PDGFR Upregulates HMGA1 Expression in the P53 Pathway to Facilitate Epidermal Differentiation of Adipose-Derived Mesenchymal Stem Cells
Yan-Hong Wu*, Jian-Wu Chen, Yu-Zhi Wang, Peng Liu, Zhong-Shan Wang, Xiao-Qiang Chen
Department of Burns and Plastic Surgery, General Hospital of Southern Theater Command of PLA, Guangzhou, Guangdong, China
*Corresponding Author: Yan-Hong Wu, Department of Burns and Plastic Surgery, General Hospital of Southern Theater Command of PLA, Guangzhou, Guangdong, China.
Received: 17 July 2025; Accepted: 22 July 2025; Published: 22 August 2025
Article Information
Citation: Yan-Hong Wu, Jian-Wu Chen, Yu-Zhi Wang, Peng Liu, Zhong-Shan Wang, Xiao-Qiang Chen. PDGFR Upregulates HMGA1 Expression in the P53 Pathway to Facilitate Epidermal Differentiation of Adipose-Derived Mesenchymal Stem Cells. Journal of Women’s Health and Development 8 (2025): 32-43.
DOI: 10.26502/fjwhd.2644-288400138
View / Download Pdf Share at FacebookAbstract
Background: During the healing process, platelet-derived growth factor (PDGF) stimulates the regeneration of damaged tissue by interacting with PDGF receptor (PDGFR). This research seeks to investigate how PDGFR promotes the differentiation of adipose-derived mesenchymal stem cells (ADMSCs) into epidermal cells. Methods: ADMSCs were initially induced to differentiate into epidermal cells using an epidermal cell induction medium. Following the induction, cell morphology was examined microscopically, and immunofluorescence was employed to assess the fluorescence intensity of epidermal cell markers CK19 and CK10. Subsequently, cells were transfected with oe-PDGFR and exposed to the PDGF inducer PDGF-BB. Western blot and qPCR were used to determine PDGFR levels in the mRNA and protein. Cell proliferation and migration were evaluated through CCK-8 and Transwell assays.The expression levels of CK19 and CK10 was further analyzed via Western blot. In addition, bioinformatics analysis of RNA sequences were conducted to find out potential downstream targets and elucidate the molecular mechanisms. Results: Upon culturing in the epidermal cell medium, ADMSCs transformed from spindle-shaped to round or oval forms, were arranged in a cobblestone pattern, and exhibited high expression levels of CK19 and CK10. PDGFR expression was significantly increased following transfection with oe-PDGFR. Notably, the increased expression of PDGFR significantly enhanced cell proliferation and migration, elevated the expression levels of CK19 and CK10, and activated the ERK and AKT signaling pathways. The study combined bioinformatics analysis with in vitro rescue experiments, confirming that PDGFR overexpression upregulated HMGA1 expression. The inhibition of p53 activation facilitates differentiation of ADMSCs into epidermal cells. Conclusion: We found that PDGFR activated HMGA1, stimulating differentiation of ADMSCs by modulating the p53 signaling pathway in a way that facilitates their differentiation into epidermal cells.
Keywords
<p>PDGFR; Adipose-derived mesenchymal stem cells; HMGA1; p53 signaling pathway; Skin damage repair</p>
Article Details
Introduction
The skin is the largest organ of the human body and serves as one of the most vital components within the body's system [1]. Given its extensive surface area exposed to external environments, the skin is particularly susceptible to physical and chemical injuries such as bruises, abrasions, burns, and other traumas that compromise its essential barrier function [2,3]. Therefore, promoting skin wound healing is crucial to ensure barrier function is restored. Traditional methods for skin repair include autografts, allografts, xenografts, and artificial substitutes [4]. However, these conventional therapies are constrained by limitations related to donor skin availability as well as significant risks associated with immune rejection and infectious diseases linked to allografts and xenografts [5]. Therefore, there is an urgent need for enhanced strategies in skin repair.
The development of skin substitutes has advanced considerably alongside innovations in tissue engineering technology. Adipose-derived mesenchymal stem cells (ADMSCs) were first characterized and isolated in the year 2001 [6]. These cells demonstrate self-renewal capabilities along with stable proliferation and multi-lineage differentiation potential [7,8]. The derivation of ADMSCs from mesodermal progenitors within adipose tissue enables their differentiate into various cell types under specific conditions [9,10]. Due to their abundant availability, ease of acquisition, low immunogenicity, and ease of in vitro expansion, ADMSCs present significant promise for application in cutaneous wound repair [11,12]. Recent studies have indicated that incorporating diverse stimulating factors during ADMSC culture can effectively mimic the native microenvironment of skin while promoting differentiation into a variety of cellular phenotypes [13]. For instance, studies have revealed that apoptotic bodies released from ADMSCs contain miRNA-21-5p which specifically targets KLF6 to promote M2 polarization in macrophages, thereby facilitating the acceleration of wound healing processes [14]. Furthermore, another study demonstrated a marked increase in epidermal cell markers following 8 weeks post-transplantation of GFP+ transfected ADMSCs into murine models [15]. This suggests a capacity for ADMSCs to differentiate into epidermal cells although precise molecular mechanisms governing this differentiation remain elusive.
Platelet-rich plasma (PRP) is rich in growth factors derived from blood plasma. It serves a significant function in the recruitment of regenerative cells and the enhancement of wound healing [16]. The regenerative efficacy attributed to PRP may be due largely to its elevated concentrations of key growth factors such as platelet-derived growth factor (PDGF) [17]. PDGF was first discovered in fibroblasts and smooth muscle cells, where it acts as a powerful mitogen capable of stimulating both cell proliferation and differentiation [18]. Research findings suggest that epidermal stem cells obtained from human dermis exhibit improved proliferation rates and migration capacties when stimulated by PDGF [19]. The PDGF receptor (PDGFR), comprising the isoforms PDGFRα and PDGFRβ, operates as a tyrosine kinase receptor. PDGF binds to PDGFR in a dimerized conformation, thereby activating the receptors. This activation subsequently triggers downstream signaling pathways that facilitate cellular proliferation and migration [20,21]. Earlier investigations have shown that ADMSCs express the PDGFR [22]. Consequently, it is hypothesized that the activation of PDGFR may promote the differentiation of ADMSCs into epidermal cells, thereby facilitating skin wound repair. This research examines how activated PDGFR may enhance the differentiation of ADMSCs into epidermal cells, while also exploring the mechanisms that underlie this process.
Materials and Methods
ADMSC culture
ADMSCs were obtained from Lonza (Walkersville, USA) and cultured in 90% L-Dulbecco’s modified Eagle’s medium (Gibco, USA) supplemented with 10% Fetal Bovine Serum (FBS, Hyclone, USA). A controlled environment of 37°C and 5% CO2 was maintained for cell cultivation. The medium was renewed every 2 days, and the cells were subcultured as necessary. Subsequent experiments utilized cells from the third passage. The characteristics of ADMSCs were evaluated through flow cytometry, employing FITC-conjugated antibodies targeting CD29, CD90, CD44, CD34, and CD45 (all from Abcam, USA) markers. And then the morphology of ADMSCs was examined using an IX73 microscope.
ADMSCs differentiation
To induce differentiation into epidermal cells, third passage ADMSCs were cultured in an epidermal cell induction medium comprising 20 ng/ml epidermal growth factor (EGF, Sigma, USA), 5 ng/ml basic fibroblast growth factor (bFGF, R&D Systems, USA), 5 µM all-trans retinoic acid (ATRA, Sigma, USA), 0.1 µM dexamethasone, and DMEM/F-12 (DF-12, Gibco, USA) supplemented with 1% Insulin/Transferrin/Selenium (ITS, Sigma, USA) [23]. The medium was refreshed every 2 days. After a 7-day induction period, the characteristics of the resulting epidermal cells were evaluated using FITC-conjugated antibodies targeting CK10 and CK19 (all from Abcam, USA) markers. Subsequently, the induction medium was replaced with a complete epidermal cell medium containing insulin, hydrocortisone, transferrin, and DF-12 supplemented with 10% FBS, followed by an additional 3 weeks of culture for the ADMSCs. Following this induction phase, ADMSCs were exposed to PDGF-BB (Sigma, USA) for a period of 24 hours.
Cell transfection
The pLV-Puro vector (Suzhou Jima Gene Co. Ltd, China) was utilized to construct the PDGFR overexpression plasmid, while small interfering RNA (si-HMGA1) from the same company was employed to downregulate HMGA1 expression. The HMGA1 sequence was 5-UGGACUUCGAGCUCGACUCAC-3, and the negative control (NC) siRNA sequence was 5-CACCGTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTTTG-3. Following the manufacturer's protocol, these vectors along with their respective negative controls were transfected into ADMSCs by means of Lipofectamine 3000 (Invitrogen, USA). Following an 8 hours incubation, a complete medium was introduced, and the cells were further cultured for 24 hours in the absence of vectors. Upon completion of this culture period, ADMSCs were harvested to assess transfection efficiency.
CCK8 assay
A total of 2000 ADMSCs were inoculated into each well of a 96-well plate, with 6 replicate wells allocated for each group. Following an incubation period of 24 hours, the ADMSCs were treated with CCK-8 solution (Beyotime Biotechnology, China) and incubated for an additional 2 hours in each well. The optical density (OD) at 450 nm was measured using a microplate reader.
Transwell assay
The Transwell assay was carried out following manufacturer’s protocols. Briefly, a solution containing ADMSCs at a concentration of 5×105 cells/ml was placed into the upper chambers of the Transwell system. Following an incubation period of 24 hours, absolute ethanol solution was used to fix the cells on the membranes, and staining with 0.1% crystal violet (Millipore, MA) was performed for an additional 10 minutes. Finally, microscopy analysis was conducted using an Olympus microscope to assess cell morphology.
Immunofluorescence staining
ADMSCs were collected and plated into 6-well plates at a concentration of 2×105 cells per well, followed by a incubation period of 24 hours. Thereafter, the cells were fixed in 4% paraformaldehyde for a duration of 20 minutes, followed by permeabilization with a 0.5% Triton X-100 solution. The cells were treated overnight with primary antibodies specific to CK10 and CK19 (Abcam, USA). Following this incubation, the cells received an additional one-hour treatment with either Alexa Fluor 488-conjugated anti-mouse IgG secondary antibodies (Abcam, USA) or without them at ambient temperature. Afterward, the nuclei of ADMSCs were labeled with DAPI, a fluorescent dye, and subsequently examined with a confocal microscope.
Quantitative PCR (qPCR)
RNA was meticulously extracted from ADMSCs utilizing TRIzol Reagent (Invitrogen, USA). Subsequently, the synthesis of complementary DNA (cDNA) was conducted utilizing the HiScript lll 1st Strand cDNA Synthesis Kit (azyme, China). qPCR was then conducted using the Taq Pro Universal SYBR qPCR Master Mix (Vazyme, China). GAPDH served as the internal control for relative quantitative analysis. qPCR analysis was repeated 3 times. The primer sequences are comprehensively presented in Table 1.
|
Gene |
Forward (5’→3’) |
Reverse (5’→3’) |
|
PDGFR |
CCATCAGCAGCAAGGCGA |
GAACGAAGGTGCTGGAGACA |
|
CK10 |
CCCTGGGCTAAACAGCATCA |
AAAGAGCCACCACTGAACCC |
|
CK19 |
GAAGGATGCTGAAGCCTGGT |
GTCAGTAACCTCGGACCTGC |
|
HMGA1 |
AGCGAAGTGCCAACACCTAAG |
TGGTGGTTTTCCGGGTCTTG |
|
GADPH |
AATGGGCAGCCGTTAGGAAA |
GCCCAATACGACCAAATCAGAG |
Table 1: Primer sequence
Western blot
The protein samples underwent sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were subsequently transferred to a PVDF membrane.The membrane was blocked and subsequently incubated all-rnight at 4°C in a solution containing the primary antibodies against PDGRR, p53, HMGBA1, ERK1/2, p-ERK1/2, AKT, p-AET (all from Cell Signaling Technology, USA ) and GAPDH (ABclonal, China).Following washing, incubation with the secondary antibodies against were performed for 2 hours, after which the target bands were visualized using a chemiluminescent substrate. Bands were detected employing Prime Western blot reagent, and band intensity values were analyzed using ImageJ software.
RNA sequencing (RNA-seq)
In accordance with previous studies, total RNA was extracted and the integrity of the RNA was evaluated using a Bioanalyzer 2100. Purified mRNA was fragmented with fragmentation buffer. A random hexamer-primed reverse transcription method was employed to synthesize first-strand cDNA, followed by the synthesis of second-strand cDNA. Subsequently, amplification of cDNA fragments was performed via PCR. The double-stranded PCR products were subjected to heat denaturation and subsequently circularized using the splint oligonucleotide sequence to generate the final cDNA library. Library quality was evaluated with the Bioanalyzer 2100 and paired-end sequencing was performed on a BGIseq500 platform.
RNA-seq data processing and analysis
Date are from triplicate RNA-seq analysis.The sequencing data were meticulously filtered utilizing SOA Pnuke (v1.5.2) in accordance with the following criteria: (1) removal of sequencing adapter; (2) a base ratio of low-quality reads exceeding 20% (base quality less than 5); (3) the ratio of unknown bases ('N' bases) exceeded 5%. The clean reads were aligned to the reference genome (hg38) utilizing HISAT2 (v2.0.4). The alignment of clean reads to the reference gene set was conducted using Bowtie2 (v2.2.5), followed by the application of RSEM (v1.2.12) to quantify the expression levels of each gene.
DESeq2 was utilized to assess differential expressions between the oe-NC and oe-PDGFR groups, with genes defined as differentially expressed genes (DEGs) if they exhibited an adjusted P-value < 0.05 and |log2 fold change (FC)| > 2. Conduct a comprehensive pathway enrichment analysis and an extensive gene set enrichment analysis employing Gene Ontology (GO) alongside the Kyoto Encyclopedia of Genes and Genomes (KEGG).
Statistical analysis
Data analysis was conducted with GraphPad Prism 9.0. Continuous variables are presented as mean±standard deviation. The t-test was used for comparisons between two groups, whereas one-way ANOVA was applied for assessing differences among multiple groups. Pairwise comparisons were performed using the LSD-T test. A P-value of less than 0.05 was considered statistically significant.
Results
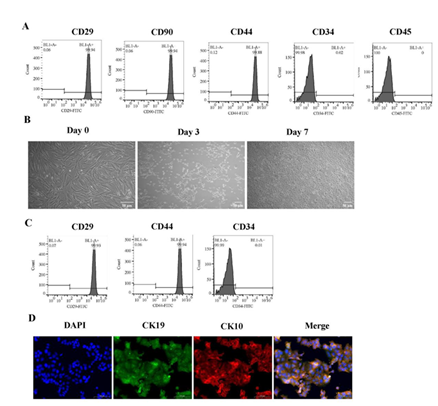
Inducing ADMSCs to differentiate into epidermal cells
The expression of surface marker on ADMSCs was evaluated using flow cytometry assay before initiating differentiation into epidermal cells. The results demonstrated a high expression of CD29, CD90, and CD44 in these cells, while CD34 and CD45 were not detected (Figure 1A). Throughout the differentiation process, cell morphology was observed under a microscope. After 3 days, the cells transitioned from a spindle shape to round or oval forms. By day 7, most cells displayed predominantly round or oval morphology and were organized in a paving stone-like arrangement (Figure 1B). At the end of the differentiation period, re-evaluation of surface markers revealed sustained high levels of CD29 and CD44 expression, however, there was an absence of CD34 (Figure 1C). Immunofluorescence analysis showed a marked increase in staining intensity for CK19 and CK10 (Figure 1D). These findings substantiate the successful differentiation of ADMSCs into epidermal cells.
(A) The expression levels of CD29, CD90, CD44, CD34, and CD45 on ADMSCs detected by flow cytometry. (B) Morphological alterations in ADMSCs were noted under microscopic examination following 0, 3, and 7 days of treatment with differentiation medium. Scale bars represent 50 μm. (C) The espression levels of CD29, CD44, and CD34 on ADMSCs after 7 days of differentiation detected by flow cytometry. (D) Immunofluorescence staining with anti-CK19 (green), anti-CK10 (red) and DAPI (blue) on ADMSCs after 7 days of differentiation. Scale bars represent 100 μm.
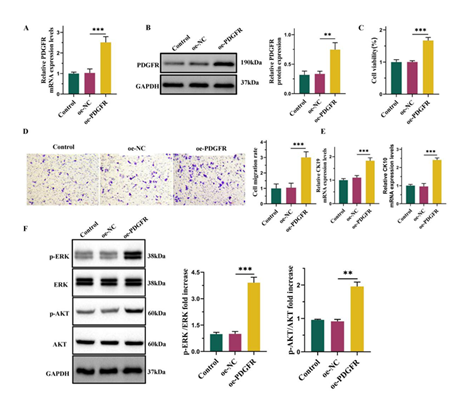
PDGFR overexpression promoted the differentiation of ADMSCs into epidermal cells
Previous studies have demonstrated that PDGF binds to PDGFR, thereby facilitating cell proliferation, migration, and invasion [24]. This study examined the function of PDGFR in facilitating epidermal differentiation of ADMSCs. Following differentiation induction, cells were exposed to PDGF-BB for a duration of 24 hours and subsequently transfected with either oe-NC or oe-PDGFR. The results revealed a marked elevation in the expression level of PDGFR following the transfection with oe-PDGFR (Figure 2A-2B). The CCK8 assay demonstrated that overexpression of PDGFR significantly increased cell viability (Figure 2C). Additionally, Transwell assay showed that PDGFR overexpression effectively promoted cell migration (Figure 2D). Furthermore, qPCR results revealed elevated expression level of CK19 and CK10 following PDGFR overexpression (Figure 2E). Previous reports have indicated that PDGF facilitates the activation of ERK and AKT phosphorylation in ADMSCs [25, 26]. We investigate whether these pathways are similarly activated following PDGFR overexpression. Our findings demonstrated a significant increase in ERK and AKT phosphorylation subsequent to PDGFR overexpression, confirming the activation of these two signaling pathways (Figure 2F).
(A) The mRNA levels of PDGFR detected by qPCR. (B) The protein levels of PDGFR detected by Western blot. (C) CCK8 assay for cell viability. (D) Transwell assay for cell migration. Scale bars represent 50 μm. (E) The mRNA levels of CK19 and CK10 detected by qPCR. (F) Western blot and semi-quantification of phosphorylated ERK(p-ERK), ERK, phosphorylated AKT(p-AKT) and AKT of ADMSCs. Date from 3 independent experiment ,** P < 0.01; *** P < 0.001.
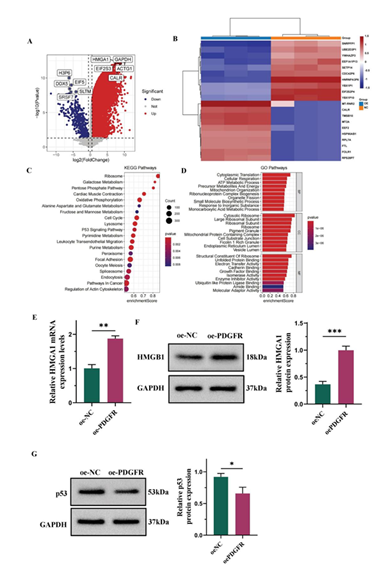
Overexpression of PDGFR upregulated HMAG1 expression and modulation the p53 signaling pathway
The molecular mechanism by which PDGFR facilitates the differentiation of ADMSC into epidermal cells was investigated though RNA sequencing of cells transfected with either oe-NC or oe-PDGFR. This analysis identified 6,742 DEGs, including 6,432 upregulated and 310 downregulated genes (Figure 3A-3B). Among these DEGs, HMGA1 emerged as a prominent factor, it is a chromatin remodeling factor recognized for its role in promoting cell proliferation and migration [27]. Subsequently, comprehensive analyses of GO and KEGG pathway were conducted on these DEGs. The functional annotation through GO classified the results into biological processes (BP), cellular components (CC), and molecular functions (MF). BP analysis indicated enrichment in cytoplasmic translation, cellular respiration, and mitochondrion organization. CC analysis revealed significant enrichment for the cytosolic ribosome, ribosome, and pigment granule. MF analysis indicated that the DEGs were significantly associated with electron transfer activity, structural constituent of ribosome, and cadherin binding. Additionally, the KEGG pathways highlighted that these DEGs are critically involved in ribosome biogenesis and the cell cycle, as well as in the p53 signaling pathway (Figure 3C-3D). Previous studies have reported the complexity of the p53 signaling network and its critical role in apoptosis regulation [28]. To validate our RNA sequencing results, qPCR and Western blot analyses were conducted. A notable elevation in HMGA1 levels was detected in the oe-PDGFR group when compared to the oe-NC group (Figure 3E-3F). Additionally, Western blot analysis revealed that the overexpression of PDGFR notably reduced the levels of p53 expression (Figure 3G).
(A) Volcano plot for RNA-seq data analysis for DEGs. (B) Heatmap for RNA-seq data analysis for DEGs. (C) Dotplot of KEGG pathway analysis. (D) Barplot of GO enrichment analysis. (E) The mRNA levels of HMGA1 detected by qPCR. (F) Western blot and semi-quantification of HMGA1. (G) Western blot and semi-quantification of p53. Date from 3 independent experiment, * P < 0.05; ** P < 0.01; *** P < 0.001.
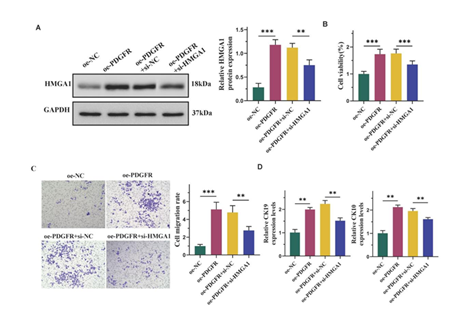
Overexpression of PDGFR facilitated the differentiation of ADMSCs into epidermal cells through upregulating of HMGA1
Subsequently, HMGA1 expression was inhibited by transfecting cells with si-HMGA1. Western blot analysis revealed that transfection with oe-PDGFR alone significantly elevated HMGA1 levels, while co-transfection with both oe-PDGFR and si-HMGA1 reversed this effect (Figure 4A). Moreover, si-HMGA1 diminished the enhanced cell viability induced by oe-PDGFR (Figure 4B). Additionally, si-HMGA1 counteracted the promoting effects of oe-PDGFR on cell migration (Figure 4C). Furthermore, si-HMGA1 also reversed the increased expression of CK19 and CK10 caused by oe-PDGFR (Figure 4D).
(A) Western blot and semi-quantification of HMGA1. (B) CCK8 assay for cell viability. (C) Transwell assay for cell migration. Scale bars represent 50 μm. (D) The mRNA levels of CK19 and CK10 detected by qPCR. Date from 3 independent experiment, * P < 0.05; ** P < 0.01; *** P < 0.001.
Overexpression of PDGFR facilitated the differentiation of ADMSCs into epidermal cells via the p53 signaling pathway
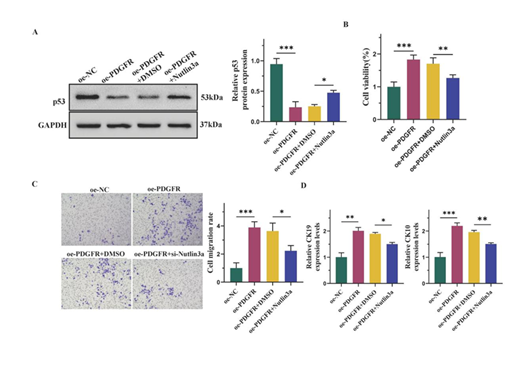
To investigate whether PDGFR promoted the differentiation of ADMSCs into epidermal cells via the p53 signaling pathway, we treated the cells with Nutlin3a, a p53 activator. The analysis resultes indicated that Nutlin3a mitigated the downregulation of p53 induced by oe-PDGFR (Figure 5A). Compared to the oe-PDGFR+DMSO group, Nutlin3a counteracted the oe-PDGFR-induced increase in cell viability (Figure 5B). Additionally, Nutlin3a diminished the enhancing effect of oe-PDGFR on cell migration (Figure 5C). Besides, Nutlin3a reversed the elevated expression of CK19 and CK10 caused by oe-PDGFR (Figure 5D).
(A) Western blot and semi-quantification of p53. (B) CCK8 assay for cell viability. (C) Transwell assay for cell migration. Scale bars represent 50 μm. (D) The mRNA levels of CK19 and CK10 detected by qPCR. Date from 3 independent experiment ,* P < 0.05; ** P < 0.01; *** P < 0.001.
Overexpression of PDGFR suppressed the activation of the p53 signaling pathway by upregulating HMGA1
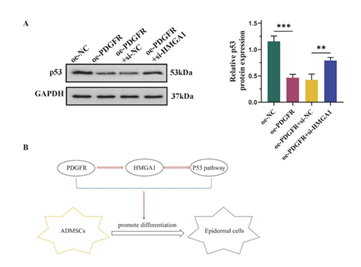
Finally, a Western blot was performed to determine whether PDGFR inhibits the p53 signaling pathway through HMGA1 upregulation. The findings indicated that the introduction of oe-PDGFR resulted in a significant decrease in p53 expression. Howerver, transfection with si-HMGA1 effectively reversed this effect (Figure 6A). Collectively, these findings suggest that PDGFR overexpression upregulates HMGA1, thereby facilitating the differentiation of ADMSCs into epidermal cells via the p53 pathway (Figure 6B).
(A) Western blot and semi-quantification of p53. (B) Molecular mechanism diagram. Date from 3 independent experiment,** P < 0.01; *** P < 0.001.
Discussion
Currently, existing skin repair methods exhibit several limitations, highlighting the urgent need for innovative strategies to enhance skin regeneration and expedite wound healing [29]. ADMSCs are increasingly acknowledged as optimal seed cells for cell transplantation and tissue engineering due to their availability, minimal invasiveness, rapid in vitro proliferation, and stable biological characteristics [30, 31]. This research intends to explore the function of PDGFR in the process by which ADMSCs differentiate into epidermal cells. Prior to inducing differentiation, the expression of surface markers on ADMSCs was assessed. Previous studies have demonstrated that ADMSCs exhibit a variety of mesenchymal stem cell markers, such as CD29, CD90 and CD105. However, they lack hematopoietic markers such as CD34 and CD45 [32]. Our investigation revealed that following culture in epidermal cell medium, the cells underwent morphological changes from spindle-shaped to round or oval forms and arranged themselves in a cobblestone-like pattern. Additionally, the expression levels of epidermal cell marker CK19 and skin stem cell marker CK10 [33] were found to be elevated. These findings suggest that ADMSCs have been effectively prompted to differentiate into epidermal cells.
In this study, cells were engineered to overexpress PDGFR and subsequently treated with the PDGF inducer PDGF-BB to elucidate the role of PDGFR in the differentiation of ADMSCs. Prior research has demonstrated that PDGF stimulates cell proliferation, migration, and angiogenesis through its interaction with PDGFR [34]. Many investigations have emphasized the critical importance of PDGF in the process of wound healing. For instance, PDGF accelerates wound repair by mitigating oxidative stress and inflammatory responses [35]. Furthermore, N. sativa seed extract has been shown to enhance wound healing by upregulating both PDGF and VEGF expression levels [36]. Our findings indicate that the overexpression of PDGFR significantly modulates cell proliferation and migration capabilities, concurrently enhancing epidermal markers expression and activating pertinent signaling pathways. These findings imply that PDGFR could promote the differentiation of ADMSCs into epidermal cells.
RNA sequencing was performed on cells transfected with either oe-NC or oe-PDGFR to identify the downstream targets of PDGFR. Differential expression analysis identified a total of 6,432 DEGs, among which HMGA1 emerged as a significant candidate. Notably, when comparing cells transfected with oe-PDGFR to those with oe-NC, HMGA1 expression was markedly elevated. The High Mobility Group (HMG) proteins are non-histone chromatin-associated proteins that indirectly modify higher-order chromatin structures to regulate transcription [37]. The HMG protein family encompasses HMGA, HMGB, and HMGN, with the HMGA subfamily further classified into HMGA1 and HMGA2 based on their encoding genes [38]. The AT-hook DNA-binding domain of HMGA proteins interacts with AT-rich regions in nuclear chromatin, facilitating DNA bending, stretching, or looping. Thus, HMGA1 is regarded as a structural transcription factor [39]. Substantial evidence indicates that HMGA1 participates in transcriptional regulation, DNA damage repair, and tumor malignancy [40,41]. Currently, research on HMGA1 primarily focuses on its role in tumors; however, its potential involvement in skin repair remains unclear. Our study demonstrates that PDGFR overexpression significantly upregulates the expression of HMGA1. To investigate whether PDGFR enhances the differentiation of ADMSCs into epidermal cells by upregulating HMGA1, we co-transfected cells with oe-PDGFR and an si-HMGA1 plasmid. Our results indicate that silencing of HMGA1 partially reverses the effects of PDGFR overexpression on cell proliferation and migration while mitigating the increased expression levels of CK19 and CK10 induced by PDGFR. These findings indicate that PDGFR promotes the differentiation of ADMSCs into epidermal cells by upregulation of HMGA1.
An analysis of the KEGG pathways was conducted on DEGs to explore the mechanisms by which PDGFR facilitates the differentiation of ADMSCs into epidermal cells. Notably, the findings indicated significant enrichment in the p53 signaling pathway. As a critical tumor suppressor, p53 enhances cellular metabolism and structural integrity by modulating cell cycle progression, apoptosis, senescence, and DNA repair processes, thus inhibiting cancer cell proliferation and metastasis. This study demonstrated that overexpression of PDGFR resulted in decreased expression levels of p53 [42]. Consequently, it is proposed that PDGFR overexpression promotes ADMSC differentiation into epidermal cells through suppression of p53 activation. To validate this hypothesis, rescue experiments were performed using Nutlin3a as a p53 pathway activator. Our results revealed that Nutlin3a not only counteracted the effects of PDGFR overexpression on cell proliferation and migration but also alleviated the increases in CK19 and CK10 levels induced by PDGFR. Furthermore, si-HMGA1 partially mitigated the inhibitory effects on p53 expression caused by PDGFR overexpression, suggesting that PDGFR inhibits activation of the p53 pathway via upregulation of HMGA1.
This study primarily investigates the molecular mechanisms by which PDGFR facilitates the differentiation of ADMSCs into epidermal cells at the cellular level. The skin healing process encompasses a diverse array of cell types, and intercellular communication involves intricate interactions among numerous signaling molecules and pathways. Studies conducted at the cellular level are limited in their ability to replicate the complexities of physiological environments. Therefore, we will advance our research by performing in vivo studies to achieve a more comprehensive and authentic representation of physiological and pathological conditions, thereby providing robust guidance for clinical applications. Furthermore, ADMSCs exhibit multidirectional differentiation potential, with the stability of their differentiation into various cell types being influenced by specific directional cues and environmental conditions. Recent studies have indicated that factors such as the composition of the culture medium, duration and density of cultivation, as well as the number of passages significantly impact the stability of ADMSCs differentiation [43]. The instability differentiation of ADMSCs into epidermal cells may result in suboptimal therapeutic outcomes and could also lead to the emergence of undesired cell phenotypes, potentially causing adverse side effects. Consequently, our subsequent investigations will focus on elucidating the factors that influence the stability of ADMSCs differentiation into epidermal cells to provide strong support for harnessing ADMSCs in wound healing therapies.
In summary, this research elucidates that the overexpression of PDGFR facilitates the differentiation of ADMSCs into epidermal cells, as demonstrated by enhanced cellular proliferation and migration, increased expression levels of epidermal markers CK9 and CK10, and the activation of ERK and AKT pathways. Mechanistic investigations revealed that PDGFR overexpression leads to an upregulation of HMGA1 expression, thereby promoting the differentiation of ADMSCs into epidermal cells via the p53 pathway. Epidermal cells constitute a critical population within the skin's epidermis and are pivotal in wound healing processes. Understanding the molecular mechanisms underlying ADMSCs' differentiation into epidermal cells may provide foundational insights for clinical applications in wound healing utilizing ADMSCs, as well as novel therapeutic strategies in skin regenerative medicine.
Contributions
Yan-Hong Wu: Conceptualization, Investigation, Formal Analysis, Funding Acquisition, Supervision, Writing-Review & Editing; Jian-Wu Chen: Visualization, Methodology, Investigation; Yu-Zhi Wang: Data Curation, Resources, Supervision; Peng Liu: Software, Validation; Zhong-Shan Wang: Visualization, Writing-Review & Editing-Original Draft; Xiao-Qiang Chen: Resources, Supervision.
Funding
This work was supported by Foundation of General Hospital of Southern Theater Command (Grant No. 2022NZB002).
Conflicts of Interest
The authors affirm that they hold no conflicts of interest.
Ethical Statement
Not applicable
Availability of data and materials
The datasets utilized and/or analyzed in the current study are available from the corresponding author upon reasonable request.
References
- Liu Y, Li C, Feng Z, et al. Advances in the Preparation of Nanofiber Dressings by Electrospinning for Promoting Diabetic Wound Healing. Biomolecules 12 (2022): 1727.
- Wang W, Lu KJ, Yu CH, et al. Nano-drug delivery systems in wound treatment and skin regeneration. J Nanobiotechnology 17 (2019): 82.
- Capanema N, Mansur A, Carvalho IC, et al. Bioengineered Water-Responsive Carboxymethyl Cellulose/Poly(vinyl alcohol) Hydrogel Hybrids for Wound Dressing and Skin Tissue Engineering Applications. Gels 9 (2023): 166.
- Bonvallet PP, Culpepper BK, Bain JL, et al. Microporous dermal-like electrospun scaffolds promote accelerated skin regeneration. Tissue Eng Part A 20 (2014): 2434-245.
- Duscher D, Barrera J, Wong VW, et al. Stem Cells in Wound Healing: The Future of Regenerative Medicine? A Mini-Review. Gerontology 62 (2016): 216-25.
- Zuk PA, Zhu M, Mizuno H, et al. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng 7 (2001): 211-28.
- Safford KM, Hicok KC, Safford SD, et al. Neurogenic differentiation of murine and human adipose-derived stromal cells. Biochem Biophys Res Commun 294 (2002): 371-379.
- Lu F, Mizuno H, Uysal CA, et al. Improved viability of random pattern skin flaps through the use of adipose-derived stem cells. Plast Reconstr Surg 121 (2008): 50-58.
- Shi Y. MEG3 regulates apoptosis of adipose-derived stem cells. Mol Med Rep 21 (2020): 2435-2442.
- Xie A, Xue J, Wang Y, et al. Kartogenin Induced Adipose-Derived Stem Cell Exosomes Enhance the Chondrogenic Differentiation Ability of Adipose-Derived Stem Cells. Dis Markers 2022 (2022): 6943630.
- Casteilla L, Planat-Bénard V, Cousin B, et al. Plasticity of adipose tissue: a promising therapeutic avenue in the treatment of cardiovascular and blood diseases. Arch Mal Coeur Vaiss 98 (2005): 922-926.
- Wang J, Wu H, Peng Y, et al. Hypoxia adipose stem cell-derived exosomes promote high-quality healing of diabetic wound involves activation of PI3K/Akt pathways. J Nanobiotechnology 19 (2021): 202.
- Ma T, Sun J, Zhao Z, et al. A brief review: adipose-derived stem cells and their therapeutic potential in cardiovascular diseases. Stem Cell Res Ther 8 (2017): 124.
- Li J, Wei C, Yang Y, et al. Apoptotic bodies extracted from adipose mesenchymal stem cells carry microRNA-21-5p to induce M2 polarization of macrophages and augment skin wound healing by targeting KLF6. Burns 48 (2022): 1893-1908.
- Ramos TV, Wang T, Maki CB, et al. Adipose stem cell side population in the mouse. J Tissue Eng Regen Med 3 (2009): 430-441.
- Perinelli DR, Bonacucina G, Pucciarelli S, et al. Rheological Properties and Growth Factors Content of Platelet-Rich Plasma: Relevance in Veterinary Biomedical Treatments. Biomedicines 8 (2020): 429.
- Boswell SG, Cole BJ, Sundman EA, et al. Platelet-rich plasma: a milieu of bioactive factors. Arthroscopy 28 (2012): 429-439.
- Hong JD, Wang X, Peng YP, et al. Silencing platelet-derived growth factor receptor-β enhances the radiosensitivity of C6 glioma cells in vitro and in vivo. Oncol Lett 14 (2017): 329-336.
- Wu Y, Fan J, Zhang B, et al. Enhanced proliferation and migration capability of epidermal stem cells by PRP and PDGF stimulation. Int J Clin Exp Pathol 10 (2017): 8804-8812.
- Demoulin JB, Essaghir A. PDGF receptor signaling networks in normal and cancer cells. Cytokine Growth Factor Rev 25 (2014): 273-283.
- Contreras O, Córdova-Casanova A, Brandan E. PDGF-PDGFR network differentially regulates the fate, migration, proliferation, and cell cycle progression of myogenic cells. Cell Signal 84 (2021): 110036.
- Mildmay-White A, Khan W. Cell Surface Markers on Adipose-Derived Stem Cells: A Systematic Review. Curr Stem Cell Res Ther 12 (2017): 484-492.
- Brzoska M, Geiger H, Gauer S, et al. Epithelial differentiation of human adipose tissue-derived adult stem cells. Biochem Biophys Res Commun 330 (2005): 142-150.
- Pan S, Hu Y, Hu M, et al. Platelet-derived PDGF promotes the invasion and metastasis of cholangiocarcinoma by upregulating MMP2/MMP9 expression and inducing EMT via the p38/MAPK signalling pathway. Am J Transl Res 12 (2020): 3577-3595.
- Capilla-González V, López-Beas J, Escacena N, et al. PDGF Restores the Defective Phenotype of Adipose-Derived Mesenchymal Stromal Cells from Diabetic Patients. Mol Ther 26 (2018): 2696-2709.
- He S, Hou T, Zhou J, et al. Endothelial Cells Promote Migration of Mesenchymal Stem Cells via PDGF-BB/PDGFRβ-Src-Akt in the Context of Inflammatory Microenvironment upon Bone Defect. Stem Cells Int 2022 (2022): 2401693.
- Chi XG, Meng XX, Ding DL, et al. HMGA1-mediated miR-671-5p targets APC to promote metastasis of clear cell renal cell carcinoma through Wnt signaling. Neoplasma 67 (2020): 46-53.
- Tang X, Amar S. p53 suppresses CCL2-induced subcutaneous tumor xenograft. Tumour Biol 36 (2015): 2801-2808.
- Buote NJ. Updates in Wound Management and Dressings. Vet Clin North Am Small Anim Pract 52 (2022): 289-315.
- Shi J, Liang J, Guo B, et al. Adipose-Derived Stem Cells Cocultured with Chondrocytes Promote the Proliferation of Chondrocytes. Stem Cells Int 2017 (2017): 1709582.
- Hao Z, Qi W, Sun J, et al. Review: Research progress of adipose-derived stem cells in the treatment of chronic wounds. Front Chem 11 (2023): 1094693.
- Suelzu CM, Conti V, Khalidy Y, et al. Xenobiotic-Free Medium Guarantees Expansion of Adipose Tissue-Derived Canine Mesenchymal Stem Cells Both in 3D Fibrin-Based Matrices and in 2D Plastic Surface Cultures. Cells 9 (2020).
- Liu J, Hu F, Tang J, et al. Homemade-device-induced negative pressure promotes wound healing more efficiently than VSD-induced positive pressure by regulating inflammation, proliferation and remodeling. Int J Mol Med 39 (2017): 879-888.
- Raica M, Cimpean AM. Platelet-Derived Growth Factor (PDGF)/PDGF Receptors (PDGFR) Axis as Target for Antitumor and Antiangiogenic Therapy. Pharmaceuticals (Basel) 3 (2010): 572-99.
- Kaltalioglu K, Coskun-Cevher S, Tugcu-Demiroz F, et al. PDGF supplementation alters oxidative events in wound healing process: a time course study. Arch Dermatol Res 305 (2013): 415-422.
- Palanisamy CP, Alugoju P, Jayaraman S, et al. Nigella sativa L. seed extracts promote wound healing progress by activating VEGF and PDGF signaling pathways: An in vitro and in silico study. F1000Res 12 (2023): 436.
- Di Marcantonio D, Galli D, Carubbi C, et al. PKCε as a novel promoter of skeletal muscle differentiation and regeneration. Exp Cell Res 339 (2015): 10-19.
- Harris HE, Andersson U, Pisetsky DS. HMGB1: a multifunctional alarmin driving autoimmune and inflammatory disease. Nat Rev Rheumatol 8 (2012): 195-202.
- Mao L, Wertzler KJ, Maloney SC, et al. HMGA1 levels influence mitochondrial function and mitochondrial DNA repair efficiency. Mol Cell Biol 29 (2009): 5426-5440.
- Sumter TF, Xian L, Huso T, et al. The High Mobility Group A1 (HMGA1) Transcriptome in Cancer and Development. Curr Mol Med 16 (2016): 353-393.
- Shen X, Li WQ. High-mobility group box 1 protein and its role in severe acute pancreatitis. World J Gastroenterol 21 (2015): 1424-1435.
- Hassin O, Oren M. Drugging p53 in cancer: one protein, many targets. Nat Rev Drug Discov 22 (2023):127-144.
- Neri S. Genetic Stability of Mesenchymal Stromal Cells for Regenerative Medicine Applications: A Fundamental Biosafety Aspect. Int J Mol Sci20 (2019): 2406.


 Impact Factor: * 3.4
Impact Factor: * 3.4 Acceptance Rate: 78.89%
Acceptance Rate: 78.89%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 