Elevated Expression of Notch 2 & Notch 3 is associated with Disease Progression in Colorectal Cancer
Abhay Kumar Sharma1, 2, Nimisha2, Apurva2, Arun Kumar2, Asgar Ali2, Sundeep Singh Saluja∧, 2, 3, Birendra Prasad*, 1
1Department of Botany/Biotechnology, Patna University, India
2Central Molecular Lab, Govind Ballabh Pant Institute of Postgraduate Medical Education and Research (GIPMER), New Delhi, India
3Department of Gastrointestinal Surgery, Govind Ballabh Pant Institute of Postgraduate Medical Education and Research (GIPMER), New Delhi, India
*Corresponding author: Dr. Birendra Prasad, Professor, Coordinator MSc. Biotech. Course, Patna University, Patna, India.
∧Co-corresponding author: Sundeep Singh Saluja, PI Central Molecular Laboratory, Professor Department ofGI surgery, Govind Ballabh Pant Institute of Postgraduate Medical Education and Research (GIPMER) 1, Jawaharlal Nehru Marg, 64 Khamba, Raj Ghat, New Delhi, India
Received: 16 September 2022; Accepted: 27 October 2022; Published: 14 November 2022
Article Information
Citation: Abhay Kumar Sharma, Nimisha, Apurva, Arun Kumar, Asgar Ali, Sundeep Singh Saluja, Birendra Prasad. Elevated Expression of Notch 2 & Notch 3 is associated with Disease Progression in Colorectal Cancer. International Journal of Applied Biology and Pharmaceutical Technology 13 (2022): 033-050.
View / Download Pdf Share at FacebookAbstract
Background: The potential involvement of Notch signaling pathway in cell fate decision, tumor heterogeneity and angiogenesis in solid tumors can be explored in colorectal cancer (CRC). This might further help to improve outcomes in CRC. Here, the promoter methylation of Notch receptor gene (Notch2 and Notch3) and their co-expression with its downstream transcription factor Hes1 has been analyzed.
Methods: Seventy-two CRC patients were enrolled to study the role of Notch2, Notch3 and Hes1 genes in colorectal cancer. Promoter methylation and mRNA expression in tumor and adjoining normal tissue were assessed by Methylation Specific PCR and quantitative Real time PCR respectively. Statistical correlation was done by using SPSS.
Results: We found that Notch2 and Notch3 were hypomethylated in 52/72 (72.22%) and 54/72 (75%) cases respectively. Hypomethylation of Notch 2 and Notch 3 showed significant association with advanced stage (p=0.001) and (p=0.003) and nodal metastasis (p=0.036) and (p=0.012) respectively. Both Notch 2 and Notch 3 showed increased mRNA expression in 49 (68.05%) and 51(70.84%) patients with a fold change of 3.37 and 5.43 respectively. Positive correlation between hypomethylation and expression was observed for both genes. High expression of Hes1 was found in 53(73.61%) of cases which was highly relatable with over expression of notch receptor genes. Upregulation of Notch 2, Notch 3 and Hes1 showed significant association with high grade tumors, advance stage and presence of LN metastasis, additionally Notch 3 and Hes1 showed significant association with distant metastasis.
Conclusion: Hypomethylation of Notch 2 and 3 receptors is playing crucial role in regulating the expression of these genes in CRC. Overexpression of Notch 2, Notch 3 and Hes1 are associated with disease progression in CRC.
Keywords
<p>Colorectal Cancer; Hypomethylation; mRNA Expression; Notch 2; Notch 3</p>
Article Details
1. Introduction
Colorectal cancer (CRC) is the third most common cancer comprising 10 % of the newly diagnosed cancer cases and second leading cause of cancer related death (9.4%) worldwide (GLOBOCAN 2020). It is the second most common cancer in women and third among men worldwide [1,2]. Basic tenets of colorectal tumorigenesis are firstly the tumor develop from activation of oncogenes and deactivation of tumor suppressor genes. The genetic and/or epigenetic changes occur at multiple sites and summation of these changes lead to malignant transformation. Understanding of these changes is imperative to find new solutions. Hence, it is important to identify molecular markers and potential drug targets to enhance the clinical decision making for treatment. There are several molecular pathways known to be involved in the development of CRC, which includes chromosomal instability (CIN), Wnt, Hedgehog and Notch signaling pathways [3,4]. Wnt and Notch signaling occurs maximum at the base of crypt. Emerging concept that cancer stem-like cells (CSCs) play a role in both chemoresistance and tumorigenesis in various cancers including solid tumours. Its role in CRC might help in improving outcomes in colorectal cancer. The Notch signaling pathway is a highly conserved signaling system involved in various biological processes, including cell differentiation, proliferation, apoptosis and stem cell maintenance [4,5]. Moreover, Notch can function as both oncogene and tumor suppressor gene. Human Notch consists of four transmembrane Notch receptors (Notch 1-4) and five Notch ligands (Delta-like-1, Delta-like-3, Delta-like-4, Jagged-1 and Jagged-2) [6]. Notch pathway gets activated by the interaction of Notch receptors with Notch ligands leading to secretion of γ-secretase protein complex. As a result, Notch receptors undergo proteolytic cleavage to release the Notch intracellular domain (NICD) which subsequently enters into the nucleus and associates with DNA binding proteins and acts as a transcription-activating factor for the regulation of Notch target genes such as Hairy/enhancer of Split 1 (Hes1) [7]. The Notch signaling pathway controls the transcriptional repressor of Hes1, a basic helix-loop-helix (bHLH) molecule that is increased upon activation of Notch [8]. Hes1 plays a crucial role in cellular development, sustaining intestinal progenitor cells and neural stem cells, and controlling apoptosis [9]. Abnormal activation of the Notch pathway plays an important role in tumor development and progression [10]. The upregulation of the Notch pathway results in cell proliferation, Epithelial-mesenchymal transition (EMT) and angiogenesis. The aggressiveness of CRC could be due to the active involvement of the oncogenic Notch pathway. Therefore, the possible approaches should be targeted to explore the alteration in various Notch receptors, ligands and Notch targets genes that cause activation of Notch pathway in colorectal cancer. Notch 2 is reported to be up-regulated in gastric cancer and brain tumors while its down-regulation was reported in renal cell carcinoma and non-small cell lung cancer [11–13].The significance of Notch 2 gene in colorectal cancer is disputed. In fact, it may play role in tumor inhibition in colon carcinogenesis as per study by Chu et al [14]. On a contrary, it could also behave as a key regulator of carcinogenesis in the Notch pathway. Moreover, the mechanism of its dysregulation is less known especially in colorectal cancer and needs more attention. Notch 3 is found to be over-expressed in cervical cancer tissues [15], lung cancer cell line [16] and Gastric cancer [17]. No clear molecular mechanism for the activation of Notch 3 overexpression has been reported. However, increased tumorigenesis was found to be associated with abnormal methylation pattern during the multistage carcinogenesis of cervical cancer [18]. Moreover, Notch 2 and 3 inhibitors Tarxetumab could have antitumor effects by blocking receptor activation thereby inhibiting tumor growth. Gliotoxin is a myotoxin, which acts by inhibiting the formation of DNA bound Notch 2 complexes, and induces apoptosis in various cancers such as melanoma, hepatocellular carcinoma and pancreatic cancer [12,19,20]. These inhibitors may help in tumor reduction in cases where Notch shows overexpression. Notch 2 and Notch 3 activation could be responsible for the elevated expression of the downstream target gene Hes1. Herein, we have investigated the methylation status of Notch 2 and 3 and the mRNA expression of Notch 2, 3 along with its downstream target gene Hes1 in CRC patients. Statistical correlation between molecular evaluation and clinical-pathological parameters was also performed.
2. Materials and Methods
2.1 Ethical Clearance
Ethical approval for the study was obtained from Institutional Ethical Committee Maulana Azad Medical College and Associated Hospital, New Delhi with file number F.1/IEC/MAMC/85/03/2021/No.430.
2.2 Patients and Sample Collection
Tumor and adjacent normal (at least 5cm away from the tumor) tissue specimen from 72 CRC patient were collected in appropriate buffer (PBS for DNA and RNA later for RNA) who underwent treatment for CRC in the Department of Gastrointestinal Surgery, GIPMER, New Delhi. The specimen was collected from patients before obtaining any neoadjuvant treatment. Among 72 patients, 46 were male and 26 were female, with mean age of 49.03 ± 14.2 years. Informed consent was taken from all participants. Detailed clinical and demographic parameters along with their management plan were recorded for all the patients enrolled in the study.
2.3 DNA Extraction and Bisulfite Modification
Genomic DNA was isolated from the tissue specimens (tumor and adjacent normal tissues) using Wizard Genomic DNA Purification Kit (Promega USA) following the manufacturer’s instructions. The purity of DNA was checked using the NanoDrop spectrophotometer (Thermo Fisher, New York) and quantified by Qubit 4 fluorometer (Invitrogen, USA). The quality of DNA was checked on 0.8% agarose gel. Bisulfite treatment of purified DNA was performed using the EZ DNA Methylation-Gold kit (Zymo Research- D5006, USA). One microgram of genomic DNA was treated with bisulfite and purified according to the manufacturer’s instructions. Bisulfite treated DNA was stored at -20°C for further analysis.
2.4 Methylation Specific Polymerase Chain Reaction (MS-PCR)
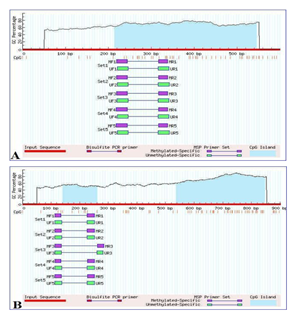
Promoter methylation status of the Notch 2 and 3 genes was determined using Methylation Specific PCR. Notch promoter regions were selected using the Eukaryotic promoter database (EPD) and their respective CpG islands were predicted using Meth Primer tools (Figure 1). The primer was synthesized commercially (Integrated DNA Technologies) according to the specific sequence of CpG island. The primer used in MS-PCR is listed in table 1. PCR was performed as the previously described protocol with some modifications [9]. The methylated and unmethylated MS-PCR condition for Notch 2 is as follows: PCR was conducted in a total reaction volume of 20 µl containing 60 ng of bisulfite treated DNA, 10 picomole of each primer, 1.5mM MgCl2, 200 µM each deoxyribonucleotide triphosphate (dNTP), 1.0-unit Hot start Taq DNA polymerase and 1X PCR buffer (Qiagen, Germany). The product was amplified in a PCR thermal cycler under the following conditions: Initial denaturation at 95°C for 10 minutes, followed by 35 cycles each, denaturation at 95°C for 30 seconds, annealing at 58°C for 30seconds, and elongation at 72°C for 30 seconds, followed by a final extension at 72°C for 7 minutes. The methylated and unmethylated MS-PCR condition for Notch 3 is as follows: PCR was conducted in a total reaction volume of 20 µl containing 60 ng of bisulfite treated DNA, 10 picomole of each primer, 2.5mM MgCl2, 300 µM each deoxyribonucleotide triphosphate (dNTP), 1.0-unit Hot start Taq DNA polymerase and 1X PCR buffer (Qiagen, Germany). The product was amplified in a PCR thermal cycler under the following conditions: Initial denaturation at 95°C for 08 minutes, followed by 35 cycles each, denaturation at 95°C for 30 seconds, annealing at 55°C for 45seconds, and elongation at 72°C for 30 seconds, followed by a final extension at 72°C for 5 minutes. Nuclease free water was used in place of bisulfite treated DNA which serves as a negative control and Universal Methylated Human DNA Standard (Zymo research- D5011, USA) was used as a positive control. The amplified PCR product for methylated and unmethylated was checked on 2% agarose gel containing 0.5µg/ml Ethidium bromide and visualized by Gel Doc Imaging system (Biorad, USA).
2.5 RNA Extraction and cDNA Synthesis
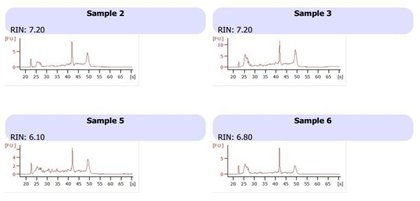
Total RNA was extracted from the stored tumor and normal tissue sample using Trizol reagent (Thermo Fisher Scientific, USA) according to previously established protocol with some modification [21]. On-column DNase treatment was performed using RNase free DNaseI enzyme (Invitrogen USA). RNA purity was checked using NanoDrop spectrophotometer (Thermo-scientific, New York) and quantified by Qubit 4 fluorometer (Invitrogen, USA). The quality of RNA was checked on 1.2% agarose gel. On the basis of RNA quality samples were selected for integrity checkup. RNA integrity number (RIN) was determined by RNA 6000 Nano kit (Cat No #5067-1511) using Bioanalyzer 2100 (Agilent Technologies Inc., USA) (Figure 2).
2.5.1 cDNA Synthesis: Reverse transcription reactions were performed to obtain cDNA using Revert Aid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). The total volume of the reaction was 20 µl and containing 500 ng of total RNA template, 5Xreaction buffer; 15 pmol random primer, 20U/μl RiboLock RNase Inhibitor, 10 mM dNTP Mix and 20U/μl Revert Aid Reverse transcriptase. The mixture was prepared in a PCR tube and reaction was performed in thermocycler under the following condition; 5 minutes at 25°C followed by 60 minutes at 42°C and reaction was stop by heating at 70°C for 5 minutes.
2.6 Quantitative Real-Time PCR (qRT-PCR)
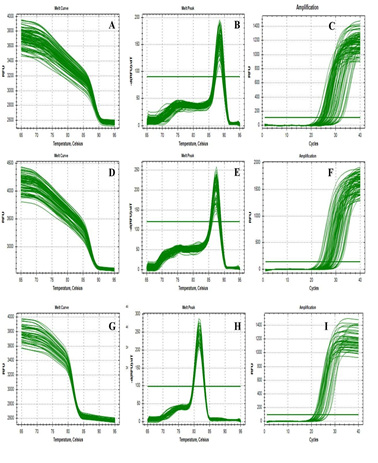
Quantitative real-time PCR were performed in triplicate for Notch 2, Notch 3 and Hes1 separately in the final reaction volume of 10 µl containing 5µl of 2X maxima SYBR Green/ROX qPCR master mix, 0.2 µl of 10 Pico moles of each primer set (forward and reverse), 2 µl of diluted (1:5X) cDNA and 2.6µl nuclease free water. Reverse transcriptase minus (RT-) negative control and no template negative (NTC) was used to access the genomic DNA contamination of the RNA sample and reagent contamination respectively. Amplification was performed in a 96 well plate in CFX Real Time Detection System (Bio-Rad), initial denaturation at 95°C for 2 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 30 seconds. Reaction specificity was checked followed by qPCR using the melting curve analysis (Figure 3). The expression of each was normalized by subtracting the Ct value of the housekeeping gene GAPDH from the Ct value of the tar-get gene. The fold increase, relative to the control, was obtained by using the formula 2−ΔΔCt. The sequences of the primers for Notch 2, Notch 3, and GAPDH are shown in (Table 1).
Table 1: Primer used in quantitative PCR (qPCR) and Methylation Specific PCR (MS-PCR), m- Methylation, um- unmethylation, * Real-Time PCR.
3. Result
Our results suggest that Notch 2 and Notch 3 could have tumor progression roles in CRC. To analyze the methylation of Notch 2 & 3 and expression of Notch 2, 3 and Hes1, we conducted MS-PCR and Real time PCR respectively.
3.1 Notch2 and Notch3 Show Hypomethylation in CRC Patients
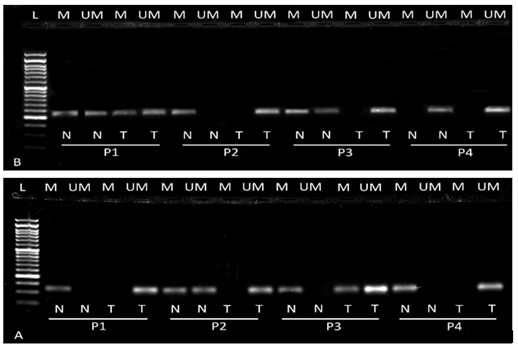
We performed methylation specific PCR to analyze the epigenetic changes in CRC tissue. Both methylation and unmethylation patterns in the promoter region were seen in both genes. However, the majority of tumor tissue showed hypomethylated patterns i.e.52/72 (72.22%), and 54/72 (75%) in Notch 2 and Notch 3 respectively compared to normal (Table 2). Representative agarose gel images of methylation and unmethylation of Notch2 and Notch3 genes are illustrated in figure 4. Further we correlated hypomethylation patterns of tumor tissue with clinico-pathological characteristics of CRC patients. Notch 2 hypomethylation showed significant association with advanced TNM stage (p= 0.001), high tumor depth (p=0.003) and presence of lymph node metastasis (p=0.036) (Table 3). The hypomethylation in Notch3 showed significant correlation with tumor site i.e. rectal tumor (p=0.041), higher TNM stage (p=0.003) and presence of lymph node metastasis (p=0.012) (Table 4).
|
Tissue type |
Notch2 methylation status (n=72) |
χ2 |
p- value |
Notch3 methylation status (n=72) |
χ2 |
p- value |
||
|
Hypo Methylation |
Hyper Methylation |
Hypo Methylation |
Hyper Methylation |
|||||
|
Normal
Tumor |
21 52 |
51 20 |
26.70 |
<0.001 |
23 54 |
49 18 |
26.82 |
<0.001 |
Table 2: Methylation status of Notch2 and Notch 3 in normal and tumor CRC tissue.
|
Clinico-pathologic parameters |
Notch2 Methylation (n= 72) |
P value |
|
|
Hypermethylation (n = 20) (27.78%) |
Hypomethylation (n = 52) (72.22%) |
||
|
Age (years) ≤ 40 > 40 |
04 16 |
16 36 |
0.558 |
|
Gender Male Female |
10 10 |
36 16 |
0.107 |
|
Site Colon Rectum |
15 05 |
34 18 |
0.173 |
|
Grade of differentiation Well Moderate Poor |
01 16 03 |
10 29 13 |
0.141 |
|
Tumor Depth T1 T2 T3 T4 |
02 09 07 02 |
07 05 29 11 |
0.003 |
|
Lymph node Metastasis Positive Negative |
04 16 |
24 28 |
0.036 |
|
Metastasis Yes No |
02 18 |
03 49 |
0.613 |
|
TNM stage I II III IV |
04 14 02 00 |
06 14 29 03 |
0.001 |
|
Lymphovascular Invasion Positive Negative |
03 17 |
11 41 |
0.744 |
|
Perineural Invasion Positive Negative |
02 18 |
06 46 |
0.851 |
|
Addiction Yes No |
05 15 |
19 33 |
0.352 |
|
Family History Yes No |
01 19 |
03 49 |
0.693 |
|
mRNA Expression Up-regualte Down-regulate |
08 12 |
41 11 |
0.004 |
Table 3: Clinicopathological correlation of Notch2 methylation in CRC patients.
|
Clinico-pathologic parameters |
Notch3 Methylation (n= 72) |
P value |
|
|
Hypermethylation (n = 18) (25%) |
Hypomethylation (n = 54) (75%) |
||
|
Age (years) ≤ 40 > 40 |
05 13 |
15 39 |
0.627 |
|
Gender Male Female |
09 09 |
37 17 |
0.129 |
|
Site Colon Rectum |
15 03 |
31 23 |
0.041 |
|
Grade of differentiation Well Moderate Poor |
04 10 04 |
07 35 12 |
0.64 |
|
Tumor Depth T1 T2 T3 T4 |
03 03 12 00 |
04 10 27 13 |
0.100 |
|
Lymph node Metastasis Positive Negative |
02 16 |
26 28 |
0.012 |
|
Metastasis Yes No |
0 18 |
5 49 |
0.226 |
|
TNM stage I II III IV |
03 13 02 00 |
07 15 29 03 |
0.003 |
|
Lymphovascular Invasion Positive Negative |
02 16 |
12 42 |
0.253 |
|
Perineural Invasion Positive Negative |
02 16 |
06 48 |
1.00 |
|
Addiction Yes No |
08 10 |
16 38 |
0.192 |
|
Family History Yes No |
00 18 |
04 50 |
0.307 |
|
mRNA Expression Up-regulate Down-regulate |
05 13 |
46 08 |
<0.001 |
Table 4: Clinicopathological correlation of Notch3 methylation in CRC patient.
3.2 Upregulated Notch2 and Notch3 is associated with Advance Disease Stage and Higher Tumor Grade
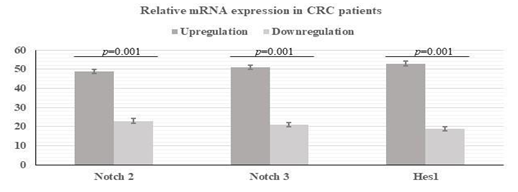
We evaluated the mRNA level of Notch 2, Notch 3 and target gene Hes1 in 72 tumor and adjacent normal tissue using quantitative real-time PCR. We observed that 49 (68.05%) patients, 51(70.84%) patients and 53(73.61%) patients showed over expression in Notch 2, Notch 3 and Hes1 gene respectively with a fold change increase of 3.37, 5.43 and 3.52 respectively (Figure 5 and 6). In addition, we correlated the mRNA expression with various clinicopathological characteristics. Notch 2 upregulation was significantly associated with higher tumor stage (p =<0.001 and presence of lymph node metastasis (p=<0.009) (Table 5) whereas Notch 3 upregulation was significantly associated with higher tumor depth (p=<0.021), presence of lymph node metastasis (p=0.005), advance tumor stage (p=0.007) and high-grade tumors (p=0.013) (Table 6). The overexpression of Hes1 showed significant correlation tumor depth, advance tumor stage and presence of lymph node metastasis (Table 7). We noticed that fold change in overexpression showed increase with worsening of tumor differentiation (p= 0.018), advancing tumor stage (p= <0.001) and presence of LN metastasis (p=0.001), lymphovascular invasion (p=<0.001), perineural invasion (p= 0.014) in Notch 2 (Table 8). In notch 3, the fold change of expression showed association with tumor grade (p= 0.021), tumor depth (p=<0.001), advance disease stage (p= <0.001), presence of LN metastasis (p=0.001), lymphovascular invasion (p=<0.001) and metastatic disease (Table 9). Similar to Notch 3, Hes1 also showed higher fold change of expression with tumor depth, advance stage, lymph node metastasis and distant metastasis (Table 10).
|
Clinico-pathologic parameters |
Notch2 mRNA expression (n= 72) |
P value |
|
|
Upregulate (n = 49) (68.05%) |
downregulate (n = 23) (31.95%) |
||
|
Age (years) ≤ 40 > 40 |
14 35 |
06 17 |
1.00 |
|
Gender Male Female |
33 16 |
13 10 |
0.435 |
|
Site Colon Rectum |
31 18 |
15 08 |
1.00 |
|
Grade of differentiation Well Moderate Poor |
06 30 13 |
05 15 03 |
0.326 |
|
Tumor Depth T1 T2 T3 T4 |
04 07 27 11 |
03 06 12 02 |
0.354 |
|
Lymph node Metastasis Positive Negative |
24 25 |
04 19 |
0.009 |
|
Metastasis Yes No |
04 45 |
01 22 |
1.00 |
|
TNM stage I II III IV |
05 13 28 03 |
05 15 03 00 |
<0.001 |
|
Lymphovascular Invasion Positive Negative |
12 37 |
02 21 |
0.114 |
|
Perineural Invasion Positive Negative |
06 43 |
02 21 |
0.655 |
|
Addiction Yes No |
18 31 |
06 17 |
0.372 |
|
Family History Yes No |
02 47 |
02 21 |
0.425 |
|
Methylation Hypermethylation Hypomethylation |
08 41 |
12 11 |
0.002 |
Table 5: Clinicopathological correlation of Notch 2 mRNA expression in CRC patient.
|
Clinico-pathologic parameters |
Notch 3 mRNA expression (n= 72) |
P value |
|
|
Upregulate (n = 51) (70.84%) |
Downregulate (n = 21) (29.16%) |
||
|
Age (years) ≤ 40 > 40 |
16 35 |
04 17 |
0.390 |
|
Gender Male Female |
33 18 |
13 08 |
0.514 |
|
Site Colon Rectum |
31 20 |
15 06 |
0.433 |
|
Grade of differentiation Well Moderate Poor |
04 33 14 |
07 12 02 |
0.013 |
|
Tumor Depth T1 T2 T3 T4 |
02 08 29 12 |
05 05 10 01 |
0.021 |
|
Lymph node Metastasis Positive Negative |
25 26 |
03 18 |
0.005 |
|
Metastasis Yes No |
05 46 |
00 21 |
0.312 |
|
TNM stage I II III IV |
03 19 26 03 |
07 09 05 00 |
0.007 |
|
Lymphovascular Invasion Positive Negative |
12 39 |
02 19 |
0.209 |
|
Perineural Invasion Positive Negative |
07 44 |
01 20 |
0.423 |
|
Addiction Yes No |
17 34 |
07 14 |
0.603 |
|
Family History Yes No |
03 48 |
01 20 |
1.00 |
|
Methylation Hypermethylation Hypomethylation |
05 46 |
13 08 |
<0.001 |
Table 6: Clinicopathological correlation of Notch3 mRNA expression in CRC patient.
|
Clinico-pathologic parameters |
Hes1 mRNA expression (n= 72) |
P value |
|
|
Upregulate (n = 53) (73.61%) |
Downregulate (n = 19) (26.38%) |
||
|
Age (years) ≤ 40 > 40 |
16 37 |
04 15 |
0.328 |
|
Gender Male Female |
34 19 |
12 07 |
0.575 |
|
Site Colon Rectum |
33 20 |
13 06 |
0.425 |
|
Grade of differentiation Well Moderate Poor |
06 34 13 |
03 11 05 |
0.269 |
|
Tumor Depth T1 T2 T3 T4 |
04 04 32 13 |
03 09 07 00 |
<0.001 |
|
Lymph node Metastasis Positive Negative |
24 29 |
04 15 |
0.054 |
|
Metastasis Yes No |
04 49 |
01 18 |
0.603 |
|
TNM stage I II III IV |
04 20 26 03 |
06 08 05 00 |
0.034 |
|
Lymphovascular Invasion Positive Negative |
12 41 |
02 17 |
0.214 |
|
Perineural Invasion Positive Negative |
08 45 |
00 19 |
0.074 |
|
Addiction Yes No |
18 35 |
06 13 |
0.544 |
|
Family History Yes No |
03 50 |
01 18 |
0.717 |
|
Notch 2 mRNA Expression Up-regulate Down-regulate |
37 16 |
12 07 |
0.397 |
|
Notch 3 mRNA Expression Up-regulate Down-regulate |
40 13 |
11 08 |
0.126 |
Table 7: Clinicopathological correlation of Hes1 mRNA expression in CRC patient.
|
Clinico-pathologic parameters |
n = 72 (%) |
Notch 2 mRNA expression mean ± SD |
P value |
|
Age (years) ≤ 40 > 40 |
20 (0.28) 52 (0.72) |
2.44±0.80 2.04±0.27 |
0.032 |
|
Gender Male Female |
46 (0.63) 26 (0.37) |
2.61±0.39 2.10±0.39 |
0.409 |
|
Site Colon Rectum |
46 (0.63) 26 (0.37) |
2.32±0.37 2.32±0.46 |
0.617 |
|
Grade of differentiation Well Moderate Poor |
11 (15.28) 45 (62.50) 16 (22.22) |
1.84±0.56 2.03±0.24 3.96±1.00 |
0.018 |
|
Tumor Depth T1 T2 T3 T4 |
07 (0.09) 13 (0.18) 39 (0.54) 13 (0.19) |
1.62±0.55 1.13±0.71 2.80±1.84 3.03±1.88 |
0.111 |
|
Lymph node Metastasis Positive Negative |
28 (38.88) 44 (61.12) |
3.84±0.61 1.53±0.18 |
<0.001 |
|
Metastasis Yes No |
05 (0.07) 67 (0.93) |
2.92±1.17 2.39±0.30 |
0.649 |
|
TNM stage I II III IV |
10 (0.13) 28 (0.39) 31 (0.43) 03 (0.45) |
1.25±0.35 1.10±0.15 3.85±0.52 4.02±1.73 |
<0.001 |
|
Lymphovascular Invasion Positive Negative |
14 (0.20) 58 (0.80) |
4.42±1.01 1.95±0.23 |
<0.001 |
|
Perineural Invasion Positive Negative |
08 (0.11) 64 (0.89) |
4.43±1.69 2.18±0.24 |
0.014 |
|
Addiction Yes No |
24 (0.33) 48 (0.67) |
2.45±0.46 2.42±0.37 |
0.961 |
|
Family History Yes No |
04 (0.06) 68 (0.94) |
3.16±1.95 2.38±0.29 |
0.547 |
Table 8: Clinicopathological correlation of Notch2 mRNA fold change in CRC patient.
|
Clinico-pathologic parameters |
n = 72 (%) |
Notch2 mRNA expression mean ± SD |
P value |
|
Age (years) ≤ 40 > 40 |
20 (0.28) 52 (0.72) |
3.16±0.70 4.23±0.57 |
0.297 |
|
Gender Male Female |
46 (0.63) 26 (0.37) |
4.18±0.60 3.50±0.69 |
0.480 |
|
Site Colon Rectum |
46 (0.63) 26 (0.37) |
3.51±0.57 4.68±0.74 |
0.221 |
|
Grade of differentiation Well Moderate Poor |
11 (15.28) 45 (62.50) 16 (22.22) |
1.54±1.54 3.94±0.54 5.61±1.20 |
0.021 |
|
Tumor Depth T1 T2 T3 T4 |
07 (0.10) 13 (0.18) 39 (0.54) 13 (0.18) |
0.73±0.25 2.62±0.67 3.82±1.59 7.29±1.19 |
<0.001 |
|
Lymph node Metastasis Positive Negative |
28 (38.88) 44 (61.12) |
5.49±0.78 2.94±0.51 |
0.006 |
|
Metastasis Yes No |
05 (0.07) 67 (0.93) |
8.92±1.68 3.56±0.44 |
0.002 |
|
TNM stage I II III IV |
10 (0.13) 28 (0.39) 31 (0.43) 03 (0.05) |
1.48±0.66 2.91±0.57 4.91±0.72 11.50±0.77 |
<0.001 |
|
Lymphovascular Invasion Positive Negative |
14 (0.20) 58 (0.80) |
6.38±1.39 3.34±0.43 |
0.008 |
|
Perineural Invasion Positive Negative |
08 (0.11) 64 (0.89) |
5.31±1.73 3.76±0.46 |
0.289 |
|
Addiction Yes No |
24 (0.33) 48 (0.67) |
3.82±0.80 3.98±0.56 |
0.869 |
|
Family History Yes No |
04 (0.05) 68 (0.95) |
4.36±2.12 3.91±0.47 |
0.821 |
Table 9: Clinicopathological correlation of Notch3 mRNA fold change in CRC patient.
|
Clinico-pathologic parameters |
n = 72 (%) |
Hes1 mRNA expression mean ± SD |
P value |
|
Age (years) ≤ 40 > 40 |
20 (0.28) 52 (0.72) |
2.78±0.49 2.40±0.26 |
0.473 |
|
Gender Male Female |
46 (0.63) 26 (0.37) |
2.45±0.28 2.61±0.37 |
0.748 |
|
Site Colon Rectum |
46 (0.63) 26 (0.37) |
2.31±0.24 2.85±0.48 |
0.276 |
|
Grade of differentiation Well Moderate Poor |
11 (15.28) 45 (62.50) 16 (22.22) |
2.49±0.88 4.48±0.27 2.60±0.44 |
0.979 |
|
Tumor Depth T1 T2 T3 T4 |
07 (0.10) 13 (0.18) 39 (0.54) 13 (0.18) |
1.57±0.42 1.09±0.48 2.77±0.34 3.65±0.50 |
0.003 |
|
Lymph node Metastasis Positive Negative |
28 (38.88) 44 (61.12) |
3.24±0.44 2.04±0.24 |
0.013 |
|
Metastasis Yes No |
05 (0.07) 67 (0.93) |
4.06±1.68 2.39±0.44 |
0.073 |
|
TNM stage I II III IV |
10 (0.13) 28 (0.39) 31 (0.43) 03 (0.05) |
1.29±0.32 1.90±0.24 3.14±0.41 5.69±0.47 |
<0.001 |
|
Lymphovascular Invasion Positive Negative |
14 (0.20) 58 (0.80) |
3.12±0.53 2.36±0.26 |
0.205 |
|
Perineural Invasion Positive Negative |
08 (0.11) 64 (0.89) |
3.54±0.74 2.38±0.24 |
0.122 |
|
Addiction Yes No |
24 (0.33) 48 (0.67) |
2.18±0.32 2.67±0.31 |
0.337 |
|
Family History Yes No |
04 (0.05) 68 (0.95) |
2.66±1.09 2.50±0.24 |
0.879 |
Table 10: Clinicopathological correlation of Hes1 mRNA fold change in CRC patient.
3.3 Correlation between Notch2 and Notch3 Overexpression with Hypomethylation in CRC Patients
The relationship between overexpression and hypomethylation of Notch 2 and Notch 3 genes was examined. The overexpression of Notch 2 showed a significant correlation with hypomethylation in tumor tissue (p=0.002) (Table 4). Similarly, Notch 3 overexpression was associated with presence of hypomethylation in tumor tissue (p=0.001) (Table 5).
3.4 Association between Notch2 and Notch3 and Target Gene Hes1
We found very strong positive correlation between Notch 2 and Notch 3 upregulation and Hes1 overexpression. We found that Hes1 was upregulated in 78% of cases where Notch 2 was overexpressed, and similar outcomes were seen in 79% of cases where Notch 3 was overexpressed (Table 7). Overall this finding suggested that both Notch signaling and Hes1 may be involved in CRC disease progression.
4. Discussion
Despite both clinical and molecular advancements in colorectal cancer, the incidence and disease progression has not shown much change. Recent colorectal staging system has included molecular markers for prognosis and use of targeted therapy, still more biomarkers are need to be explored. In colonic crypts, Notch signaling pathway plays major role in maintaining the balance between cell proliferations and cell fate determination by regulating colon stem cell behaviour and differentiation [22,23]. Each Notch receptor has a distinct function at cellular level. Notch2 and Notch3 are key members of the Notch family that regulate cellular functions. The functions of these depend upon the type of tumor, suggesting that they may not always play the same roles in different malignancies. [24–26]. Notch3 signaling regulates cellular activities during the progression of cancer and maintains the stemness of cancer stem cells (CSCs) [27]. Another key characteristic of Notch3 is the development of tumor resistance to several chemotherapy agents such as platinum, doxorubicin, gemcitabine, and EGFR tyrosine kinase inhibitors (TKIs).[27–29]. We analyzed the methylation pattern of Notch 2 &3 and expression of Notch 2, Notch 3 and Hes1 in 72 colorectal cancer patients. Consequently, we correlated the findings with clinicopathological characteristics to decipher its role in colorectal cancer. Epigenetic instability plays a crucial role in the development of cancer by the blocking of cell proliferation and cell cycle arrest [30,31]. Recent studies have focused on DNA methylation that plays a vital role in the regulation of gene expression by the blocking of transcription factor binding sites on the promoter region. We report hypomethylation of Notch 2 and 3 genes in tumor tissue in 72% and 75% patients respectively. The hypomethylation showed significant correlation with clinical parameters including higher tumor depth, advance stage and presence of lymph node metastasis, suggesting their association in tumor progression. The hypomethylation also showed significant correlation (Notch 2; p=0.004, Notch 3; p=0.001) with expression in approx. 90 % cases suggesting it could be one of the regulatory mechanisms controlling notch 2 & 3 gene expression in colorectal cancer. Previous reports suggest distinct miRNA such as 23b and 133a regulate notch 2 gene at translation level in gastric cancer [12]. miRNA-23b's capacity to directly bind to the Notch2 mRNA and prevent its translation made it possible to stop tumor development by restoring its expression [12] As a substitute for the activation of the γ -secretase complex, miRNA-133a can target and inhibit the translation of presenilin 1, inhibiting the release of NICD and obstructing the pro-oncogenic function of Notch signaling. Our results indicate that mRNA expression was upregulated in 68% and 70.84% in Notch 2 and Notch3 genes respectively in CRC patients. Notch 2 overexpression results in abnormal proliferation and dedifferentiation resulting in tumor development in gastric cancer. Similar findings are observed in our study in colorectal cancer where Notch 2 expression was higher in poorly differentiated tumors and advance stages. Though the notch 2 overexpression showed significant correlation with perineural and lymphovascular invasion, however it should be further evaluated as the number of patients are less. Moreover, previous study by Chu et al showed Notch 2 function is opposite to Notch 1 and inversely associated with differentiation and tumor stage. Similar role of Notch 2 as oncogene is reported in gastric cancer (71.4%) laryngeal squamous cell carcinoma (87.3%) and medulloblastoma (74.4%) [13,19,32]. In gastric cancer Notch 2 intracellular domain (N2ICD) activation has been shown to encourage the increased expression of cyclooxygenase-2 (COX-2) and promotes the epithelial-mesenchymal transition (EMT) in tumor cells [33]. The increased expression of Notch 3 showed significant association with tumor grade, tumor depth, TNM stage and presence of lymph node metastasis. Unlike Notch 2, Notch 3 expression was significantly higher in fold change in patients with metastatic disease compared to non-metastatic disease (8.92±1.68 vs 3.56±0.44 p = 0.002). Similar results were reported in the CRC tissue and mice model indicating that increased Notch 3 expression was associated with distant metastasis and poor prognosis [34,35]. In ovarian cancer, Notch 3 expression was linked to tumor grade, lymph node, distant metastasis, and clinical stage [36–38]. However, there is disagreement concerning the function of Notch 3 in breast tumors, although some researchers reported that Notch 3 encourages tumor aggressiveness by triggering EMT, other researchers showed that Notch 3 actually inhibits it. Moreover, it has been reported that the pathogenesis of HCC may be aided by the activation of Notch 3 signaling, which reduces the Wnt/-catenin signaling and enhances the expression of the protein Nanog linked to stemness. [39]. Similar association of Notch3 and Wnt pathway should be looked at in CRC to further explore its effect on CSCs. Hes1 is a well-known Notch signaling target gene. It is a novel bHLH transcriptional repressor which is overexpressed in colorectal cancer [21,40]. It can be activated when cleaved fragments of Notch intracellular domain enters into the nucleus, connect with DNA-binding protein, and transform DNA-binding protein into Hes1 activator. Therefore, Hes1 expression has been considered as a marker for Notch activation. The Notch signaling regulates Hes1 in several cancers including colorectal, oral squamous cell carcinoma and pancreatic [38,41,42]. We found a substantial relationship between activation of Notch 2 and 3 with Hes1 expression. Hes1 showed high expression in 73.61% cases. This finding suggests the activation of Notch 2 and 3 could have resulted in raised Hes1 as notch target gene in colorectal cancer. This could be one of the mechanisms by which the Notch pathway can result in tumor progression in colorectal cancer. Candy et al found overexpression of Hes1 in 60% patients in colorectal tumors. They found its value in predicting survival with chemotherapy in combination with other Notch induced transcription factors HEY1 and SOX 9 [43]. However, Reedijk et al analyzed Notch activation using Hes1 expression as surrogate marker [44]. Although the expression was observed in all patients, raised expression was seen in 31% tumor tissue compared to normal and did not correlate with survival. Our results suggest that Notch signaling activation may occur due to aberrant hypomethylation of Notch promoter and its activation can result in overexpression of Hes1. Both Notch 2 & 3 played a role as an oncogene in CRC. These findings support to explore use of Notch receptor inhibitors in reducing tumor burden and improving survival.
5. Limitation and Future Prospective
Although our study has pointed towards the role of Notch signaling pathway in CRC management; however, the sample size and availability of metastatic samples were the major limitations. Moreover, the activation of Notch signaling depends on the interaction between Notch receptor and ligands which regulate the downstream target gene. Therefore, it is important to analyze all the members of Notch family to decode their actual role in disease progression. Further, the cell line-based study should be targeted which can aid in novel therapeutic approaches.
6. Conclusion
Our results suggest that the promoter hypomethylation influences the mRNA upregulation of Notch 2 and Notch 3 in CRC. The aberrant methylation pattern and dysregulated gene expression of Notch 2 and Notch 3 was found to be associated with advanced stage, nodal metastasis and increased tumor depth in CRC. Our additional finding indicates that the gene expression of Hes1 transcription factor may be regulated by Notch 2 and 3 receptor genes Therefore, it can be stated that the DNA methylation of Notch 2 and Notch 3 promoters may have great potential in clinical applications. It may also prove as useful indicator for the development and progression of CRC. But before coming to a definitive conclusion this research needs further exploration to replicate our significant findings.
Acknowledgement
This research was financially supported by the Department of Biotechnology (DBT) Government of India, with grant no. BT/INF/22/SP33063/2019.We would like to also thank the Central Molecular Lab, Govind Ballabh Pant Institute of Postgraduate Medical Education and Research, New Delhi, for allowing us to conduct our research.
Conflicts of Interest
No competing interests exist.
References
- Jin L, Vu T, Yuan G, et al. STRAP promotes stemness of human colorectal cancer via epigenetic regulation of the NOTCH pathway. Cancer Research 77 (2017): 5464-5478.
- Shaik JP, Alanazi IO, Pathan AAK, et al. Frequent activation of notch signaling pathway in colorectal cancers and its implication in patient survival outcome. J Oncology (2020): 6768942.
- Sonbol MB, Ahn DH, Bekaii-Saab T. Therapeutic targeting strategies of cancer stem cells in gastrointestinal malignancies. Biomedicines 1 (2019): 17
- Colussi D, Brandi G, Bazzoli F, et al. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int J Mol Science 14 (2013): 16365-16385.
- Hu J, Yu J, Gan J, et al. Notch1/2/3/4 are prognostic biomarker and correlated with immune infiltrates in gastric cancer. Aging (Albany NY) 12 (2020): 2595-2609.
- Lee SH, Jeong EG, Yoo NJ, et al. Mutational analysis of NOTCH1, 2, 3 and 4 genes in common solid cancers and acute leukemias. APMIS 115 (2007): 1357-1363.
- Wang MM. Notch signaling and Notch signaling modifiers. Int J Biochem Cell Biology 43 (2011): 1550-1562.
- Shang Y, Coppo M, He T, et al. The transcriptional repressor Hes1 attenuates inflammation by regulating transcription elongation. Nat Immunol 17 (2016): 930-937.
- Kadian LK, Gulshan G, Ahuja P, et al. Aberrant promoter methylation of NOTCH1 and NOTCH3 and its association with cervical cancer risk factors in North Indian population. Am J Transl Research 12 (2020): 2814-2826.
- Aithal MGS, Rajeswari N. Role of Notch signalling pathway in cancer and its association with DNA methylation. J Genetics 92 (2013): 667-675.
- Pancewicz J. A brief overview of clinical significance of novel Notch2 regulators. Mol Cell Oncology 7 (2020): 1776084.
- Huang TT, Ping YH, Wang AM, et al. The reciprocal regulation loop of Notch2 pathway and miR-23b in controlling gastric carcinogenesis. Oncotarget 6 (2015): 18012-1826.
- Zou Y, Fang F, Ding YJ, et al. Notch 2 signaling contributes to cell growth, anti-apoptosis and metastasis in laryngeal squamous cell carcinoma. Mol Med Report 14 (2016): 3517-3524.
- Chu D, Zheng J, Wang W, et al. Notch2 expression is decreased in colorectal cancer and related to tumor differentiation status. Ann Surg Oncol 16 (2009): 3259-3266.
- Osanyingbemi-Obidi J, Dobromilskaya I, Illei PB, et al. Notch signaling contributes to lung cancer clonogenic capacity in vitro but may be circumvented in tumorigenesis in vivo. Mol Cancer Research 9 (2011): 1746-1754.
- Talora C, Sgroi DC, Crum CP, et al. Specific down-modulation of Notch1 signaling in cervical cancer cells is required for sustained HPV-E6/E7 expression and late steps of malignant transformation. Genes Development 16 (2002): 2252-2263.
- Wang Q, Wen YG, Li DP, et al. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncology 29 (2012): 77-83.
- Virmani AK, Muller C, Rathi A, et al. Aberrant methylation during cervical carcinogenesis. Clin Cancer Research 7 (2001): 584-589.
- Fiaschetti G, Schroeder C, Castelletti D, et al. NOTCH ligands JAG1 and JAG2 as critical pro-survival factors in childhood medulloblastoma. Acta Neuropathol Communication 2 (2014): 39.
- Pancewicz J, Bernatowicz PL. Differential Notchl and Notch2 expression in non-small cell lung cancer. Progress in Health Sciences 9 (2019): 1
- Wu Y, Gong L, Xu J, et al. The clinicopathological significance of HES1 promoter hypomethylation in patients with colorectal cancer. Onco Targets Therapy 10 (2017): 5827-5834.
- Bellavia D, Checquolo S, Palermo R, et al. The Notch3 Receptor and Its Intracellular Signaling-Dependent Oncogenic Mechanisms. Adv Exp Med Biology 1066 (2018): 205-222.
- Suman S, Das TP, Ankem MK, et al. Targeting notch signaling in colorectal cancer. Curr Colorectal Cancer Reports 10 (2014): 411-416.
- Fan X, Mikolaenko I, Elhassan I, et al. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Research 64 (2004): 7787-7793.
- Hubmann R, Schwarzmeier JD, Shehata M, et al. Notch2 is involved in the overexpression of CD23 in B-cell chronic lymphocytic leukemia. Blood 99 (2002): 3742-3747.
- Massi D, Tarantini F, Franchi A, et al. Evidence for differential expression of Notch receptors and their ligands in melanocytic nevi and cutaneous malignant melanoma. Mod Pathology 19 (2006): 246-254.
- Xiu M, Wang Y, Li B, et al. The role of notch3 signaling in cancer stemness and chemoresistance: molecular mechanisms and targeting strategies. Front Mol Bioscience 8 (2021): 694141.
- Arasada RR, Amann JM, Rahman MA, et al. EGFR blockade enriches for lung cancer stem-like cells through Notch3-dependent signaling. Cancer Research 74 (2014): 5572-5584.
- Giovannini C, Gramantieri L, Chieco P, et al. Selective ablation of Notch3 in HCC enhances doxorubicin’s death promoting effect by a p53 dependent mechanism. J Hepatology 50 (2009): 969-979.
- Lakshminarasimhan R, Liang G. The role of DNA methylation in cancer. Adv Exp Med Biolology 945 (2016): 151-172.
- Romero-Garcia S, Prado-Garcia H, Carlos-Reyes A. Role of DNA methylation in the resistance to therapy in solid tumors. Front Oncology 10 (2020): 1152.
- Du X, Cheng Z, Wang YH, et al. Role of Notch signaling pathway in gastric cancer: a meta-analysis of the literature. World J Gastroenterol 20 (2014): 9191-9199.
- Tseng YC, Tsai YH, Tseng MJ, et al. Notch2-induced COX-2 expression enhancing gastric cancer progression. Mol Carcinogenesis 51 (2012): 939-951.
- Koch U, Radtke F. A third Notch in colorectal cancer progression and metastasis. J Exp Medicine 10 (2020):
- Huang K, Luo W, Fang J, et al. Notch3 signaling promotes colorectal tumor growth by enhancing immunosuppressive cells infiltration in the microenvironment. Res Square (2022): 1974731.
- Liu Z, Yun R, Yu X, et al. Overexpression of Notch3 and pS6 Is Associated with Poor Prognosis in Human Ovarian Epithelial Cancer. Mediators Inflammation (2016): 5953498.
- Zhang TH, Liu HC, Zhu LJ, et al. Activation of Notch signaling in human tongue carcinoma. J Oral Pathol Medicine 40 (2011): 37-45.
- Jin HY, Zhang HY, Wang X, et al. Expression and clinical significance of Notch signaling genes in colorectal cancer. Tumour Biology 33 (2012): 817-824.
- Gramantieri L, Giovannini C, Lanzi A, et al. Aberrant Notch3 and Notch4 expression in human hepatocellular carcinoma. Liver International 27 (2007): 997-1007.
- Yuan R, Ke J, Sun L, et al. HES1 promotes metastasis and predicts poor survival in patients with colorectal cancer. Clin Exp Metastasis 32 (2015):169-179.
- Gao F, Zhang Y, Wang S, et al. Hes1 is involved in the self-renewal and tumourigenicity of stem-like cancer cells in colon cancer. Sci Report 4 (2014): 3963.
- Lee SH, Hong HS, Liu ZX, et al. TNFα enhances cancer stem cell-like phenotype via Notch-Hes1 activation in oral squamous cell carcinoma cells. Biochem Biophys Res Communication 425 (2012): 58-64.
- Candy PA, Phillips MR, Redfern AD, et al. Notch-induced transcription factors are predictive of survival and 5-fluorouracil response in colorectal cancer patients. Br J Cancer 109 (2013): 1023-1030.
- Reedijk M, Odorcic S, Zhang H, et al. Activation of Notch signaling in human colon adenocarcinoma. Int J Oncology 33 (2008): 1223-1229.


 Impact Factor: * 3.0
Impact Factor: * 3.0 Acceptance Rate: 76.32%
Acceptance Rate: 76.32%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
