Metadichol Treatment of Fibroblasts and Embryonic Stem Cells Regulates Key Cardiac Progenitors
Palayakotai R. Raghavan1*
1Nanorx Inc., PO box 131, Chappaqua, NY 10514, USA.
*Corresponding author: Palayakotai R. Raghavan. Nanorx Inc., PO box 131, Chappaqua, NY 10514, USA
Received: 09 July 2023; Accepted: 12 August 2023; Published: 22 August 2023
Article Information
Citation: Palayakotai R. Raghavan. Metadichol Treatment of Fibroblasts and Embryonic Stem Cells Regulates Key Cardiac Progenitors. Cardiology and Cardiovascular Medicine. 7 (2023): 302-310.
View / Download Pdf Share at FacebookAbstract
Background: Heart disease has been identified as one of the main causes of heart attack; moreover, it is known to result in the death of billions of cardiomyocytes, which cannot be reproduced and replaced. The remaining cells are often faced with a significant increase in hemodynamic burden. Repairing the heart after an attack or other cardiovascular disease has eluded medical science. The ability to repair the heart muscles using the own cells of a patient who had suffered from a heart attack is a long-sought goal to regenerate healthy new tissues. Cell sources for cardiac disease treatment include human embryonic stem cells (hESCs), which are known to have the capacity to differentiate into chondrocytes, osteoblasts, adipocytes, and cardiomyocytes. Cardiac fibroblasts are present in large numbers in the heart; they are known to be involved in the repair of the structural, biochemical, mechanical, and electrical properties of the myocardium.
Methods: Here, we present the use of Metadichol for the treatment of hESCs and human cardiac fibroblasts, wherein it resulted in the increase in the expression of the key cardiac progenitors ISL1, NKX2-5, and GATA4, WT-1, KIT, determined to be crucial in heart development. We characterized their mRNA expression by qRT-PCR and protein expression by Western blot.
Results: Treating hESCs with Metadichol at a concentration of 1 nanogram for 24 h has led to the enhanced gene expression of ISL1 (67-fold), GATA4 (7.5-fold), NXK2-5 (4.35-fold), Kit (10.41-fold) and WT1(4.99-fold). In human cardiac fibroblasts treated with 100 picogram of Metadichol, increases of ISL1 (1.77-fold), NKX2-5 (19.82-fold), and GATA4 (19.9- fold) kit (12.23-fold) and WT1(1.87-fold) were seen.
Conclusions: These cardiac progenitors play a role in heart diseases, especially ISL1, which has been suggested to be a single source for the heart lineages and its development. Given the fact that Metadichol is nontoxic and safe it can be directly tested in humans skipping other procedures being employed today and improvements monitored non-invasively. possibility of the direct use of Metadichol in patients with heart disease, as it has no known toxicity and is commercially available and marketed worldwide.
Keywords
<p>Fibroblasts; Cardiac progenitor markers; GATA4; Cardiomyocytes; NKX2-5; WT1; ISL1; c-kit; Metadichol VDr; Inverse agonist; Vitamin C.</p>
Article Details
1. Introduction
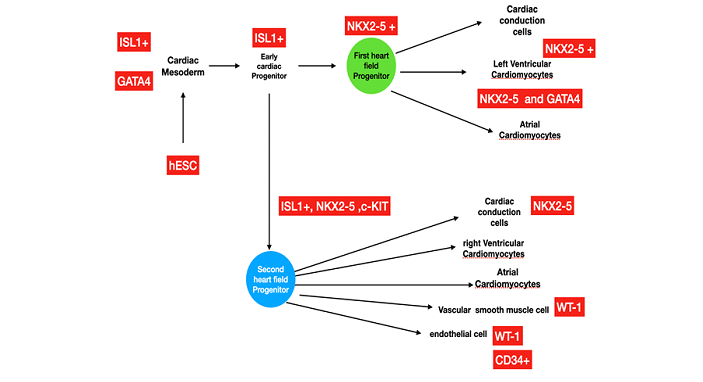
More than 12 million people are reported to die annually from heart failure worldwide. Moreover, 2.5 million children are born with congenital heart disease yearly [1], and many of them will eventually develop heart failure. When the heart muscle is damaged and unable to cope with metabolic challenges, this may lead to a heart attack. Unlike some tissues, the heart muscle has a limited capacity for self-repair. Scar tissue often cannot transmit electric impulses and contract, often leading to heart failure or even death. The heart cannot replace cardiomyocytes that are lost due to cardiac injury. Heart transplantation is the only treatment for end-stage heart failure. Patients who suffer from other severe medical conditions are not candidates for this therapy. Additionally, there remains an insufficient supply of donated hearts to meet the demand of the growing number of patients with end-stage disease, and this procedure is not universally available. Therefore, alternative treatments to stimulate heart regeneration are significantly needed to treat end-stage heart disease and congenital defects. Approaches to increase the number of cardiomyocytes in the adult heart could involve stimulation and proliferation of existing cardiomyocytes, activation of endogenous cardiac progenitor cells, or transplantation of in vitro derived cardiomyocytes into the injured area. Converting cardiac fibroblasts by reprograming them into cardiomyocytes has been identified as one option for patients with heart diseases [2]. Cardio-differentiating factors are needed to balance the suppression of gene expression and the induction of angiogenic factors in fibroblasts. Some well-documented cardiac stem or progenitor cell markers are NKX2-5, c-kit (CD117), GATA-4, ISL1 (Islet-1), and WT1 [3]. Heart development (Figure 1) involves two kinds of cardiac progenitor cells, namely, the first heart field (FHF) and the second heart field (SHF). Malfunction of any of these heart field progenitors can lead to congenital heart defects. The two heart fields express unique biomarkers. ISL1 is the key marker for the differentiation of SHF cells, whereas NKX2.5 is a marker for both the FHF and SHF cells.
The transcription factors c-kit, GATA4, WT1, ISL1, and NKX2.5 drive the morphogenic events involved in the formation of the multi-chambered heart. Progenitor cells are involved in the replacement of dead cardiomyocytes at a low basal level. The regulation of early factors such as NANOG, OCT4, and SOX2 is needed for pluripotency [4]. Metadichol already expresses these factors with stem as well as fibroblast cells [5]. Endothelial cells created during heart development are key for vessel formation, cardiac muscle cells for contractility, and cardiac conduction cells for the overall electrical activity of the heart [6]. Dysregulation of cardiac progenitor cells may lead to heart problems. Cells needed to repair the damaged heart muscle and other tissues could be derived from stem cells. Recent research has demonstrated the role of multipotent progenitor cells in the heart; it also demonstrated the processes on how pluripotent cells may aid in the expression of types of cells needed to treat certain human heart diseases. Transcription factors from cells at sites of damage can protect tissue from being damaged. [7]. Committed cells like fibroblasts can be programmed into induced pluripotent stem cells (iPSCs) [8]. Reprogramming can convert cardiac fibroblasts in the myocardium into cardiomyocytes [9] with the GATA4, MEF2C, and TBX5 (GMT) [10] transcription factors. Somatic cells are also found useful since a patient’s own fibroblasts can be converted for specific use in cardiac diseases [11,12]. Cardiomyocytes derived from iPSCs or ESCs have been shown to improve cardiac function after transplantation of (c-kit+ enriched cardiac stem cells treatment in an ischemic heart led to formation of blood-carrying new vessels) [13,14]. Cardiac resident stem cells (CRSCs) can be another source of cells to treat heart diseases. CRSCs express GATA4, but not c-kit, NKX2.5, ISL1, or WT1. CRSCs can produce cardiovascular lineages and release cytokines implicated in angiogenesis, inflammation, and cell survival [15].
1.1 OVERVIEW OF CARDIAC PROGENITOR CELL BIOMARKERS
1.1.1 ISL1
One of the key cardiac progenitors is ISL1, which is known to generate diverse cardiovascular lineages [16,17,18]. ISL1 is an important heart progenitor factor, as it induces the development of the heart and cardiac muscle, smooth muscle, and endothelium. ISL1+ progenitors are present in different regions of the embryonic and adult heart, leading to the theory that it is a stem cell factor for the differentiation of various lineages in the heart [19,20].
1.1.2 WT1
Wilms’ tumor-1 (WT1) has been determined to play a major role in cardiac development [21] WT1 expression is seen in the proepicardium and the epicardium [22]. WT1 expression is key for the development of the coronary vessels and fibroblasts. WT1 is expressed exclusively via the endothelial cells in the heart. WT1 expression is seen to increase when cardiac injury is present; it is often localized in the area where the infraction occurred, demonstrating the importance of WT1 in cardiac repair and regeneration. WT1 expression is also needed for the development of the kidneys, gonads, and spleen [23].
1.1.3 C-KIT
In 2003, Beltrami et al. [24] reported the discovery of c-kit expression in rodent’ cardiac cells that could potentially lead to all cardiac lineages, including cardiomyocytes. C-kit was co-expressed with Nkx2.5 and GATA-4 and has positive effects on cardiac repair and regeneration .
1.1.4 GATA4
GATA4 along with NKX2-5 can regulate cardiac gene expression and convert cardiac fibroblasts into functional cardiac cells in vitro and in vivo [25-28]. GATA4 plays a key role in cardiac signaling pathways with NKX2-5 in the regulation of cardiac development and hypertrophy.
1.1.5 NKX2-5
Nkx2-5 has been determined as one of the earliest markers of the cardiac lineage, and its role in embryonic development has been examined extensively [reviewed in [29-30]. It is expressed in the FHF and the SHF [31]. The role of Nkx2-5 has also been examined at later stages of heart development using Nkx2-5 conditional null mice. These animals exhibited heart chamber dilatation and progressive heart failure [32]. After birth, Nkx2-5 is known to be expressed in the post-natal heart, and its expression is significantly increased in hypertrophied hearts [33]. Mutations in the human NKX2-5 gene may lead to congenital heart disease. Despite much progress in the field, major hurdles remain for human heart regeneration [34-36]. Based on previous knowledge, we used Metadichol [37], which is nontoxic [38-40], at concentrations of 1 picogram to 100 ng in stem cells and human cardiac fibroblast cells to establish the overexpression of the cardiac transcription factors NKX2.5, WT1, KIT, GATA4, and ISL1[41]. We characterized their mRNA expression by qRT-PCR and protein expression by Western blot.
2. Materials and Methods
All work was performed by a service provider, Skanda Life Sciences (Bangalore, India).
2.1 CHEMICALS AND REAGENTS
A549 cells, Colo-205 cells, and human cardiac fibroblast cells were obtained from ATCC (USA). Primary breast cancer cells were obtained from BIOIVT (Detroit, Michigan, USA). Primary antibodies were purchased from ABclonal (Woburn, MA, USA) and Elabscience (Houston, Texas USA). Primers (Table 1) were sourced from SahaGene (Hyderabad, India).. Additional reagents were obtained from Sigma-Aldrich, Bangalore, India.
Table 1: Primers
|
Gene Name |
Sequence |
|
|
ISL-1 |
Forward |
TACAAAGTTACCAGCCACC |
|
Reverse |
GGAAGTTGAGAGGACATTGA |
|
|
Gata4 |
Forward |
CCTGTCATCTCACTACGG |
|
Reverse |
GCTGTTCCAAGAGTCCTG |
|
|
Nkx2-5 |
Forward |
CACCGGCCAAGTGTGCGTCT |
|
Reverse |
GCAGCGCGCACAGCTCTTTC |
|
|
c-kit |
Forward |
TGGGCCACCGTTTGGAAAGCT |
|
Reverse |
AGGGTGTGGGGATGGATTTGCTCT |
|
|
WT-1 |
Forward |
CAGCTTGAATGCATGACCTG |
|
Reverse |
TATTCTGTATTGGGCTCCGC |
2.2 MAINTENANCE AND SEEDING
The cells were maintained in the appropriate medium, with or without the required supplements and 1% antibiotics, in a humidified atmosphere of 5% CO2 at 37°C. The medium was changed every other day until the cells reached confluency. The viability of the cells was assessed using hemocytometer.
When the cells reached 70%–80% confluence, single-cell suspensions containing 106 cells/mL were prepared and seeded into 6-well plates at a density of 1 million cells per well. The cells were incubated for 24 h at 37°C in 5% CO2. After 24 h, the cell monolayer was rinsed with serum-free medium and treated with Metadichol concentrations as has been described below.
2.3 CELL TREATMENTS
Different concentrations of Metadichol (1 pg/mL, 100 pg/mL, 1 ng/ml, and 100 ng/mL) were prepared in serum-free medium. Subsequently, a Metadichol-containing medium was added to predesignated wells. Control cells received the medium without the drug. The cells were then incubated for 24 h. After treatment, the cells were gently rinsed with sterile PBS.
2.4 QUANTITATIVE REAL-TIME PCR (QRT-PCR)
RNA ISOLATION
Total RNA was isolated from each sample using TRIzol. Approximately, 1 × 106 cells were collected in 1.5-mL microcentrifuge tubes. The cells were thereafter centrifuged at 5000 rpm for 5 min at 4°C, and the cell supernatant was discarded. Then, 650 µL of TRIzol was added to the pellet, and the contents were mixed well and incubated for 20 min on ice. Next, 300 µL of chloroform was added, and the samples were mixed for 1–2 min by gentle inversion and incubated for 10 min on ice. Then, the samples were centrifuged at 12,000 rpm for 15 min at 4°C. The upper aqueous layer was transferred into a sterile 1.5-mL centrifuge tube, an equal amount of prechilled isopropanol was added, and the samples were incubated at −20°C for 60 min. After incubation, the mixture was centrifuged at 12,000 rpm for 15 min at 4°C. The supernatant was thereafter carefully discarded. The pellet containing RNA was washed with 1.0 mL 100% ethanol, followed by 700 µL of 70% ethanol via centrifugation, as described above. The RNA pellet was air-dried at room temperature for 15–20 min. Then, it was resuspended in 30 µL of DEPC-treated water. The RNA concentration was quantified using a SpectraDrop (SpectraMax i3x, USA) spectrophotometer (Molecular Devices). Finally, cDNA was synthesized using reverse-transcription PCR (RT-PCR).
cDNA Synthesis
cDNA was synthesized from 2 µg RNA using the PrimeScript cDNA synthesis kit (Takara) and oligo-dT primers according to the manufacturer’s instructions. A 20 μL reaction volume was used, and cDNA synthesis was performed on an Applied Biosystems instrument (Veriti). Then, qPCR was performed (50°C for 30 min followed by 85°C for 5 min).
Primers and QPCR
The PCR mixture (at a final volume of 20 µL) contained 1 µL of cDNA, 10 µL of SYBR Green Master Mix, and 1 µM specific forward and reverse primers for the respective target genes. PCR was performed under the following conditions: an initial denaturation at 95°C for 5 min, followed by 30 cycles of secondary denaturation at 95°C for 30 s, annealing at the optimized temperature for 30 s, and extension at 72°C for 1 min. The number of cycles amplifying in the exponential range without reaching a plateau was selected as the optimal cycle number. The results were then analyzed using CFX Maestro software. Fold change was calculated using the fold change was calculated using the following equation.
(ΔΔCT Method)
The comparative CT method determined the relative expression of target genes to the housekeeping gene (β-actin) and untreated control cells.
The delta CT for each treatment was calculated using the formula.
Delta Ct = Ct (target gene) – Ct (reference gene)
To compare the delta Ct of individually treated samples with the untreated control sample, the Ct was subtracted from the control to obtain the delta delta CT.
Delta delta Ct = delta Ct (treatment group) – delta Ct (control group)
The fold change in target gene expression for each treatment was calculated using the formula. Fold change = 2^ (−delta delta Ct)
Protein Isolation
Total protein was isolated from 106 cells with RIPA buffer supplemented with the protease inhibitor phenylmethyl sulfonyl fluoride. The cells were lysed for 30 min at 4°C while gently inverted. Next, the cells were centrifuged at 10,000 rpm for 15 min. The supernatant was transferred to a new tube. The Bradford method was used to determine the protein concentration, and 25 µg of protein was mixed with 1× sample loading dye containing SDS and loaded onto a gel. Proteins were then separated in Tris-glycine buffer under denaturing conditions.
The proteins were then transferred onto methanol activated PVDF membranes (Invitrogen) using the Trans-Blot Turbo system (Bio-Rad, USA). Nonspecific binding to the membranes was blocked via incubation in 5% BSA for 1 h. The membranes were then incubated overnight with the respective primary antibodies at 4°C and then with a species-specific secondary antibody for 1 h at room temperature. The blots were washed and incubated with ECL substrate (Merck) for 1 min in the dark. The images showing the results were captured at appropriate exposure settings using the ChemiDoc XRS system (Bio-Rad, USA).
3. Results and discussion
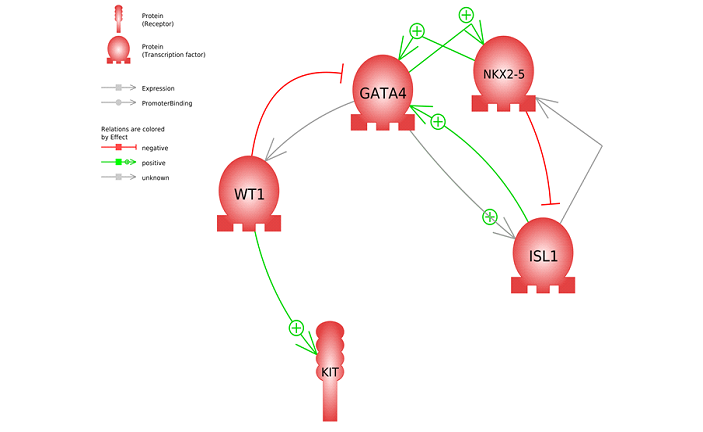
The fibroblasts and hESCs treated with Metadichol results are shown in Tables 2-5. All five targeted transcription factors were expressed. Differences can be noted between the cell types; for example, in hESCs, ISL1 at 1 ng of Metadichol showed a 67-fold increase, but it showed only a 1.77-fold increase in fibroblasts. Other than ISL1 and WT1, there was increased expression of GATA4, NKX2-5, and c-kit in fibroblasts at 100 pg compared to the 1 ng result from hESCs cells. Next, we have carried out gene network analysis in order to characterize the interactions among these five genes and assess their importance in heart regeneration. The analysis of gene networks using Pathway Studio [42,43] and protein-protein interaction maps is shown in Figure 2. The top 18 most highly significant (P < e−9) cell processes regulated by the gene set are shown in Table 6. A complete list of the diseases in the network maps regulated by these genes is available in the Supplementary Material. Figure 2 shows that WT1 regulates GATA4, which has been observed in studies demonstrating that GATA4/WT1 cooperation regulates the transcription of genes required for mammalian sex determination [44]. Maximum GATA4/WT1 synergism was dependent on WT1, but not GATA4 [45]. Additionally, NKX2-5 regulates ISL1, and this is necessary for proper specification and maturation of the ventricular myocardial lineage [46]. WT1 plays a key role in controlling the expression of GATA4 and c-kit [47].
Table 2: Mean Cq values in human cardiac fibroblasts after treatment with Metadichol
|
Treatment |
Actin |
ISL1 |
WT1 |
c-KIT |
NKX2-5 |
GATA4 |
|
Control |
20.85 |
27 |
12.86 |
16.71 |
22.85 |
22.94 |
|
1 pg |
20.53 |
26.39 |
12.6 |
14.34 |
21.49 |
21.28 |
|
100 pg |
21.34 |
26.67 |
12.45 |
13.59 |
19.82 |
19.91 |
|
1 ng |
20.62 |
26.56 |
11.69 |
14.89 |
21.9 |
20.2 |
|
100 ng |
20.75 |
27.99 |
12.34 |
16.7 |
23.26 |
22.56 |
Table 3: Relative fold expression of various biomarkers
|
Treatment |
ISL1 |
WT1 |
c-KIT |
NKX2-5 |
GATA4 |
|
Control |
1 |
1 |
1 |
1 |
1 |
|
1 pg |
1.23 |
0.96 |
4.16 |
2.06 |
2.54 |
|
100 pg |
1.77 |
1.87 |
12.23 |
11.48 |
11.51 |
|
1 ng |
1.16 |
1.91 |
3.01 |
1.65 |
5.72 |
|
100 ng |
0.47 |
1.33 |
0.94 |
0.7 |
1.22 |
Table 4: Mean Cq values in human embryonic stem cells after treatment with Metadichol
|
Treatment |
GAPDH |
ISL1 |
WT-1 |
c-Kit |
NKx-2.5 |
GATA |
|
Control |
21.18 |
28.54 |
21.75 |
25.55 |
24.67 |
26.42 |
|
1 pg |
21.65 |
25.48 |
21.25 |
25.43 |
25.05 |
26.68 |
|
100 pg |
19.09 |
22.07 |
20.71 |
21.69 |
21.82 |
24.94 |
|
1 ng |
22.43 |
23.72 |
20.68 |
23.42 |
23.8 |
24.76 |
|
100 ng |
22.25 |
25.57 |
21.7 |
24.26 |
25.55 |
28.33 |
Table 5: Fold expression of target genes post treatment
|
Treatment |
ISL1 |
WT-1 |
c-Kit |
NKx-2.5 |
GATA |
|
|
Control |
1 |
1 |
1 |
1 |
1 |
|
|
1 pg |
11.55 |
1.96 |
1.51 |
1.06 |
1.16 |
|
|
100 pg |
20.82 |
0.48 |
3.41 |
1.69 |
0.66 |
|
|
1 ng |
67.18 |
4.99 |
10.41 |
4.35 |
7.52 |
|
|
100 ng |
16.45 |
2.17 |
5.13 |
1.14 |
0.56 |
Table 6: Cell processes regulated by input genes
A sample of over 300 cellular processes regulated by these genes is shown in Table 6. Most of the processes are found to be involved in cell differentiation, pluripotency maintenance, cardiomyocyte differentiation, heart looping, and other processes related to development of the heart and vital cellular processes. These genes have more interactions among themselves than expected for a random set of genes of the same size and degree distribution drawn from the genome. Such enrichment indicates that the group of five genes shares a significant biological connection through their involvement in heart development processes [48]. Based on these results, the expressed genes and their related mechanisms pave the way in reproducing these findings in vivo in humans. All patients had normalized blood pressure in 12 weeks on treatment with Metadichol, as shown in our previous study [49]. Endothelial dysfunction [50] is a leading cause of hypertension; thus, it is likely that Metadichol treatment led to overexpression of genes like WT1 and replacement of the damaged endothelial cells, thereby leading to normalized diastolic and systolic blood pressure. Because Metadichol is nontoxic, it allows one avoid in vitro cell approaches; thus, avoiding harvesting, and other cumbersome methods that are in use today and can be tested directly in patients to potentially repair their damaged heart tissues noninvasively.
4. Conclusions:
Our results show that the vital cardiac progenitors that play a critical role in heart diseases are expressed in the stem, and cardiac dermal cells without grafting that are in use today. Direct cardiac reprogramming [51] is what this work can potentially bring us closer to cardiac regeneration therapy given the non-toxic nature of Metadichol® and avoiding the cumbersome methods in operation today in gene therapy.
Abbrevations:
Gene Description
ISL1 Insulin Gene Enhancer Protein ISL-1
WT1 WT1 transcription factor
NKX2-5 NK2 homeobox 5
KIT KIT proto-oncogene, receptor tyrosine kinase
GATA4 GATA binding protein 4
Declarations
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author Contributions
All work was planned and supervised by the author (PRR) who is solely responsible for its content.
Competing Interests:
NONE
Availability of Data and Material:
All data is in manuscript and in the supplementary material provided.
Funding
Funding was provided by Nanorx Inc. R&D Budget.
Acknowledgments
This work has been published as a pre-print. P.R. Raghavan (2022. https://doi.org/10.2120/rs.3.rs-19111993/v1.
References
- Mozaffarian D, Benjamin EJ, Go AS, et al. Heart disease and stroke statistics-2016 Update: A Report From the American Heart Association. Circulation 133 (2016): 38-360.
- Kuhn EN, Wu SM. Origin of cardiac progenitor cells in the developing and postnatal heart. J Cell Physiol 225 (2010): 321-325.
- Patel VK, Mathison M, Singh VP, et al. Direct cardiac cellular reprogramming for cardiac regeneration. Curr Treat Options Cardiovasc Med 18 (2016): 58.
- Rizzino A. Concise review: the Sox2-Oct4 connection: critical players in a much larger interdependent network integrated at multiple levels. Stem Cells 31 (2013): 1033-1039.
- R. Raghavan. Metadichol, a natural ligand for the expression of Yamanaka reprogramming factors in somatic and primary cancer cell lines 4 (2022).
- Später D, Hansson EM, Zangi L, et al. How to make a cardiomyocyte. Development 141 (2014): 4418-4431.
- Zhang Y, Sivakumaran P, Newcomb AE, et al. Cardiac repair with a novel population of mesenchymal resident in the human heart, 2015. Stem Cells 33 (2015): 3100-3113.
- Srivastava D, Ivey KN. Potential of stem-cell-based therapies for heart disease. Nature 441 (2006): 1097-1099.
- Chen Y, Yang Z, Zhao ZA, et al. Direct reprogramming of fibroblasts into cardiomyocytes. Stem Cell Res Ther 8 (2017): 118.
- Ieda M, Fu JD, Delgado-Olguin P, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142 (2010): 375-386.
- Davis RP, Casini S, van den Berg CW, et al. Cardiomyocytes derived from pluripotent stem cells recapitulate electrophysiological characteristics of an overlap syndrome of cardiac sodium channel disease. Circulation 125 (2012): 3079-3091.
- Nelson TJ, Martinez-Fernandez A, Yamada S, et al. Repair of acute myocardial infarction by human stemness factors induced pluripotent stem cells. Circulation 120 (2009): 408-416.
- Hosoda T. C-kit-positive cardiac stem cells and myocardial regeneration 2. Am J Cardiovasc Dis 2 (2012): 58-67.
- Bolli R, Chugh AR, D’Amario D, et al. Cardiac stem cells in patients with ischaemic cardiomyopathy (scipio): initial results of a randomised phase 1 trial. Lancet 378 (2011): 1847-1857.
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51 (1987): 987-1000.
- Laugwitz K-L, Moretti A, Caron L, et al. Islet1 cardiovascular progenitors: a single source for heart lineages? Development 135 (2008): 193-205.
- Genead R, Danielsson C, Andersson AB, et al. Islet-1 cells are cardiac progenitors present during the entire lifespan: from the embryonic stage to adulthood. Stem Cells Dev 19 (2010): 1601-1615.
- Fonoudi H, Yeganeh M, Fattahi F, et al. ISL1 protein transduction promotes cardiomyocyte differentiation from human embryonic stem cells. Plos one 8 (2013): e55577.
- Moretti A, Caron L, Nakano A, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 127 (2006): 1151-1165.
- Bu L, Jiang X, Martin-Puig S, et al. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature 460 (2009): 113-117.
- Wagner N, Wagner KD. Every beat you take-the Wilms’ tumor suppressor WT1 and the heart. Int J Mol Sci 22 (2021): 7675.
- Sjoerd N Duim, Marie-José Goumans, and Boudewijn PT, et al. Cardiac Development and Disease van den Heuvel-Eibrink MM (2016).
- Kreidberg JA, Sariola H, Loring JM, et al. WT-1 is required for early kidney development. Cell 74 (1993): 679-691.
- Beltrami AP, Barlucchi L, Torella D, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 114 (2003): 763-776.
- Addis RC, Ifkovits JL, Pinto F, et al. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J Mol Cell Cardiol 60 (2013): 97-106.
- Srivastava D, Ieda M. Critical factors for cardiac reprogramming. Circ Res 111 (2012): 5-8.
- Xin M, Olson EN, Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat Rev Mol Cell Biol 14 (2013): 529-541.
- Välimäki MJ, Ruskoaho HJ. Targeting GATA4 for cardiac repair. IUBMB Life 72 (2020): 68-79.
- Lyons I, Parsons LM, Hartley L, et al. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev 9 (1995): 1654-1666.
- Tanaka M, Kasahara H, Bartunkova S, et al. Vertebrate homologs of tinman and bagpipe: roles of the homeobox genes in cardiovascular development. Dev Genet 22 (1998): 239-249.
- Kasahara H, Bartunkova S, Schinke M, et al. Cardiac and extra cardiac expression of Csx/Nkx2.5 homeodomain protein. Circ Res 82 (1998): 936-946.
- Pashmforoush M, Lu JT, Chen H, et al. Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell 117 (2004): 373-386.
- Abdul Samad F, Suliman BA, Basha SH, et al. A comprehensive in silico analysis on the structural and functional impact of SNPs in the congenital heart defects associated with NKX2-5 gene-A molecular dynamic simulation approach. PLoS One 11 (2016): 0153999.
- Cao N, Huang Y, Zheng J, al. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science 352 (2016): 1216-1220.
- Steinhauser ML, Lee RT. Regeneration of the heart. EMBO Mol Med 3 (2011): 701-712.
- Weinberger F, Eschenhagen T. Cardiac regeneration: New Hope for an old dream. Annu Rev Physiol 83 (2021): 59-81.
- Raghavan PR. Policosanol nanoparticles. Policosanol Nanoparticles;B2:B2 Metadichol Liquid and Gel Nanoparticle formulations (2015).
- Alemán CL, Más R, Hernández C, Rodeiro I, et al. A 12-month study of policosanol oral toxicity in Sprague Dawley rats. Toxicol Lett 70 (1994): 77-87.
- Alemán CL, Ferreiro RM, Puig MN, et al. Carcinogenicity of policosanol in Sprague Dawley rats: a 24 month study. Teratog Carcinog Mutagen 14 (1994): 239-249.
- Alemán CL, Puig MN, Elías EC, et al. Carcinogenicity of policosanol in mice: an 18-month study. Food Chem Toxicol 33 (1995): 573-578.
- R. Raghavan. Metadichol tretment of Fibrobalsts and embryonic cells that regulate key cardiac Progenitors (2022).
- Nikitin A, Egorov S, Daraselia N, et al. Pathway studio--the analysis and navigation of molecular networks. Bioinformatics 19 (2003): 2155-2157.
- Sivachenko AY, Yuryev A, Daraselia N, et al. J Bioinform Comput Biol 5 (2007): 429-456.
- Miyamoto Y, Taniguchi H, Hamel F, et al. A GATA4/WT1 cooperation regulates transcription of genes required for mammalian sex determination and differentiation. BMC Mol Biol 9 (2008): 44.
- Miyamoto Y, Taniguchi H, Hamel F, et al. A GATA4/WT1 cooperation regulates transcription of genes required for mammalian sex determination and differentiation. BMC Mol Biol 9 (2008): 44.
- Dorn T, Goedel A, Lam JT, et al. Direct Nkx2-5 transcriptional repression of Isl1 controls cardiomyocyte subtype identity. Stem Cells 33 (2015): 1113-1129.
- Wagner K-D, Cherfils-Vicini J, Hosen N, et al. The Wilms’ tumor suppressor, Wt1 is a major regulator of tumor angiogenesis and progression. Nat Commun 5 (2014): 5852.
- Tomczak A, Mortensen JM, Winnenburg R, et al. Interpretation of biological experiments changes with evolution of the Gene Ontology and its annotations. Sci Rep 8 (2018): 5115.
- Raghavan PR. Systolic and diastolic BP control in metabolic syndrome patients with Metadichol® a novel nano emulsion lipid. J Cardiol & Cardiovasc Ther 5 (2017): 555660.
- Gallo G, Volpe M, Savoia C. Endothelial dysfunction in hypertension: current concepts and clinical implications. Front. Med 8 (2022): 798958.
- Yamakawa H, Ieda M. Cardiac regeneration by direct reprogramming in this decade and beyond. Inflamm Regen 41 (2021): 20.


 Impact Factor: * 5.6
Impact Factor: * 5.6 Acceptance Rate: 74.36%
Acceptance Rate: 74.36%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks