Function and Characterization of the Argonaute2 Gene in the RNA Interference Pathway in the Diamondback Moth
Muhammad Salman Hameed1, 2, Xiaodong Jing2, 3-6, Wei Chen2, 3- 6, Jinzhi Chen2, 3-6, Liette Vasseur2, 7, Gefu Wang-Pruski4, 8, Guang Yang2, 3-6*
1Department of Plant Protection, Faculty of Agricultural Sciences, Ghazi University, Dera Ghazi Khan 32200, Pakistan; mhameed@gudgk.edu.pk.
2State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, (Fujian Province University) Fujian Agriculture and Forestry University, Fuzhou 350002, China.
3State Key Laboratory of Ecological Pest Control for Fujian and Taiwan crops, Institute of Applied Ecology (Fujian Province University) Fujian Agriculture and Forestry University, Fuzhou 350002, China.
4Joint International Research Laboratory of Ecological Pest Control, Ministry of Education, Fuzhou 350002, China
5Key Laboratory of Integrated Pest Management for Fujian-Taiwan Crops, Ministry of Agriculture, Fuzhou 350002, China.
6Key Laboratory of Green Pest Control (Fujian Province University), Fujian Agriculture and Forestry University, Fuzhou 350002, China.
7Department of Biological Sciences, Brock University, 1812 Sir Isaac Brock Way, St. Catharines, Ontario L2S 3A1, Canada.
8Department of Plant, Food and Environmental Sciences, Faculty of Agriculture, Dalhousie University, PO Box 550, Truro, Nova Scotia B2N 5E3, Canada.
*Corresponding Author: Guang Yang, State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, (Fujian Province University) Fujian Agriculture and Forestry University, Fuzhou 350002, China
Article Information
Citation: Muhammad Salman Hameed, Xiaodong Jing, Wei Chen, Jinzhi Chen, Liette Vasseur, Gefu Wang-Pruski, Guang Yang. Function and Characterization of the Argonaute2 Gene in the RNA Interference Pathway in the Diamondback Moth. Arch Biochem Mol Biol 10 (2019): 035-051.
View / Download Pdf Share at FacebookAbstract
Argonaute2 (AGO2) is a core catalytic component of the RNA-induces silencing complex (RISC) that binds to small guide RNAs containing small interfering RNA (siRNA) and microRNA (miRNA). The guide RNA leads RISC to the complementary mRNA for gene suppression. We cloned the full length cDNA (2193 bp) of the Ago2 gene (PxAgo2) from diamondback moth (DBM, Plutella xylostella). The predicted PxAgo2 protein had an 83 kDa molecular weight with a theoretical isoelectric point of 9.39. The phylogenetic tree showed a high similarity of PxAgo2 with Bombyx mori Ago2 (BmAgo2). Western blot and RT-qPCR analyses showed a clear increase in the PxAgo2 mRNA and protein expression levels in the egg, 4th instar larva, pre-pupa, pupa and adult. The double-stranded RNA-mediated RNAi of PxAgo2 in DBM larvae was found 3 h after dsRNA injection, and the knockdown level was increased over time up to 36 h. PxAgo2 silencing recovered the expression of PxBurs-α (Bursicon-α in DBM) to the normal expression level, which was suppressed by dsPxBurs-α in a DBM cell line. The overexpression of PxAgo2 fundamentally enhanced the PxBurs-α silencing efficiency in DBM cells. Our findings reveal that PxAgo2 is involved in the dsRNA-regulated gene silencing mechanism and performs a crucial function in the RNAi process of DBM.
Keywords
<p>Plutella xylostella; PxAgo2; double-stranded RNA; RNAi mechanism</p>
Article Details
1. Introduction
RNA interference is a process initiated by double-stranded RNA (dsRNA) that leads to post-transcriptional gene silencing by the RISC [1-3]. RISC comprises Argonaute family member genes, including Ago2 [3]. Based on its functional domains, the Ago2 protein can load small RNAs, regulate translational processes, degrade mRNA and participate in the generation of Piwi-interacting RNAs [4]. The crystalline structure of the Ago2 protein shows two major domains, a Piwi argonaute zwile (PAZ) motif and a P element-induces whimpy testes (PIWI) motif [5]. The PAZ domain is involved in the identification of the ribose nucleic acid 3' overhang, which is a nucleotide (nt) connecting site [5], while the PIWI motif contains a catalytic triad of amino acid residues DDH, involved in slicing the target mRNA [6, 7].
The Ago2 protein is an siRNA-related protein [7, 8] that acts as a core element of RISC in the gene silencing pathways of different insects [9], such as Drosophila melanogaster [10], Bombyx mori [11], Tribolium castaneum [12] and Leptinotarsa decemlineata [13]. In D. melanogaster, Ago2 is essential for unwinding the siRNA duplex [10]. In Manduca sexta larvae, Ago2 is upregulated in response to dsRNA injection [14]. Ago2-mediated suppression by dsRNA of two distinct insect RNA viruses, Norovirus and Cricket paralysis virus (CrPV), demonstrates the significance of Ago2 in antiviral defense mechanisms [15]. Ago2 is also involved in the RNAi pathway of L. decemlineata [13], B. mori [16], and its overexpression significantly increases larval RNAi efficiency [17].
Diamondback moth (DBM, Plutella xylostella) is an important insect worldwide that causes significant damage to cruciferous crops [18, 19], with the yearly cost of loss and control assessed at US$ 4-5 billion throughout the world [19]. RNAi works well in P. xylostella. For instance, specific suppression by RNAi can cause insect mortality [20-24], sensitivity to insecticide [25], and sex ratio changes [20]. However, the role of Ago2 in P. xylostella is still unclear. In this study, we analyzed the sequence characteristics of PxAgo2, examined its expression profile with Western blot analysis and RT-qPCR in different developmental stages of P. xylostella and investigated PxArgo2 suppression by dsRNA in larvae. We hypothesized that the dsRNA-mediated silencing of PxAgo2 would block or reduce the RNAi efficiency when targeting the endogenous gene PxBurs-α, which is involved in the post-ecdysial tanning of the adult cuticle and wing expansion, with dsPxBurs-α. The effect of the suppression or overexpression of PxAgo2 on the dsRNA-mediated silencing of PxBurs-α in a DBM cell line was determined. Our findings confirmed the vital role of PxAgo2 in the gene silencing process of P. xylostella.
2. Materials and methods
Selection of insect and cell line cultureAn insecticide-free strain (S) of P. xylostella Fuzhou-S was selected for our experimental study, which was used for complete DNA sequencing [26]. For rearing, the immature larvae of P. xylostella were fed by Raphanus raphanistrum seedling plants and incubated in a climate chamber (temperature of 25 ± 1 °C; relative humidity of 65% ± 5%; and the photoperiod of 16 h light: 8 h dark). A stable DBM embryonic cell line was cultured [27] in the Grace’s medium (Invitrogen, Waltham, United States of America) including 10% fetal bovine serum at 27 °C.
Gene cloningThree 4th instar P. xylostella larvae were used for total RNA purification by using the TRIzol-REAGENT (Fisher Scientific, Massachusetts, United States of America). RNA quantity was observed with a NanoDrop 1000 spectrophotometer (Fisher Scientific, Massachusetts, United States of America). Electrophoresis in 1% gel was used to evaluate the quality of the RNA. The purified RNAs were dissolved in the nuclease-free water (Fisher Scientific, Massachusetts, United States of America) at the concentration of 250 ng/μl and then immediately stored at -80 °C. Additionally, complementary DNA was synthesized using 2 μL of extracted RNA as the template according to the GoTaqTM 2 Step RT-qPCR System (Fisher Scientific, Massachusetts, United States of America).
For PxAgo2 gene sequence amplification, two primers (PxAgo2-forward and PxAgo2-reverse) (Table 1) were designed with the Primer Premier 6 software using the DNA sequence of PxAgo2 (gene ID: Px012073) (Accessible online: http://iae.fafu.edu.cn/DBM/). PCR was performed using the High-Fidelity PCR (Vazyme Biotech, Shanghai, China) according to the production guidelines at the following settings: at 94 °C for 3 min for denaturation; 40 rounds of 95 °C for 30 s, 56 °C for 30 s, and 72 °C for 3 min for amplication; at 72 °C for 5 min for the final extension; and then at 10 °C for storage. PCR product was purified in 1% gel by following the manufacturer’s instructions for the Gel Purification kit (200) (Omega, Bienne, Switzerland). The purified PCR product was ligated into the PJET vector using the Thermo ClonjetTM PCR Cloning kit (Fisher-Scientific, Massachusetts, United States of America) with the T4 DNA ligase (Promega, Madison, United States of America).
The ligated product was directly diluted with 100 μL high efficiency E. coli DH5α cells (Fisher Scientific, Massachusetts, United States of America) and then stored at -20 °C for 30 min. These cells were then incubated at 42 °C for 90 s and maintained in a mixture of media to obtain E. coli silver colonies. For confirmation, PCR was performed using the GoTaq G2 DNA polymerase (Promege, Fitchburg, United States of America) with the Ago2-based primers listed in Table 1. Sixteen transformed colonies were selected for the identification of the PxAgo2 expression vector. Only one transformed colony was cultured in the Thermostatic Microplate Thermo Linear Orbital Rotary Shaking Incubator (Senova Biotch, Shanghai, China) at 36 °C for 6 h, and then 1 ml of the cultivated solution of positive silver colony was sent to the Life Sciences (China) for sequencing. Furthermore, the nucleotide sequence of PxAgo2 was evaluated in the NCBI database (Accessible online; https://www.BLAST.ncbi.nlm.nih.gov/blast.cgi) to determine its similarity. The whole PxAgo2 gene sequence was then submitted to the GenBank (Accessible online: https://submit.ncbi.nlm.nih.gov/subs/genbank/) to obtain an accession number.
To prepare the PxAgo2 expression plasmid (Pizt/v5+PxAgo2), two restriction enzymes, KpnI and EcoRI (Fisher Scientific, Massachusetts, United States of America), were used to separately cleave the PCR product and plasmid Pizt/v5 at 37 °C overnight (Table 1). Both of the digested products were ligated and then directly transform to E. coli cells. The subsequent cells were verified by colony PCR using one pair of primers (forward sense: PIZT-F CGCAACGATCTGGTAAACA and reverse antisense: PIZT-R CTGACTAAATCTTAGTTTGT ATTGT) and 1% gel electrophoresis following the method described in the previous section. The PxAgo2 expression vector was extracted from the transformed cells following the manufacturer’s instructions using the Hispeed Plasmid Midi kit (Qiagen, Venlo, Netherlands).
The domains were identified from the PxAgo2 sequence using the online NCBI-CDD (conserved-domain-database) (Accessible online; https://www.ncbi.nlm.nih.GOV/structure/ CDD/wrpsb.cgi) and compared with Ago1, Ago2 and Ago3 sequences from D. melanogaster and B. mori, which were downloaded from the online GenBank database. The molecular weight and isoelectronic point of the amplified PxAgo2 were also analyzed using the online Expasy tool (http://web.expasy.org/cgi-bin/computepi/pitool). Multiple-sequence alignment was performed at https://www.Genome.jp/Tools-bin/CLASTALW. The phylogenetic tree was constructed with the PxAgo2 and 19 eukaryotic Ago family member proteins using the Molecular-Evolutionary-Genetics-Analysis Version 6.0.6 (Tokyo, Japan) with the neighbor/joining method and a bootstrap rate of 1000 replicates.
The specific sequences of each gene were used as templates for dsRNA synthesis, e.g., 200 base pairs of PxAgo2, 419 base pairs of PxBurs-α, and 457 base pairs of enhanced green fluorescence protein (eGFP). The dsRNAs of the three gene segments were synthesized with the Ambion MEGAscript RNAi kit (Invitrogen, Waltham, United States of America) by following the manufacturer’s instructions with the specific primers containing the T-7 promoter (taatacgactcactataggg) sequence in Table. The reaction was conducted at the following conditions: at 36 °C for 4 h, at 76 °C for 4 min for heat shock, and at room temperature for 90 min to anneal the double strands. Next, the dsRNAs were treated with DNase-I to eliminate the template DNA, purified by using the phenol-chloroform method and finally dissolved in the appropriate amount of distilled water. The dsRNA purity and quantity were determined according to the method previously described.
Sixty 4th instar embryos of P. xylostella were selected for dsRNA injection. A borosilicate capillary (Thomas Scientific, Swedesboro, United States of America) was pulled by heat set at 64 °C in PC-10 (Malcomtech, Hayward, United States of America) for injection. A total of 30 μL (1 μg) of the PxBurs-α, PxAgo2 or eGFP dsRNAs was injected directly into the hemolymph of each larva using a microinjection apparatus (World Precision Instrument, Sarasota, Florida, United States of America). After the injection, the larvae were kept in a plastic box and nourished with fresh R. raphanistrum leaves. The extracted dsRNAs were also used to treat a DBM cells by using the Cellfectin II Reagent (Fisher Scientific, Massachusetts, United States of America) according to the manufacturer’s instructions.
DBM cells were maintained in 3.5 ml of Gibco SF-900 SFM (Thermo Fisher Scientific, Detroit, United States of America) with 10% Gibco FBS (Fetal Bovine Serum) (Thermo Fisher Scientific, Detroit, United States of America) at a quantity of 6 × 106 DBM cells/1000 μL in 12-well plates (Thermo Fisher Scientific, Detroit, United States of America) and then stored at 27 °C overnight or until 60% – 70% confluence. Two types of dsRNA were individually transfected into DBM cells in order. First, 1 μg of dsPxAgo2 was transfected into DBM cells according to the manufacturer’s instructions for the Cellfectin II Reagent (Fisher-Scientific, Massachusetts, United States of America). The treated DBM cells were cultivated at 25 ± 2 °C for 24 h in a locked chamber to avoid infection. A total of 1 μg of dsPxBurs-α was then transfected using the same Cellfectin II Reagent (described as before) into the treated DBM cells 24 h after the first dsPxAgo2 transformation. The dsEGFP was used as a control. The PxAgo2 expression level was determined by the RT-qPCR assay. For the overexpression of PxAgo2, plasmid Pizt/v5+PxAgo2 was transfected into a DBM cells. The expression of PxAgo2 was determined by the RT-qPCR. The presence of green fluorescence was quantified using an inverted microscope (Olympus, Greece). These cells were collected 48 h after the second transfection in sterilized 1.5 mL tubes (Thomas Scientific, Swedesboro, United States of America) by centrifugation at 1000 G and 5 °C for 6 min. The methods of the entire RNA purification, quantity and quality verification and complementary DNA synthesis were the same as previously described. PxAgo2 and PxBurs-α expression levels were determined by the RT-qPCR using the primers displayed in Table 1. All experiments were repeated by three times.
The egg, 1st, 2nd, 3rd, 4th instar larva, pre-pupa, pupa and adult of DBM were separately homogenized and liquefied in the phosphate-buffered saline (1xPBS). The protein quantification was measured with the BCA Protein Assay kit (YEAβSEN, Shanghai, China). Samples were separated with an 8% SDS-PAGE and then transferred to PVDF membranes (Thermo Fisher Waltham, United States of America). The polyclonal rabbit anti-mAgo2 (1:300 dilutions; ABclonal) and anti-actin (1:1000 dilution; Vazyme) primary antibodies were used to identify the PxAgo2 and PxActin proteins, respectively. The secondary goat anti-rabbit IgG antibody (1:5000 dilutions; BOSTER, Wuhan, China) was conjugated with HRP.
GoTaq® qPCR master mix (Promega, Madison, United States of America) was used for the RT-qPCR with the following settings: denaturation at 94 °C for 3 min, followed by 35 cycles at 94 °C for 6 s and 70 °C for 1 min. The amount of amplification was quantified at the end of each cycle by measuring the fluorescence strength. The quantitative mRNA measurements were performed in triplicate and included an internal control of RL-32 (Ribosomal Protein 32). All the data were evaluated using the GraphPAD Prism version 8.0. All of the RT-qPCR primers are in Table 1.
|
Gene Name |
Sequence (5′ - 3′) |
Use |
|
PxAgo2-F |
ATGTCTTTTAAAGGAACGGGAATCG |
PCR |
|
PxAgo2-R |
TTAAACGAAGAACATGGGGTTTCTC |
PCR |
|
PxAgo2-KpnI-F1 |
GGTACCATGTCTTTTAAAGGAACGGGAATCG |
PCR |
|
PxAgo2-EcoRI-R1 |
GAATTCAACGAAGAACATGGGGTTTCTC |
PCR |
|
PxAgo2-F1 |
AACTACTCTGAACCCGCCTACT |
RT-qPCR |
|
PxAgo2-R1 |
TCCACATTTCTAATCCATCGC |
RT-qPCR |
|
PxBurs-α-F2 |
TGTTCCAAAGCCAATACCT |
RT-qPCR |
|
PxBurs-α-R2 |
TTTACTTCCCGAAACCTGA |
RT-qPCR |
|
RL32-F |
CAATCAGGCCAATTTACCGC |
RT-qPCR |
|
RL32-R |
CTGGGTTTACGCCAGTTACG |
RT-qPCR |
|
dsAgo2-F |
CTAGAAGTATAGGGTTGT |
dsRNA synthesis |
|
dsAgo2-R |
AGATATGTCTTTTAAGGA |
dsRNA synthesis |
|
dsPxBurs-α-F |
GAGAAGGCTCAGGAGGTCCAAGTA |
dsRNA synthesis |
|
dsPxBurs-α-R |
AGAGTTGTGAAGTGGTGATTCC |
dsRNA synthesis |
|
dseGFP-F |
GTGTTCAATGCTTTTCCCGTTATCC |
dsRNA synthesis |
|
dseGFP-R |
ACCATGTGGTCACGCTTTTCG |
dsRNA synthesis |
Table 1: Description of primers used for PxAgo2 vector construction, gene expression and dsRNA preparation.
Statistical analysesThe statistical analyses were performed using the SPSS software (IBM SPSS Statistics version 24.0, Armonk, New York, United States of America). The gene expression data were analyzed by one-way analysis of variance (ANOVA), followed by multiple comparisons among treatments with Tukey’s test at P < 0.05. For the comparison of low and high concentrations of dsRNA treatments, the data were analyzed by the Wilcoxon test.
3. Results
Identification and characterization of the deduced amino acid sequence of PxAgo2The Ago2 gene was identified in P. xylostella genomic data (Gene ID: Px012073) by using CDS sequence of Ago2 of B. mori (BmAgo2, GenBank: NM_001043530.2). The specific pair of primers, PxAgo2-forward and PxAgo2-reverse (Table 1), were designed to clone PxAgo2, and the PCR product was sequenced (Fig. 1). The sequence was then submitted to the GenBank under the succession number of MH444596. To identify the conserved domains, the deduced amino acid sequence of PxAgo2 was analyzed using the online NCBI-CDD to identify the domains of PxAgo2, including the Piwi-like subfamily (799-2046 aa), PAZ (304-666 aa) and the Argonaute linker 1 (157-294 aa) (Figure 1).
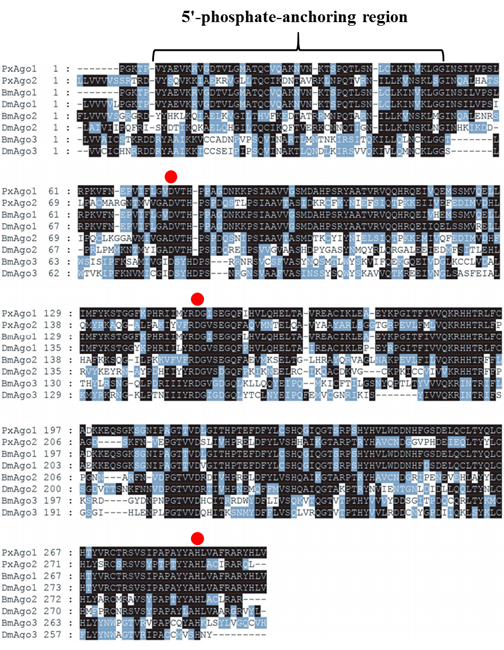
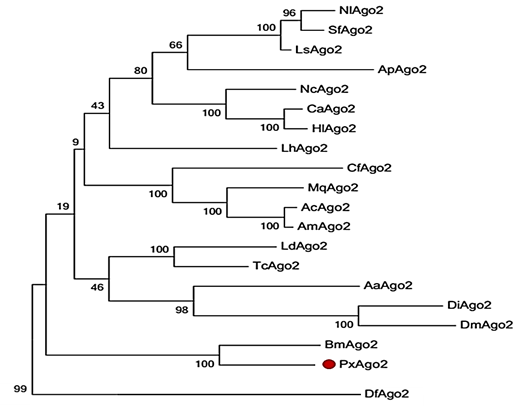
The similarity of PxAgo2 with Pectinophora gossypiella Ago2 was 63.2%, with Bombyx mori Ago2 58.5%, and with Pararge aegeria Ago2 59.8%. The molecular weight of PxAgo2 was 83 kDa and its isoelectronic point was 9.39. To examine the functional parts of the sequence, the deduced amino acid sequence of PxAgo2 and some AGO protein sequences were compared based on the highly preserved PIWI domain. Multiple sequence alignment was directed to the recognition of one 5′-phosphate anchoring area and three cleavage residues (DDH indicated by red circle in Figure 2) in the region of the PIWI domain. Nineteen Ago2 protein sequences from different insect species and Dermatophagoides farinae were downloaded from the NCBI website to make the phylogenetic tree, which revealed that the insect Ago2 could be classified into different groups based on order and PxAgo2 is closely related to BmAgo2 in the Lepidoptera group (Figure 3).
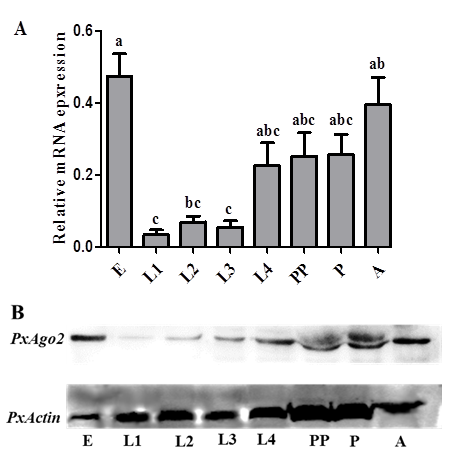
PxAgo2 mRNA and protein expression profiles in P. xylostellaThe PxAgo2 mRNA expression levels were low in the 1st, 2nd, and 3rd instar larva, but high in egg, 4th instar larva, pre-pupa, pupa and adult of P. xylostella (F=6.69, df1= 7, df2= 16, P= 0.0008) (Figure 4A). The protein expression profile of PxAgo2 was similar to the mRNA expression profile of all developmental stages of P. xylostella, with the highest expression levels in egg and adult (Figure 4B).
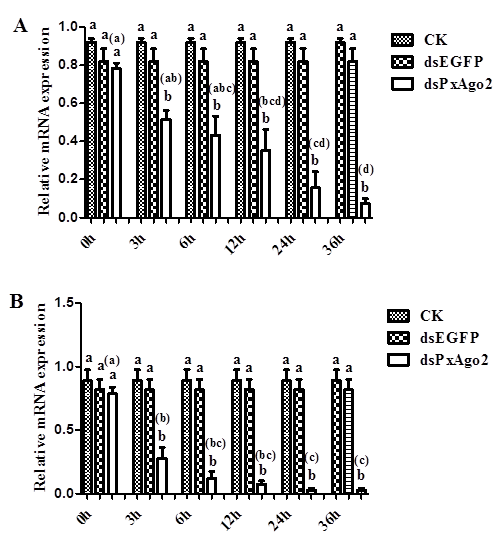
DsRNA-mediated suppression of PxAgo2 in P. xylostella embryoThe mRNA expression levels of PxAgo2 were considerably lower in the 3rd instar larvae 3 h after dsPxAgo2 injection at the low concentration of 1 μg/μL than those of CK and dsEGFP (Figure 5A). The silencing effect of PxAgo2 was significantly increased with time over the 36 h period (F=11.80, df1= 5, df2= 12, P=0.0003). DsRNA at the high concentration of 2 μg/μL also suppressed PxAgo2 expression (Figure 5B), but there was an increased silencing of PxAgo2 over the time period of 36 h by the high dsRNA concentration compared with the low concentration of 1 μg/μL dsRNA (Z = -3.419, P = 0.001).
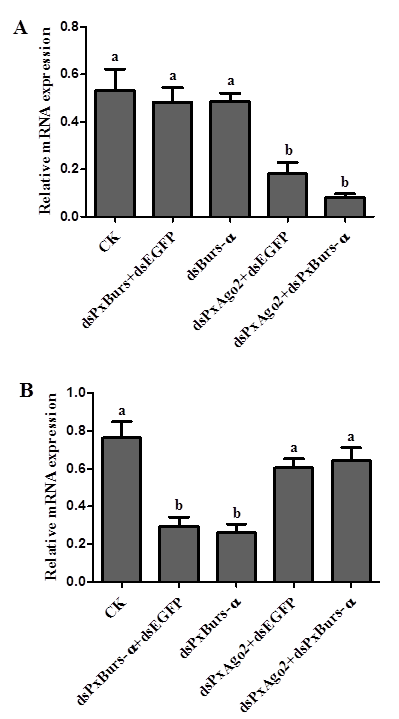
PxAgo2 suppression reduced the RNAi response in DBM cellsPxAgo2 was significantly suppressed by dsPxAgo2 transfection at the mRNA level (dsPxAgo2+dsEGFP and dsPxAgo2+dsPxBurs-α) in DBM cells (F=13.25, df1=4, df2=10, P=0.0005) (Figure 6A). The PxBurs-α expression level was significantly reduced by dsPxBurs-α and dsPxBurs-α+dsEGFP but not by dsPxAgo2+dsPxBurs-α compared with dsEGFP and dsPxAgo2+dsEGFP (F=13.17, df1= 4, df2= 10, P= 0.0005) (Figure 6B). Therefore, PxAgo2 knockdown by dsPxAgo2 decreased the gene silencing efficiency by dsRNA in DBM cells.
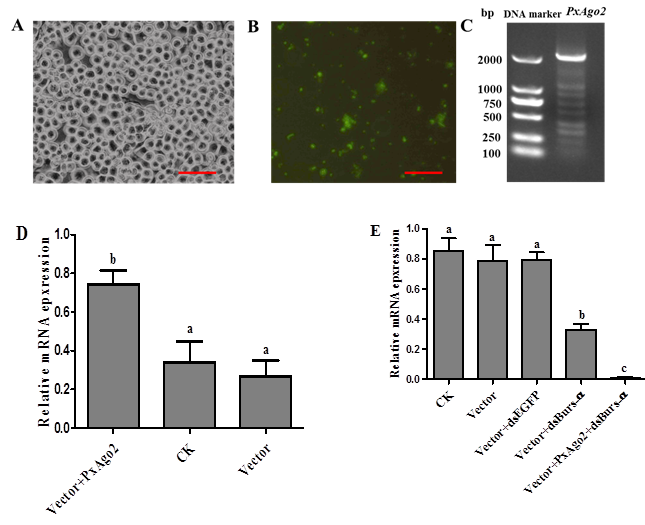
PxAgo2 overexpression improved the RNAi response in DBM cellsTo overexpress PxAgo2 in DBM cells, the PxAgo2 sequence was introduced into the plasmid Pizt/v5 and then transfected into a DBM cells. The transfection effectiveness was confirmed by eGFP expression, with the detection of a green color under the florescence microscope (Figure 7A and 7B) and by PCR product using the Pizt/v5-based primers displayed in Table 1 (Figure 7C). The PxAgo2 expression level was clearly increased in transfected DBM cells (F=8.53, df1=2, df2=6, P=0.0176) (Figure 7D). RNAi efficiency was evaluated in PxAgo2-overexpressing cells by silencing PxBurs-α with dsPxBurs-α. PxBurs-α expression was downregulated in wild type cells, while the silencing effect was markedly improved in the PxAgo2-overexpressing cells (F=30.65, df1=4, df2=10, P<0.0001) (Figure 6E). Therefore, the overexpression of PxAgo2 enhanced RNAi effectiveness in DBM cells.
4. Discussion
In this study, we determined the complete sequence of Ago2 (PxAgo2) from P. xylostella. The nucleotide sequence of PxAgo2 contained two specific domains, a PAZ motif and a PIWI motif. Both the PAZ and PIWI motifs are highly conserved in many insect species, including D. melanogaster [5, 28], B. mori [29], P. xylostella [30], and Diabrotica virgifera [31]. The interaction of the 3' end of siRNA with the PAZ domain is needed for RNAi response [30, 32]. One of the functions of the PIWI domain is to bind to the 5' end of the gRNA [33]. The cleavage motif of DDH was also found in the PIWI domain of PxAgo2. This type of motif (DDH) within the PIWI domain of Ago2 is associated with catalytic action in the Ago2 of H. sapiens [6], D. melanogaster [7, 8], and B. mori [11].
The PxAgo2 mRNA and protein expression levels were high in egg, 4th instar larva, pre-pupa, pupa and adult of P. xylostella. Recently, the mRNA and protein expression levels of Ago2 have been reported in cancer cell lines [34] and in melanoma cell lines [35]. Increased mRNA expression of Ago2 has been observed in hepatocellular carcinoma cells [36] and D. melanogaster [37, 38]. In B. mori, increased Ago2 protein expression has been reported in the 5th instar larvae [17]. Thus, Ago2 expression levels may differ among animal developmental stages, cells and tissues.
PxAgo2 was suppressed following dsPxAgo2 exposure and showed a high suppression rate by 36 h. In mammals, Ago2 can be silenced by dsAgo2 in 36 h and 72 h [39, 40]. Ago2 expression is decreased due to the induction of dsAgo2 at 24 h in Bombyx mori [41], 36 h in H. sapiens A549 cells [42], and 72 h in HeLa cells [43]. The silencing effect was increased using a high dsRNA concentration in P. xylostella larvae. High dsRNA concentrations enhance the dsRNA efficacy in D. melanogaster [44] and Dugesia japonica [45]. The dsRNA-mediated RNAi by Ago2 might be effective with a high dsRNA concentration in eukaryotes.
The PxAgo2 knockdown induced by dsPxAgo2 reduced the gene silencing efficiency in DBM cells. Ago2 participates in dsRNA-mediated RNAi in B. mori Bm5 cells [17] as well as in D. melanogaster [10] and L. decemlineata [13]. The overexpression of PxAgo2 improved the gene silencing efficiency in DBM cells. Ago2 overexpression in B. mori larvae [17], H. sapiens [46], and in mammals [47] can increase the RNAi efficiency. Ago2 has been described as a slicer element in the siRNA pathway involved in the gene silencing process of insects [5, 48]. We found that PxAgo3 can also play an essential role in the gene silencing process of P. xylostella [30]. The relationship of PxAgo2 and PxAgo3 should be further investigated in the RNAi pathway of P. xylostella. In addition, further research should focus on the identification of proteins that interact with Ago2 in RISC and clarify their functions in the RNAi pathway.
Acknowledgement
The authors would like to thank Ms. Xuejiao Xu and Mr. Wei Chen for their technical help in the PxAgo2 gene cloning and Western blot analysis. I am also thankful to Dr. Xiaoli Ma for providing a DBM cells. This study was supported by the NSFC (Natural Science Foundation of China) Project #31772237 and the Fujian Science and Technology Major Project #2018NZ01010013.
Author Contributions
Guang Yang and Muhammad Salman Hameed designed all the experiments. Muhammad Salman Hameed, Guang Yang, Gefu Wang-Pruski and Liette Vasseur wrote the research article; Muhammad Salman Hameed performed all experiments, while Xiaodong Jing, Jinzhi Chen and Wei Chen participated in gene cloning and Western blotting; Muhammad Salman Hameed and Xiaodong Jing, and Wei Chen performed statistical analysis.
Conflict of interests
The authors declare no conflict of interest.
References
- Fire A. RNA-triggered gene silencing. Trends in Genetics 15 (1999): 358-363.
- Marcel T, F Rene and H Ronald. The genetics of RNA silencing. Annual Review Genetics 36 (2002): 489-519.
- Chendrimada TP, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436 (2005): 740-744.
- Hutvagner G and MJ Simard. Argonaute proteins: key players in RNA silencing. Nature Reveiw Molecular Cell Bioliology 9 (2008): 22-32.
- Lingel A, et al. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426 (2003): 465-469.
- Rivas FV, et al. Purified Argonaute2 and an siRNA form recombinant human RISC. Nature Structural and Molecular Biology 12 (2005): 340-350.
- Kim K, YS Lee and RW Carthew. Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes. RNA 13 (2007): 22-29.
- Miyoshi K., et al. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes & Development 19 (2005): 2837-2848.
- Wynant N, D Santos and JV Broeck. The evolution of animal Argonautes: evidence for the absence of antiviral AGO Argonautes in vertebrates. Scientific Reports 7 (2017): 1-13.
- Okamura K, et al. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes & Development 18 (2004): 1655-1666.
- Wang GH, et al. Characterization of Argonaute family members in the silkworm, Bombyx mori. Insect Science 20 (2013): 78-91.
- Miller SC. RNA interference in the red flour beetle Tribolium castaneum. 2009: Kansas State University.
- Yoon JS, et al. RNA interference in the Colorado potato beetle, Leptinotarsa decemlineata: Identification of key contributors. Insect Biochemisty and Molecular Biology 78 (2016): 78-88.
- Garbutt JS and SE Reynolds. Induction of RNA interference genes by double-stranded RNA; implications for susceptibility to RNA interference. Insect Biochemistry and Molecular Biology 42 (2012): 621-628.
- Van Mierlo JT, et al. Convergent evolution of argonaute-2 slicer antagonism in two distinct insect RNA viruses. PLoS Pathogens 8 (2012): 1-14.
- Zhu L, et al. A MC motif in silkworm Argonaute 1 is indispensible for translation repression. Insect Molecular Biology 22 (2013): 320-330.
- Li Z, et al. Enhancement of larval RNAi efficiency by over-expressing Argonaute2 in Bombyx mori. International Journal of Biological Sciences 11 (2015): 176-185.
- Talekar N and A Shelton. Biology, ecology, and management of the diamondback moth. Annual Review of Entomology 38 (1993): 275-301.
- Furlong MJ, DJ Wright, and LM Dosdall. Diamondback moth ecology and management: problems, progress, and prospects. Annual Review of Entomology 58 (2013): 517-541.
- Yang ZX, et al. Effects of injecting cadherin gene dsRNA on growth and development in diamondback moth Plutella xylostella (Lep.: Plutellidae). Journal of Applied Entomology 133 (2009): 75-81.
- Mohamed AA and Y Kim. A target?specific feeding toxicity of β1 integrin dsRNA against diamondback moth, Plutella xylostella. Archives of Insect Biochemistry and Physiology 78 (2011): 216-230.
- Gong L, et al. Silencing of Rieske iron–sulfur protein using chemically synthesised siRNA as a potential biopesticide against Plutella xylostella. Pest Management Science 67 (2011): 514-520.
- Gong L, et al. Testing insecticidal activity of novel chemically synthesized siRNA against Plutella xylostella under laboratory and field conditions. PLOS ONE 8 (2013): 1-8.
- Wang H, et al. Molecular cloning and characterization of a SID?1?like gene in Plutella xylostella. Archives of Insect Biochemistry and Physiology 87 (2014): 164-176.
- Bautista MAM, et al. RNA interference-mediated knockdown of a cytochrome P450, CYP6BG1, from the diamondback moth, Plutella xylostella, reduces larval resistance to permethrin. Insect Biochemistry and Molecular Biology 39 (2009): 38-46.
- You M, et al. A heterozygous moth genome provides insights into herbivory and detoxification. Nature Genetics 45 (2013): 220-225.
- Ma XL, et al. Cell lines from diamondback moth exhibiting differential susceptibility to baculovirus infection and expressing midgut genes. Insect Science 2017: 1-37.
- Song JJ. et al. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nature Structural and Molecular Biology 10 (2003): 1026-1032.
- Santos D, et al. Insights into RNAi-based antiviral immunity in Lepidoptera: acute and persistent infections in Bombyx mori and Trichoplusia ni cell lines. Scientific Reports 8 (2018): 1-10.
- Hameed MS, et al. Molecular Characterization and the Function of Argonaute3 in RNAi Pathway of Plutella xylostella. International Journal of Molecular Sciences 19 (2018): 1-13.
- Vélez AM, et al. Knockdown of RNA interference pathway genes in western corn rootworms (Diabrotica virgifera virgifera Le Conte) demonstrates a possible mechanism of resistance to lethal dsRNA. PloS ONE 11 (2016): 1-12.
- Kandeel M and Y Kitade. Computational analysis of siRNA recognition by the Ago2 PAZ domain and identification of the determinants of RNA-induced gene silencing. PLoS ONE 8 (2013): 1-10.
- Parker JS, SM Roe and D Barford. Structural insights into mRNA recognition from a PIWI domain–siRNA guide complex. Nature 434 (2005): 663-666.
- Völler D, et al. Argonaute family protein expression in normal tissue and cancer entities. PLoS ONE 11 (2016): 1-14.
- Völler D, et al. Strong reduction of AGO2 expression in melanoma and cellular consequences. British Journal of Cancer 109 (2013): 3116-3124.
- Zhang J, et al. MiRNA-99a directly regulates AGO2 through translational repression in hepatocellular carcinoma. Oncogenesis 3 (2014): 1-10.
- Wang Z, et al. Drosophila Dicer-2 has an RNA interference–independent function that modulates Toll immune signaling. Science Advances 1 (2015): 1-13.
- Förstemann K, et al. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell 130 (2007): 287-297.
- Ruda VM, et al. The roles of individual mammalian argonautes in RNA interference in vivo. PLoS one 9 (2014): 1-11.
- McManus MT, et al. Small interfering RNA-mediated gene silencing in T lymphocytes. The Journal of Immunology 169 (2002): 5754-5760.
- Czech B, et al. An endogenous small interfering RNA pathway in Drosophila. Nature 453 (2008): 798-802.
- Puissegur M, et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death and Differentiation 18 (2011): 1-49.
- Chu CY and TM Rana. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biology 4 (2006): 1-15.
- Shih JD and CP Hunter. SID-1 is a dsRNA-selective dsRNA-gated channel. RNA 17 (2011): 1057-1065.
- Rouhana L, et al. RNA interference by feeding in vitro–synthesized double?stranded RNA to planarians: Methodology and dynamics. Developmental Dynamics 242 (2013): 718-730.
- Börner K, et al. Robust RNAi enhancement via human Argonaute-2 overexpression from plasmids, viral vectors and cell lines. Nucleic Acids Research 41 (2013): 1-22.
- He J, et al. A new design of a lentiviral shRNA vector with inducible co-expression of Argonaute-2 for enhancing gene silencing efficiency. Cell & Bioscience 5 (2015): 1-12.
- Liu F, et al. Silencing the HaAK gene by transgenic plant-mediated RNAi impairs larval growth of Helicoverpa armigera. International Journal of Biological Sciences 11 (2015): 67-74.


 Impact Factor: * 3.0
Impact Factor: * 3.0 Acceptance Rate: 76.32%
Acceptance Rate: 76.32%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
