877,.
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D. Biol. Crystallogr 68 (2012): 352-367.
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Cryst 66 (2010): 486-501.
- Hopkins JB, Gillilan RE, Skou S. BioXTAS RAW: Improvements to a free open- source program for small-angle X-ray scattering data reduction and analysis. J. Appl. Crystallogr 50 (2017): 1545-1553.
- Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr 25 (1992): 495-503.
- Keller S, Vargas C, Zhao H, Piszczek G, Brautigam CA, Schuck P. High- precision isothermal titration calorimetry with automated peak-shape analysis. Anal. Chem 84 (2012): 5066-5073.
- Zhao H, Piszczek G, Schuck P. SEDPHAT - A platform for global ITC analysis and global multi-method analysis of molecular interactions. Methods76 (2015): 137-148.
- Brautigam CA. Calculations and Publication-Quality Illustrations for Analytical Ultracentrifugation Data. Methods Enzymol 562 (2015): 109-133.
- Madhavi Sastry G, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. Comput. Aided. Mol. Des 27 (2013): 221-234.
- Lu C, Wu C, Ghoreishi D, Chen W, Wang L, Damm W, et al. OPLS4: Improving force field accuracy on challenging regimes of chemical space. J. Chem. Theory Comput 17 (2021): 4291-4300.
- Abascal JLF, Vega C. A general purpose model for the condensed phases of water: TIP4P/2005. J. Chem. Phys 123 (2005).
- Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA, et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. ACM/IEEE SC (2006): 43.
Supplementary
Supplementary Figure 1: Molecular Dynamics Stability Analysis of SePBP3 Over 100 ns. a) Trajectories of RMSD values for Cα atoms, the backbone, side chains, and heavy atoms across the 100 ns simulation period. b) Evolution of SePBP3's secondary structure content throughout the 100 ns simulation, depicted both in terms of overall percentage and per-residue distribution. Blue sections represent α-helices, while red sections denote β-sheets.


 Impact Factor: * 3.0
Impact Factor: * 3.0 Acceptance Rate: 76.32%
Acceptance Rate: 76.32%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
