Anthraquinones: A Promising Multi-target Therapeutic Scaffold To Treat Covid-19
Safae El Mazouria, Tarik Aanniza*, Jihane Touhtouha, Ilham Kandoussia, Mohammed Hakmia, Lahcen Belyamanib, Azeddine Ibrahimia, Mouna Ouadghiria
aMedical Biotechnology Laboratory (MedBiotech), Bioinova Research Center, Medical and Pharmacy School, Mohammed Vth University, Rabat, Morocco
bEmergency Department, Military Hospital Mohammed V, Medical and Pharmacy School, Mohammed Vth University, Rabat, Morocco
*Corresponding Author: Tarik Aanniz, Medical Biotechnology Laboratory (MedBiotech), Bioinova Research Center, Medical and Pharmacy School, Mohammed Vth University, Rabat, 10102, Morocco
Received: 25 February 2021; Accepted: 08 March 2021; Published: 18 March 2021
Article Information
Citation:
Safae El Mazouri, Tarik Aanniz, Jihane Touhtouh, Ilham Kandoussi, Mohammed Hakmi, Lahcen Belyamani, Azeddine Ibrahimi, Mouna Ouadghiri. Anthraquinones: A Promising Multi-target Therapeutic Scaffold To Treat Covid-19. International Journal of Applied Biology and Pharmaceutical Technology 12 (2021): 338-355.
View / Download Pdf Share at FacebookAbstract
The coronavirus disease 19 (Covid-19) pandemic caused by the SARS-CoV-2 virus has become a humanitarian crisis. Considering the severity of the situation we have performed a virtual screening of anthraquinones derivative drugs and phytochemicals targeting simultaneously multiple essential proteins of SARS-CoV-2 namely Mpro, PLpro, RdRp and the spike.
Among the 9 screened anthraquinones derivative drugs, valrubicin, idarubicin, daunorubicin, doxorubicin, epirubicin and diacerein were the most potent inhibitors of SARS-CoV-2 Mpro, PLpro, RdRp and Spike simultaneously. Valrubicin has the best affinity towards the spike protein (-9.5 kcal/mol), RdRp (-8.2 kcal/mol) and PLpro (-7.9 kcal/mol) while idarubicin and doxorubicin were the most effective against Mpro (-8.3 kcal/mol). No toxicity measurements are required for these drugs since they were tested prior to their approval by the FDA. Of the 140 screened phytochemicals anthraquinones were the most potent candidates. Hypericin and rhein were able to bind to the active site of all four targets, while chrysophanol, aloesaponarin II, emodine, aloe-emodine, physcion and danthron simultaneously bound to the active site of SARS-CoV-2 Mpro, Spike and RdRp. Hypericin showed the best affinity towards the spike protein (-9.7 kcal/mol), RdRp (-10.2 kcal/mol) and PLpro (-7.8 kcal/mol), while chrysophanol was the most effective one against Mpro (-8.4 kcal/mol).
Our overall prediction findings indicate that anthraquinones may inhibit the activity of the four essential proteins of SARS-CoV-2 and those results can pave the way in drug discovery.
Keywords
<p>Anthraquinones; Virtual screening; Spike; RdRp; PLpro; Mpro</p>
Article Details
This work was carried out under national funding from the Moroccan Ministry of Higher Education and Scientific Research (COVID-19 program) to A.I. This work was also supported by a grant from the Moroccan Institute of Cancer Research and the PPR-1 program to A.I.
1. Introduction
The world is facing a pandemic named Covid-19 caused by the novel coronavirus known as SARS-CoV-2 that emerged in the city of Wuhan, China by the end of 2019. Coronaviruses have repeatedly evolved during the past 1000 years [1]. They represent a diverse family of positive-sense RNA viruses capable of infecting the pulmonary system, the intestines, the liver and the nerve cells in human and animal hosts [2]. Almost a year after the first case detected, there are still no approved specific antiviral drugs against Covid-19. Notably, many antiviral, anti-malaria and anti-inflammatory molecules have been tested, with very limited success [3;4].
Even in the presence of approved vaccines, antivirals are needed in some specific circumstances. Many people are not allowed to be vaccinated such as pregnant women, immunocompromised and people with severe allergic reactions. Manufacturing and distributing a vaccine at large scale will present significant challenges. The use of monoclonal antibodies is a promising outlook and provide an effective and specific therapeutic option. Nevertheless, large-scale production is complex, time consuming and expensive, meaning poor countries might be priced out and only wealthy countries could afford them [5]. Monoclonal antibodies are administrated relatively at high doses and are difficult for generic-drug makers to duplicate.
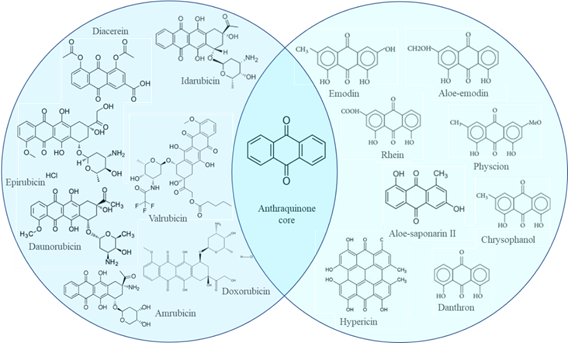
The process of a new drug development is cost and time consuming, scientists are trying to solve the puzzle through drug repurposing, the most realistic in the present pandemic, or the use of compounds from natural sources, offering novel testable hypotheses [6;7]. Anthraquinones, structurally related to 9,10-dioxoanthracene, are an important privileged structure in medicinal chemistry for more than a millennium. They have been proven to serve as a suitable starting point for optimization leading to the drug development. Anthraquinones have been known and used as laxatives for centuries and currently are employed as antiarthritics, anti-inflammatory, antifungal, antiviral, antimalarial, antimicrobial, antiplatelet, antidiabetic, neuroprotective, antioxidants, antibacterial, for the treatment of multiple sclerosis, as immune boosters [8-10] and as anticancer agents. Marketed drugs containing anthraquinone scaffold include amrubicin, daunorubicin, diacerein, doxorubicin, epirubicin, idarubicin, mitoxantrone, and valrubicin [10].
Anthraquinones may produce potential damage to cells because of the close similarity with the toxic analogue, anthracene [11]. However, there is no clear-cut correlation, and the substitution pattern appears to be an important determinant and all serious side effects have typically been the result of abuse and chronic use [9]. Few studies indicated that some anthraquinones may inhibit coronavirus activity by targeting different steps of the infection process [12-14]. Anthraquinones are well known for their immunity boosting properties, making them valuable to tackle the Covid-19 [8]. Nevertheless, few studies attempted to identify simultaneous inhibitors of these targets.
Overall, these observations prompted us to investigate the potential effect of anthraquinones on SARS-CoV-2 infection. The present work concentrates on the effectiveness of anthraquinones as simultaneous multi-target binders to the Mpro, PLpro, RdRp and Spike of the SARS-CoV-2.
2. Methods
2.1. Data sources
A dataset of 9 anthraquinones derivative drugs and 140 phytochemicals were screened against 4 crucial SARS-CoV-2 targets namely Mpro, PLpro, RdRp and Spike proteins. Phytochemicals database was obtained based on Lipinski’s rule of five. Molecular docking was performed using Autodock Vina to predict the ability of small molecules to bind to the active site of the used targets to identify potent inhibitors.
2.2. Preparation of receptors
The three-dimensional crystal structure of SARS-CoV-2 Mpro (PDB ID: 6lu7), PLpro (PDB ID: 6xaa), RdRp (PDB ID: 7bv2) and the spike protein (PDB ID: 6lzg) were collected from the RCSB Protein Data Bank (PDB). The crystal structures were prepared according to Pitsillou et al. (2020) [15]. The solvation parameters, charges assignment, fragmental volumes, and protein optimization were checked [16-18]. Kollman united atom charges, solvation parameters and polar hydrogens were added to the targets using AutoDockTools [19].
2.3. Preparation of ligands
The ligand molecules were retrieved from PubChem database [20] and were converted to PDBQT format [21]. Gasteiger charges were assigned and then non-polar hydrogens were merged.
2.4. Virtual screening and molecular docking protocols
For all of the targets, a grid box (X: 20 Å, Y: 20 Å and Z: 20 Å) was generated around the active site residues. The center of the grid box for the spike protein was X: -26.84, Y: 18.32 and Z: -14.06, for Mpro was X: -26.28, Y: 12.59, Z: 58.96, for PLpro was X: -24.83, Y: 20.63, Z: -1.87 and for RdRp was X: 98.76, Y: 98.79, Z: 98.98. During the docking process using Autodock Vina, the protein was considered to be rigid and the ligand to be flexible [7;21]. The binding affinities of the drugs were measured in kcal/mol unit and sorted according to the higher negative values, which imply the best binding affinities [22].
2.5. Analysis and visualization
Based on calculated binding affinities, the ligands with the lowest binding energy (less than -7 kcal/mol) were chosen for the interactions analysis using PyMol for 3D visualization and BIOVIA Discovery Studio Visualizer for 2D visualization.
3. Results
The rapid development and identification of efficient therapies against SARS-CoV-2 remains a major challenge. Repurposing of available drugs and the use of natural compounds always offer a great promise [7;8;23-25]. Currently, in silico studies are providing lots of preliminary data about potential drugs, which can be a great help in further in vitro and in vivo studies [15].
In this study, we screened 9 anthraquinones drugs and 120 phytochemicals against 4 targets of SARS-CoV-2 using structure based virtual screening approach towards four targets of the SARS-CoV-2 i.e., PLpro, Mpro, RdRp and the spike protein. A potent effect leading to more efficiency against SARS-CoV-2 could be the result of the multi-target inhibition by anthraquinones.
Anthraquinones derivative drugs repurposing
Considering -7 kcal/mol as a cutoff value, anthraquinones drugs i.e., valrubicin, idarubicin, daunorubicin, doxorubicin, epirubicin and diacerein exhibited the best binding affinity simultaneously towards all of the four targets. The binding energy of all the potent anthraquinones drugs are shown in Table 1 and their structures are shown in Figure 1. Valrubicin is the drug with the best affinity towards the spike protein (-9.5 kcal/mol), RdRp (-8.2 kcal/mol) and PLpro (-7.9 kcal/mol) while idarubicin and doxorubicin were the most effective against Mpro (-8.3 kcal/mol). Whereas, mitoxantrone and pixantrone showed low affinities towards all of the four targets. No toxicity measurements are required for these drugs since they were tested prior to their approval by the FDA.
Table 1: The binding energy and H-bonded interactions between anthraquinones derivative drugs and the SARS-CoV-2 targets
Table 2: The binding energy and H-bonded interactions between natural anthraquinones and the SARS-CoV-2 targets
Natural anthraquinones screening
Among the 140 phytochemicals screened, anthraquinones were the most potent inhibitors of the used targets. Anthraquinones namely rhein and hypericin also exhibited good binding affinities simultaneously towards all of the four targets, while chrysophanol, aloesaponarin II, emodin, aloe-emodin, danthron and physcion docked all the targets except PLpro (Figure 1; Table 1). Hypericin showed the best affinity towards the spike protein (-9.7 kcal/mol), RdRp (-10.2 kcal/mol) and PLpro (-7.8 kcal/mol), while chrysophanol exhibited the best affinity towards Mpro (-8.4 kcal/mol). Details of the binding energy of the best natural anthraquinones are tabulated in Table 2 and their structures are shown in Figure 1. The remaining phytochemicals showed low affinities. All of the prioritized natural anthraquinones are obeying Lipinski’s rule of five. The toxicity of anthraquinones is well documented and discussed [9;26].
Molecular interaction studies
All of the screened molecules occupied the active site of the selected targets. Researchers suggest that non-covalent interactions with the active site of the protein may lead to possible inhibition and blockage of the target.
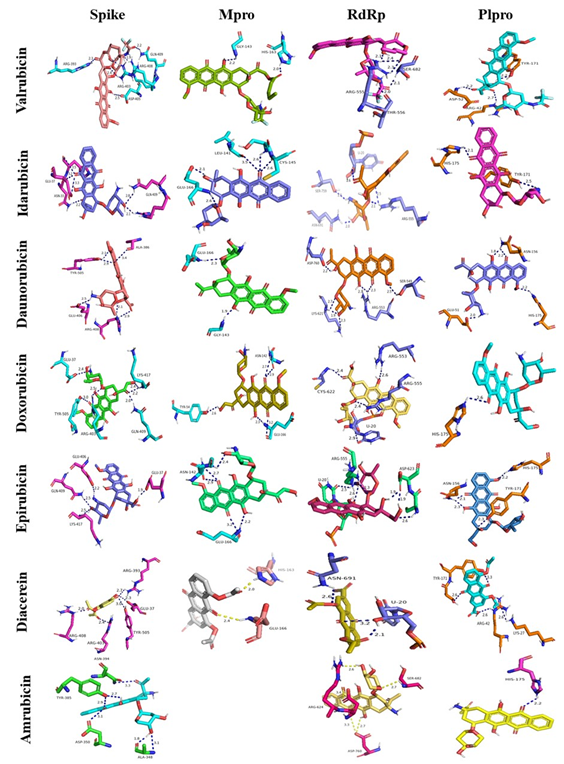
Valrubicin, idarubicin, daunorubicin, doxorubicin and amrubicin have shown numerous interactions with the active site of the spike protein including 6 H-bonds, diacerein formed 5 H-bonds, while epirubicin formed 4 H-bonds. Idarubicin, doxorubicin and epirubicin formed 5 H-bonds with the active site of Mpro, valrubicin, daunorubicin, diacerein formed 2 H-bonds while the complex amrubicin-Mpro lacked hydrogen bonding interactions. Daunorubicin was interestingly locked into the binding site of RdRp, forming 7 H-bonds, while epirubicin formed 6 H-bonds, followed by idarubicin, valrubicin and amrubicin that formed 5 H-bonds, doxorubicin formed 4 H-bonds while the complex diacerein-Mpro was stabilized with 3 hydrogen bonding interactions. The interactions of diacerein, daunorubicin and epirubicin with the active site of PLpro were maintained by 4 H-bonds, valrubicin formed 3 H-bonds, idarubicin formed 2 H-bonds while doxorubicin and amrubicin formed one H-bond.
Details regarding polar interactions involving hydrogen-bonds are shown in the 3D interaction plots of Figure 2 and are tabulated in Table 1.
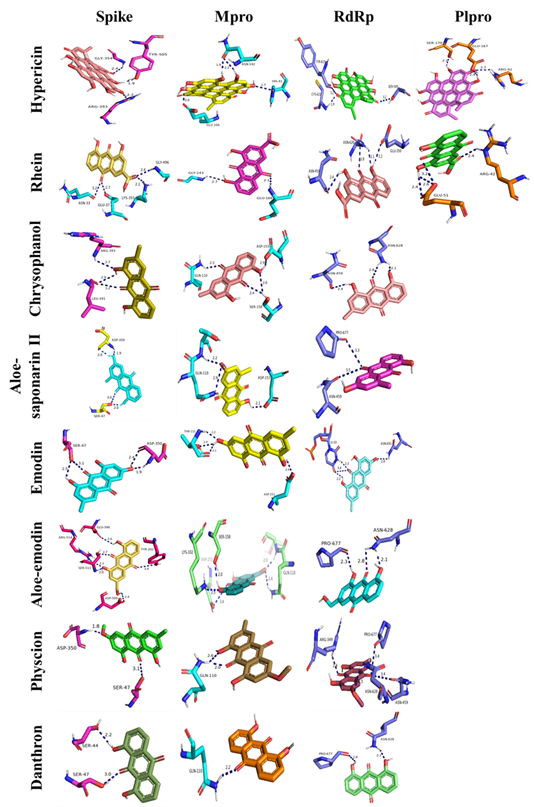
As regards to natural anthraquinones, 6 H-bonds can be seen between aloe-emodin and the active site of the spike protein, rhein, aloe-saponarin II, and emodin formed 4 H-bonds, hypericin and chrysophanol formed 3 H-bonds while physcion and danthron formed 2 H-bonds. The interactions between the active site of the Mpro and aloe-emodin and hypericin consisted of 5 H-bonds, chrysophanol and emodin formed 4 H-bonds, aloe-saponarin II formed 3 H-bonds, rhein and physcion formed 2 H-bonds while the complex danthron-Mpro was stabilized with one H-bond. Rhein, physcion and emodin formed 4 H-bonds with the active site of the RdRp, aloe-emodin, hypericin and chrysophanol formed 3 H-bonds, while aloe-saponarin II and danthron formed 2 H-bonds. 4 H-bonds were formed between rhein and the active site of the PLpro, while hypericin formed 3 H-bonds. The interactions between the remaining natural anthraquinones and PLpro were not considered as they show low affinities.
Details regarding polar interactions involving hydrogen-bonds are shown in the 3D interaction plots of Figure 3 and are tabulated in Table 2.
4. Discussion
The use of drugs with known mechanisms of action has emerged as a valid tool to identify compounds that can contribute to contain Covid-19 [7;27-29]. Assessing evidences from molecular docking studies, it was clearly seen that anthraquinones derivative drugs i.e. valrubicin, idarubicin, daunorubicin, doxorubicin, epirubicin and diacerein exhibited good binding affinity towards PLpro, Mpro, RdRp and the spike simultaneously indicating their competency of inhibiting SARS-CoV-2.
Valrubicin, an antitumoral, was identified as potent inhibitor of SARS-CoV-2 Mpro [30]. Our study showed that valrubicin has a good affinity towards the spike protein as well as RdRp, PLpro and Mpro respectively, which allows us to suggest that valrubicin could be a promising inhibitor of SARS-CoV-2. In another study, idarubicin exhibited strong affinity towards endoribonuclease (EndoU) [31]. Here we showed that idarubicin can also bind to PLpro, Mpro, RdRp and the spike protein indicating its multi-target effect against SARS-CoV-2. Daunorubicin was reported to be a potential inhibitor of SARS-CoV-2 Mpro based on docking studies performed by Jimenez-Alberto et al. (2020) [32]. The current study showed that daunorubicin has also high affinity towards RdRp, Spike and PLpro. Al- Motawa et al. (2020) [33] reported that doxorubicin, an antitumor drug with historical antiviral activity, could be used to defeat SARS-CoV-2 by increasing cellular methylglyoxal (MG) concentration to virucidal levels. Relatively short-term treatment with doxorubicin increasing cellular MG may be beneficial in patients with Covid-19. Our finding brings additional supports and mechanisms to these results by proving that doxorubicin exhibits a very good affinity towards the four crucial SARS-CoV-2 proteins (PLpro, Mpro, RdRp and the spike protein). Epirubicin used for chemotherapy to treat breast cancer was identified as a potential inhibitor of SARS-CoV-2 Mpro in a study conducted by Khan et al. (2020) [28] which promotes our docking results showing that epirubicin has an inhibitory effect on SARS-CoV-2 Mpro, in addition to RdRp, Spike and PLpro. de Oliveira et al. (2020) [34] hypothesized that diacerein is a multi-target drug useful for Covid-19 treatment. By inhibiting the IL-1β production, this drug may have an impact on viral infection, disease onset, progression and outcome by controlling hyperinflammatory conditions [35, 36]. Amrubicin, used in the treatment of lung cancer was identified as an inhibitor of SARS-CoV-2 Mpro by Jimenez-Alberto et al. (2020) [32]. Moreover, in the present work we found that amrubicin has a good affinity towards Spike, RdRp and PLpro.
Many studies showed that anthraquinones like hypericin, rhein, emodin, aloe-emodin and chrysophanol could hinder the entry, replication and release of many viruses including coronaviruses. The antiviral activity of emodin and rhein has well been documented. Rhein from Rheum palmatum ethanol extract inhibited hepatitis B virus (HBV) replication [37]. Barnard et al. (1992) [38] reported that emodin, rhein and hypericin inhibited human cytomegalovirus (HCMV) while Chang et al. (2014) [39] reported the antiviral activity of aloe-emodin, chrysophanol, rhein, emodin and physcion against Japanese encephalitis virus (JEV). Lin et al. (2005) [40] showed also that aloe-emodin, emodin, and chrysophanol inhibited the SARS-CoV 3CLpro.
Interest in hypericin is ascribed to the discovery that it possesses extremely high activity towards enveloped viruses including Hepatitis C virus, herpes simplex virus, murine cytomegalovirus, sindbis virus, hepatitis, some leukemic viruses, and avian infectious bronchitis virus (IBV). Additionally, it has also been used in the clinical treatment of AIDS patients [41-47]. Balmeh et al. (2020) [48] showed that hypericin has a high affinity towards ACE2, GRP78, AT1R, and TMPRSS2 making it a promising compound to prevent the Covid-19 infection. Hypericin, could also bind the N-terminus and C-terminus of the SARS-CoV-2 Nsp14 [49]. Here we report that hypericin could effectively bind to the active site of SARS-CoV-2 Spike, RdRp Mpro and PLpro which corroborates its antiviral potential.
Rhein suppresses lung inflammatory injury induced by human respiratory syncytial virus, through inhibiting NLRP3 inflammasome activation via NF-κB pathway in mice [50]. Influenza A virus (IAV) adsorption and replication was inhibited by rhein through decreasing IAV-induced oxidative stress and downregulating the activation of the TLR4, Akt, p38, JNK MAPK, and NF- κB-mediated signaling pathways and the production of inflammatory cytokines and matrix metalloproteinases [50]. The inhibitory effect of this molecule on IL-6 and TNF-α has been reported [34]. Importantly, rhein inhibits the interaction between the SARS-CoV-2 S protein and ACE2 suggesting that rhein is a potential therapeutic agent for the treatment of SARS-CoV-2 [12]. Preincubation of rhein with biotinylated S protein inhibited S protein-ACE2 interaction. Savarino et al. (2007) [13] reported the effect of rhein on the activity of the human liver enzyme cathepsin B within endosomes which is a critical step in SARS-CoV-2 infection [14]. Rhein was also reported for exhibiting anti-coagulant effects [34]. In our study, we showed that rhein was also able to dock the active site of SARS-CoV-2 PLpro, Mpro, RdRP and the spike protein. All these findings make rhein advantageous as an inhibitor of SARS-CoV-2.
Different antiviral activities of chrysophanol towards poliovirus type 2 and 3, Coxsackie virus type A21 and B4, human rhinovirus type 2, Ross River virus and herpes simplex virus type 1, have been investigated [51]. Subbaiyan et al. (2020) [52] found that this compound has one of the highest binding affinities towards the viral spike protein which is consistent with our findings.
The antiviral properties of emodin have also been confirmed, mainly on enveloped viruses. Emodin protected against H1N1 [53]. Batista et al. (2019) [54] showed that emodin triggers a strong virucidal effect on ZIKA by reducing the hydrodynamic radius of virus particle in solution. Emodin could hinder the SARS-CoV by blocking both viral entry and release [12]. Luo et al. (2009) [55] showed that emodin and aloe-emodin blocked the interaction between S protein of SARS-CoV and ACE2. In another study, emodin inhibited the SARS-CoV 3a protein and S protein [56; 57]. Furthermore, here we report that emodin could inhibit PLpro, Mpro, RdRP and the spike protein which offers a potential therapeutic approach for SARS-CoV-2 [12].
Li et al. (2014) [58] reported the antiviral activity of aloe-emodin against influenza A virus by up-regulating galectin-3, and thioredoxin as well as down-regulating nucleoside diphosphate kinase A while Lin et al. (2008) [59] reported that it acted as an interferon-inducing agent with antiviral activity against Japanese encephalitis virus and enterovirus 71. In addition, here we show that aloe-emodin could bind perfectly to the active site of Mpro, RdRp and the spike protein making it a potent inhibitor of SARS-CoV-2.
Otherwise, little is known about the antiviral activity of physcion, danthron and aloesaponarin II. Here we report also that they are able to perfectly dock Mpro, and RdRp proteins suggesting them as potent inhibitors of SARS-CoV-2.
Safety issues of anthraquinones should definitely be considered while developing derivatives for medical use, but it should be borne in mind that toxicity is based on the entire structure of a molecule. Moreover, the cytotoxicity was found to be related to the dose and the duration of treatment [60;61]. Drug treatment of Covid-19 may be shorter than cancer chemotherapy: for example, median hospitalization time of patients surviving severe symptoms of Covid-19 was 28 days [62] while a typical course of cancer chemotherapy is 6 months or longer [60;61].
5. Conclusion
To sum up, the present study corroborates that anthraquinones are promising scaffolds for anti-SARS-CoV-2 antivirals development. Further validation by in vitro and in vivo studies are needed. If a high potency antiviral effect of these agents is found, low dose and short duration of treatment are expected to decrease the risk of adverse effects.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was carried out under national funding from the Moroccan Ministry of Higher Education and Scientific Research (COVID-19 program) to A.I. This work was also supported by a grant from the Moroccan Institute of Cancer Research and the PPR-1 program to A.I.
References
- Forni D, Cagliani R, Clerici M, et al. Molecular evolution of human coronavirus genomes. Trends in Microbiology 25 (2017): 35-48.
- Zhang Y, Geng X, Tan Y, et al. New understanding of the damage of SARS-CoV-2 infection outside the respiratory system. Biomedicine & Pharmacotherapy (2020): 110195.
- Asif M, Saleem M, Saadullah M, et al. COVID-19 and therapy with essential oils having antiviral, anti-inflammatory, and immunomodulatory properties. Inflammopharmacology (2020): 1-9.
- Singh AK, Singh A, Shaikh A, et al. Chloroquine and hydroxychloroquine in the treatment of COVID-19 with or without diabetes: A systematic search and a narrative review with a special reference to India and other developing countries. Diabetes & Metabolic Syndrome: Clinical Research & Reviews 14 (2020): 241-246.
- Jahanshahlu L, Rezaei N. Monoclonal antibody as a potential anti-COVID-19. Biomedicine & Pharmacotherapy (2020): 110337.
- Pushpakom S, Iorio F, Eyers PA, et al. Drug repurposing: progress, challenges and recommendations. Nature Reviews Drug Discovery 18 (2019): 41-58.
- Hakmi M, El Mehdi Bouricha IK, El Harti J, et al. Repurposing of known anti-virals as potential inhibitors for SARS-CoV-2 main protease using molecular docking analysis. Bioinformation 16 (2020): 301.
- Khanal P, Patil BM, Chand J, et al. Anthraquinone derivatives as an immune booster and their therapeutic option against COVID-19. Natural Products and Bioprospecting 10 (2020): 325-335.
- Malik EM, Müller CE. Anthraquinones as pharmacological tools and drugs. Medicinal Research Reviews 36 (2016): 705-748.
- Siddamurthi S, Gutti G, Jana S, et al. Anthraquinone: a promising scaffold for the discovery and development of therapeutic agents in cancer therapy. Future Medicinal Chemistry 12 (2020): 1037-1069.
- Sendelbach LE. A review of the toxicity and carcinogenicity of anthraquinone derivatives. Toxicology 57 (1989): 227-240.
- Ho TY, Wu SL, Chen JC, et al. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antiviral Research 74 (2007): 92-101.
- Savarino L, Fioravanti A, Leo G, et al. Anthraquinone-2, 6-disulfonic acid as a disease-modifying osteoarthritis drug: an in vitro and in vivo study. Clinical Orthopaedics and Related Research® 461 (2007): 231-237.
- Simmons G, Gosalia DN, Rennekamp AJ, et al. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proceedings of the National Academy of Sciences 102 (2005): 11876-11881.
- Pitsillou E, Liang J, Karagiannis C, et al. Interaction of small molecules with the SARS-CoV-2 main protease in silico and in vitro validation of potential lead compounds using an enzyme-linked immunosorbent assay. Computational Biology and Chemistry 89 (2020): 107408.
- Chandel V, Sharma PP, Raj S, et al. Structure-based drug repurposing for targeting Nsp9 replicase and spike proteins of severe acute respiratory syndrome coronavirus 2. Journal of Biomolecular Structure and Dynamics (2020): 1-4.
- Kumar D, Chandel V, Raj S, et al. In silico identification of potent FDA approved drugs against Coronavirus COVID-19 main protease: A drug repurposing approach. Chemical Biology Letters 7 (2020): 166-175.
- Raj S, Chandel V, Rathi B, Kumar D. Understanding the Molecular Mechanism (s) of SARS-CoV2 Infection and Propagation in Human to Discover Potential Preventive and Therapeutic Approach. Coronaviruses 1 (2020): 73-81.
- Prasanth DS, Murahari M, Chandramohan V, et al. In silico identification of potential inhibitors from Cinnamon against main protease and spike glycoprotein of SARS CoV-2. Journal of Biomolecular Structure and Dynamics (2020): 1-5.
- Kim S, Thiessen PA, Bolton EE, et al. PubChem substance and compound databases. Nucleic Acids Research 44 (2016): D1202-D1213.
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry 31 (2010): 455-461.
- Rahman MM, Saha T, Islam KJ, et al. Virtual screening, molecular dynamics and structure–activity relationship studies to identify potent approved drugs for Covid-19 treatment. Journal of Biomolecular Structure and Dynamics (2020): 1-11.
- Elfiky AA. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sciences 248 (2020): 117477.
- Elfiky AA. Natural products may interfere with SARS-CoV-2 attachment to the host cell. Journal of Biomolecular Structure and Dynamics (2020): 1-10.
- Islam MT, Sarkar C, El-Kersh DM, et al. Natural products and their derivatives against coronavirus: A review of the non-clinical and pre-clinical data. Phytotherapy Research 34 (2020): 2471-2492.
- Chien CH, Chiang-Hsieh YF, Chen YA, et al. AtmiRNET: a web-based resource for reconstructing regulatory networks of Arabidopsis microRNAs. Database (2015): 2015.
- Cherian SS, Agrawal M, Basu A, et al. Perspectives for repurposing drugs for the coronavirus disease 2019. The Indian Journal of Medical Research 151 (2020): 160.
- Khan MA, Mahmud S, Alam AR, et al. Comparative molecular investigation of the potential inhibitors against SARS-CoV-2 main protease: a molecular docking study. Journal of Biomolecular Structure and Dynamics (2020): 1-7.
- Muralidharan N, Sakthivel R, Velmurugan D, et al. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics (2020): 1-6.
- Wang L, He W, Yu X, et al. Coronavirus disease 2019 in elderly patients: characteristics and prognostic factors based on 4-week follow-up. Journal of Infection 80 (2020): 639-645.
- Chandra A, Gurjar V, Qamar I, et al. Identification of potential inhibitors of SARS-COV-2 endoribonuclease (EndoU) from FDA approved drugs: a drug repurposing approach to find therapeutics for COVID-19. Journal of Biomolecular Structure and Dynamics (2020): 1-11.
- Jiménez-Alberto A, Ribas-Aparicio RM, Aparicio-Ozores G, et al. Virtual screening of approved drugs as potential SARS-CoV-2 main protease inhibitors. Computational Biology and Chemistry 88 (2020): 107325.
- Al-Motawa MS, Abbas H, Wijten P, et al. Vulnerabilities of the SARS-CoV-2 virus to proteotoxicity—opportunity for repurposed chemotherapy of COVID-19 infection. Frontiers in Pharmacology 11 (2020).
- de Oliveira PG, Termini L, Durigon EL, et al. Diacerein: A potential multi-target therapeutic drug for COVID-19. Medical Hypotheses 144 (2020): 109920.
- Yaron M, ShirazI I, Yaron I. eAnti-interleukin-1 effects of diacerein and rhein in human osteoarthritic synovial tissue and cartilage cultures. Osteoarthritis and Cartilage 7 (1999): 272-280.
- Martel-Pelletier J, Mineau F, Jolicoeur FC, et al. In vitro effects of diacerhein and rhein on interleukin 1 and tumor necrosis factor-alpha systems in human osteoarthritic synovium and chondrocytes. The Journal of Rheumatology 25 (1998): 753-762.
- Li Z, Li LJ, Sun Y, et al. Identification of natural compounds with anti-hepatitis B virus activity from Rheum palmatum ethanol extract. Chemotherapy 53 (2007): 320-326.
- Barnard DL, Huffman JH, Morris JL, et al. Evaluation of the antiviral activity of anthraquinones, anthrones and anthraquinone derivatives against human cytomegalovirus. Antiviral Research 17 (1992): 63-77.
- Chang SJ, Huang SH, Lin YJ, et al. Antiviral activity of Rheum palmatum methanol extract and chrysophanol against Japanese encephalitis virus. Archives of Pharmacal Research 37 (2014): 1117-1123.
- Lin CW, Tsai FJ, Tsai CH, et al. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antiviral Research 68 (2005): 36-42.
- Verebová V, Beneš J, Stanicová J. Biophysical Characterization and Anticancer Activities of Photosensitive Phytoanthraquinones Represented by Hypericin and Its Model Compounds. Molecules 25 (2020): 5666.
- Chen H, Feng R, Muhammad I, et al. Protective effects of hypericin against infectious bronchitis virus induced apoptosis and reactive oxygen species in chicken embryo kidney cells. Poultry Science 98 (2019): 6367-6377.
- Chen H, Muhammad I, Zhang Y, et al. Antiviral Activity Against Infectious Bronchitis Virus and Bioactive Components of Hypericum perforatum L. Frontiers in Pharmacology 10 (2019): 1272.
- Miskovsky P. Hypericin-a new antiviral and antitumor photosensitizer: mechanism of action and interaction with biological macromolecules. Current Drug Targets 3 (2002): 55-84.
- Jacobson JM, Feinman L, Liebes L, et al. Pharmacokinetics, safety, and antiviral effects of hypericin, a derivative of St. John's wort plant, in patients with chronic hepatitis C virus infection. Antimicrobial Agents and Chemotherapy 45 (2001): 517-524.
- Weber ND, Murray BK, North JA, et al. The antiviral agent hypericin has in vitro activity against HSV-1 through non-specific association with viral and cellular membranes. Antiviral Chemistry and Chemotherapy 5 (1994): 83-90.
- Hudson JB, Lopez-Bazzocchi I, Towers GH. Antiviral activities of hypericin. Antiviral Research 15 (1991): 101-112.
- Balmeh N, Mahmoudi S, Mohammadi N, et al. Predicted therapeutic targets for COVID-19 disease by inhibiting SARS-CoV-2 and its related receptors. Informatics in Medicine Unlocked 20 (2020): 100407.
- Liu C, Zhu X, Lu Y, et al. Potential Treatment of Chinese and Western Medicine Targeting Nsp14 of SARS-CoV-2. Journal of Pharmaceutical Analysis (2020).
- Wang QW, Su Y, Sheng JT, et al. Anti-influenza A virus activity of rhein through regulating oxidative stress, TLR4, Akt, MAPK, and NF-κB signal pathways. PLoS One 13 (2018): e0191793.
- Semple SJ, Pyke SM, Reynolds GD, et al. In vitro antiviral activity of the anthraquinone chrysophanic acid against poliovirus. Antiviral Research 49 (2001): 169-178.
- Subbaiyan A, Ravichandran K, Singh SV, et al. In silico molecular docking analysis targeting SARS-CoV-2 spike protein and selected herbal constituents. J Pure Appl Microbiol 14 (2020): 989-998.
- Dai JP, Wang QW, Su Y, et al. Emodin inhibition of influenza A virus replication and influenza viral pneumonia via the Nrf2, TLR4, p38/JNK and NF-kappaB pathways. Molecules 22 (2017): 1754.
- Batista MN, Braga AC, Campos GR, et al. Natural products isolated from oriental medicinal herbs inactivate Zika virus. Viruses 11 (2019): 49.
- Luo W, Su X, Gong S, et al. Anti-SARS coronavirus 3C-like protease effects of Rheum palmatum extracts. Bioscience Trends 3 (2009).
- Fuzimoto AD, Isidoro C. The antiviral and the coronavirus-host protein pathways inhibiting properties of herbs and natural compounds-Additional weapons in the fight against the COVID-19 pandemic?. Journal of Traditional and Complementary Medicine (2020).
- Schwarz S, Wang K, Yu W, et al. Emodin inhibits current through SARS-associated coronavirus 3a protein. Antiviral Research 90 (2011): 64-69.
- Li SW, Yang TC, Lai CC, et al. Antiviral activity of aloe-emodin against influenza A virus via galectin-3 up-regulation. European Journal of Pharmacology 738 (2014): 125-132.
- Lin CW, Wu CF, Hsiao NW, et al. Aloe-emodin is an interferon-inducing agent with antiviral activity against Japanese encephalitis virus and enterovirus 71. International Journal of Antimicrobial Agents 32 (2008): 355-359.
- Barrett-Lee PJ, Dixon JM, Farrell C, et al. Expert opinion on the use of anthracyclines in patients with advanced breast cancer at cardiac risk. Annals of Oncology 20 (2009): 816-827.
- Rowinsky, MD EK. The development and clinical utility of the taxane class of antimicrotubule chemotherapy agents. Annual Review of Medicine 48 (1997): 353-374.
- Wang J. Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. Journal of Chemical Information and Modeling 60 (2020): 3277-3286.


 Impact Factor: * 3.0
Impact Factor: * 3.0 Acceptance Rate: 76.32%
Acceptance Rate: 76.32%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
