Recent Advances on the Structure, Genomic Arrangement, Life Cycle, and Virus-Host Proteins Interactome of SARS CoV-2
Endeshaw Chekol Abebe1*, Tadesse Asmamaw Dejenie2
1Department of Medical Biochemistry, College of Health Sciences, Debre Tabor University, Debre Tabor, Ethiopia 2Department of Medical Biochemistry, College of Medicine and Health Sciences, University of Gondar, Gondar, Ethiopia
*Corresponding author: Endeshaw Chekol Abebe, Department of Medical Biochemistry, College of Health Sciences, Debre Tabor University, P. O. Box: 272; Debre Tabor Town, 6300, Ethiopia.
Received: August 20, 2022; Accepted: September 22, 2022; Published: September 30, 2022
Article Information
Citation: Endeshaw Chekol Abebe, Tadesse Asmamaw Dejenie. Recent Advances on the Structure, Genomic Arrangement, Life Cycle, and Virus-Host Proteins Interactome of SARS CoV-2. International Journal of Applied Biology and Pharmaceutical Technology 13 (2022): 019-032.
View / Download Pdf Share at FacebookAbstract
Novel coronavirus disease of 2019 (COVID-19) is a highly contagious disease that has been recorded as a third global pandemic caused by the coronavirus (CoV) family in the past twenty years in the aftermath of severe acute respiratory syndrome (SARS) and middle east respiratory syndrome (MERS). COVID 19 is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that is transmitted by person-to-person transmission and it remains asymptomatic or presented with mild flu-like symptoms in most occasions, while in some instances, it may progress to severe life threatening and potentially fatal illnesses. This disease is now imposing immense negative influences across the world due to the highly contagious nature of the disease as well as due to the absence of effective treatment targeting the disease. This review addresses the recent advances on the structure and genomic arrangement of SARS-CoV-2 as well as the viral entry, replication and virus-host protein interactome that potentially contribute to cell infectivity, immune evasion, and viral spread. Unveiling the details of such aspects of SARS-CoV-2, therefore, possibly has paramount importance for discovering therapeutic targets.
Keywords
<p>Genomic Arrangement; Life Cycle; SARS CoV-2; Structure; Virus-Host Protein Interactome</p>
Article Details
1. Introduction
Coronaviruses (CoV) are the largest family of RNA viruses that belong to the order Nidovirales, family Coronaviridae and subfamily Coronavirinae. These group of viruses involve four genera, namely alpha-, beta-, gamma-, and delta CoV that may cause illness ranging from common cold to potentially fatal severe illnesses [1]. In the past two decades, the world has faced three pandemic respiratory diseases caused by CoV family. The two regional epidemics results from CoV were severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) in 2001 to 2003 and 2012 to 2015, respectively. These are serious and potentially fatal illnesses caused by pathogenic beta CoVs known as SARS-CoV and MERS-CoV, respectively [2]. Since December 2019 the third pandemic from novel CoV, now officially named novel coronavirus infectious disease of 2019 (COVID-19), has been identified in Wuhan, China. COVID 19 was identified to be caused by a novel CoV known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). SARS-CoV-2 is an enveloped and crown shaped viruses with a positive-sense single-stranded RNA genome grouped under the genus beta CoV, subgenus Sarbecovirus, and species SARS-related CoV along with SARS-CoV and MERS-CoV [1,3,4]. Despite many attempts to contain its spread, COVID-19 continues to become a significant public health concern and inflict enormous burdens of morbidity and mortality while seriously challenging the globe to unprecedented strain on health system, economies and social life [5,6]. Hence, this review will provide insights on the structure, genomics and life cycle, virus-host protein interactome of SARS CoV-2, which is important to find out more effective vaccine and drug.
2. The Structure of SARS-CoV-2
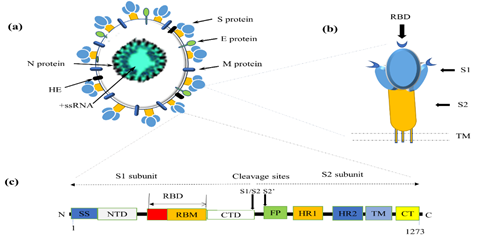
SARS-CoV-2 is a spherical, non-segmented enveloped virus with diameter ranging between 50–200 nm [7]. It resembles the conventional CoV structure with five structural proteins, several non-structural proteins (nsp), and accessory proteins [8]. Structurally, the SARS-CoV-2 has a double-layered lipid envelope, including four viral surface proteins, namely spike (S), envelope (E), membrane (M), and hemagglutinin-esterase (HE) proteins that are embedded into the lipid bilayer, and the nucleocapsid (N) protein situated within the envelope and associated with a single stranded positive sense viral RNA genome (Figure 1a) [4,9-11]. Due to the lipid nature of the viral envelope, enveloped CoVs including SARS-CoV-2 are typically susceptible to destruction following exposure to heat, detergents, and organic solvents [12]. Thus, SARS-CoV-2 can only live for a limited period outside the host environments and usually need to be moved directly from one to another host to continue survival [11].
2.1 Spike (S) Protein
S protein is a large homo-trimeric transmembrane glycoprotein of 1273 amino acids long and 126-168kDa molecular weight, comprising a very large N-terminus ectodomain, a transmembrane (TM) domain, and a small C-terminus endodomain [10]. It is synthesized as single-chain precursors that oligomerize in the endoplasmic reticulum (ER) and undergoes post translational modification such as glycosylation and folding into S1 and S2 domains in Golgi to eventually form long, club-shaped spikes emanating from the outer surface of virions that provide the name CoV [13].
S protein plays a key role in the early steps of SARS-CoV-2 infection in host cell receptor recognition, viral attachment, and membrane fusion [14]. S protein exhibits as an inactive precursor in native state but it undergoes substantial conformational rearrangement when it interacts with the host cell. The host cell proteases activate S protein by trimming it into two distinct subunits: S1 subunit that serve for viral attachment, and S2 subunits that allow membrane fusion (Figure 1b) [15].
2.2 S1 Subunit
The S1subunit is 685 amino acids long segment of S protein involving a 13 amino acid long signal sequence (SS) at the N terminus to direct the transport of the S protein to its membrane destination, an N-terminal domain (NTD) involved in sugar binding, receptor-binding domain (RBD), and C terminal domains (CTD) (Figure 1c) [15]. RBD is 223 amino acids long and 21kDa weight structure situated near the C-terminus of S1 to mediate the docking of SARS-CoV-2 with the host cell surface receptors. It possesses three domains where one lying upward while two lying downward in most beta-CoVs including SARS-CoV-2. It contains a core structure and a receptor-binding motif (RBM) that make direct contact with the host cell receptor, such as the angiotensin-converting enzyme 2 (ACE2) [16,17]. The RBD undergoes conformational changes that transiently mask or unmask the receptor binding determinants, known as the “down” and the “up” conformation respectively, where “down” corresponds the inaccessible or buried (closed) state of the receptor and “up” corresponds to the accessible (open) and less stable state of the receptor. The RBD of SARS-CoV-2 shares 73%–76% sequence homology with that of SARS-CoV. However, unlike the down conformation of SARS-CoV RBD which packed tightly against the NTD of the nearby protomer, the down conformation of SARS-CoV-2 RBD angled closer to the central core of the trimer [17]. RBD is a highly immunogenic component producing effective neutralizing antibodies and hence it is a promising therapeutic target for anti-viral drug and vaccine development. Nonetheless, the highly variable sequence and hidden location of RBD may challenge to develop effective RBD based drugs and vaccines [15-17].
Figure 1: Schematic representation of SARS-CoV-2 (a) Structural proteins. The spike (S) protein, envelope (E) protein, membrane (M) protein, and hemagglutinin esterase (HE) are found embedded in the lipid bilayer, whereas the nucleocapsid (N) protein is associated with the viral RNA. (b) A three-dimensional structure of S protein. S protein consists of S1 and S2 subunits with three small size RBD in S1 subunit where one lying up and two lying down. (c) A one-dimensional structure of S protein. S protein has 1273 amino acids with two cleavage sites: S1/S2 and S2’ sites (shown with arrows). The S1subunit contains a signal sequence (SS), N-terminal domain (NTD), receptor-binding domain (RBD) possessing a core structure and receptor-binding motif (RBM, and C terminal domains (CTD). Whereas the S2 subunit composed of a large ectodomain involving fusion peptide (FP) and heptad repeats (HR1 and HR2), a single pass transmembrane domain (TM) and a small cytoplasmic tail (CT).
2.3 S2 Subunit
The S2 subunit of S protein is 588 amino acids containing type I integral membrane protein, which composed of a large N-terminal ectodomain involving a fusion peptide (FP) and two highly conserved heptad-repeats (HR), a single TM domain anchored to the viral membrane, and a C-terminal cytoplasmic tail (CT) [18]. FP is a 15–20 amino acids long fusogenic peptide anchored to the target cell membrane when the S protein assumes the pre-hairpin conformation. It plays an essential role in inducing viral and cell membrane fusion in late infection and facilitate SARS-CoV-2 entry by disrupting and connecting the membrane lipid bilayers of the target cell [15,19]. HR is a highly conserved alpha helical coiled coil structure that composes repeated heptapeptides (HPPHCPC) richly containing hydrophobic (H)-, polar (P)-, and charged (C)-amino acid residues [20]. Crystallographic studies indicated that HR involves three HR1 (912th-984th residues)and three HR2 (1163rd-1213rd residues) to form a stable six-helical bundle structure involved in close apposition of the viral and cellular membranes and thus, in facilitating membrane fusion and subsequent SARS-CoV-2 entry [21,22]. The HR1, which is located adjacent to the N terminus of S2 and hence also known as HRN, forms the central trimeric core in parallel manner. Whereas HR2, which is situated closer to the C-terminus immediately preceding the TM domain and thus also known as HRC, is packed into three antiparallel hydrophobic grooves on the surface of the central trimeric core [22]. Based on prior data from other enveloped viruses, antibodies against the HR domains are broadly neutralizing and targeting HR could be a promising target for repurposing fusion inhibitors against SARS-CoV-2 infection [14].
2.4 Hemagglutinin-Esterase (HE)
Additionally, there is a dimeric structural protein located on the surface of beta CoVs such as SARS-CoV-2 called hemagglutinin-esterase (HE). The HE of SARS-CoV-2 acts as a typical glycan-binding lectin and receptor-degrading enzyme having acetyl-esterase activity [23]. HE contains O-acetylated sialic acids that bind with the lectin-like S protein of SARS-CoV-2 for the initial attachment of SARS CoV-2 to get into the host cells. These activities are suggested to facilitate S protein-mediated cell entry and mucosal virus spread albeit not needed for viral replication [4,23-25].
2.5 Membrane (M) Protein
M protein is a type III transmembrane structural glycoprotein possessing nearly 230 amino acids and has 25–35kDa molecular weight. It is the most abundant viral surface protein responsible for viral assembly and determining the characteristic shape of virion [26]. The M protein of SARS-CoV-2 has been reported to have 39.2% and 90.1% structural identity with MERS-CoV and SARS-CoV, respectively [27]. It is a polytopic protein comprising three domains: an ectodomain, a TM domain, and an endodomain [28]. The ectodomain is a small domain in N-terminus of M protein, whereas the endo-domain is a large domain in the C-terminus of M protein and located in the interior of the virion or on the cytoplasmic part of intracellular membranes that has the ability to bind with RNA. Besides, M protein of SARS-CoV-2 has a triple helix bundle or a single 3-TM domain that resembles the sugar transporter semi-SWEET, suggesting that this protein may play a role in the viral entry into the host cell and viral RNA maturation though further confirmatory studies are required [29]. M protein is synthesized in rough ER and largely modified by N-linked glycosylation at α and σ with a limited extent by O-linked glycosylation at β to form this glycoprotein [26, 30].
2.6 Envelope (E) Protein
E protein is a small hydrophobic membrane protein containing 76 to 109 amino acids and weighting 8 to 12kDa [10]. This protein is a minor component of CoV having a common architecture of a short negatively charged (hydrophilic) N-terminal ectodomain, a large uncharged (hydrophobic) TM region, and a large negatively charged (hydrophilic) C-terminal tail. E protein has PDZ-binding motif in the C-terminal region that plays a critical role in viral pathogenicity by disturbing cell signaling. The PDZ-binding motif is considered as a pathologic factor of SARS-CoV [31,32]. Similarly, available evidence indicated that E protein is reported to play an important role in COVID 19 pathogenesis as it interacts with the tight junction related protein, protein associated with Lin Seven 1(PALS1) [33]. This protein was also observed to have ion channel activity, viroporin activity and assembling into homo-oligomers, ranging from dimers through a pentameric to hexameric α-helical structure [30,34,35]. Thus, E protein has key roles in viral assembly and release, host cell membrane permeability and virus-host cell interaction, but it is not required for viral replication [4,36]. Taken together, the ion channel activity and the pathogenic role of PDZ-binding motif of E protein make SARS-CoV and SARS-CoV-2 more pathogenic than other CoVs by triggering cytokine storm, inflammasome, subsequent pulmonary edema, and ultimately acute respiratory distress syndrome and death [33]. Due to these crucial roles of E protein, impairing this mechanism using E protein blockers was reported as appropriate therapeutic target for SARS-CoV-2 treatment as per the report of animal model studies [37].
2.7 Nucleocapsid (N) Protein
N protein is 43–50kDa weight and 419 amino acids long viral protein enclosing viral nucleocapsid within the viral protein shell (capsid) or inside the infected cells [38]. It has an independently folded N-terminal domain which serve as a RNA-binding site via its lysine and arginine amino acids, and a C-terminal (dimerization) domain [10,11]. This protein coats the single strand positive sense viral RNA that allows the virus to hijack human cells and turn them into factories of viruses. N protein therefore plays critical roles in RNA synthesis (replication and transcription), transcription regulation, cell signaling pathway as well as in encapsidated viral RNA genome packaging into a long helical nucleocapsid structure or matrix of the ribonucleoprotein to form new virions [4,39,40]. N protein is a phosphoprotein that is found phosphorylated at many serine and arginine-rich positions in SARS-CoV-2 and other CoVs but the role of phosphorylation is still elusive [41-43]. Different studies indicated that N protein is a highly immunogenic viral protein that can induce immune response and could be considered as a possible vaccine candidate for SARS-CoV-2 [27,44]. In contrast, other studies reported that, due to the biological function of N protein and its hidden location from antibodies, either by viral or cell membranes, antibodies against N protein are less likely to directly neutralize SARS-CoV-2 [45,46]. Therefore, antibodies produced against N protein may not provide protective immunity, and it could not be effective vaccine candidate. However, N protein could serve as a potent diagnostic and therapeutic target for COVID 19 [38]. This is supported by an in silico study that proposed N protein as a possible therapeutic target against COVID 19 after further thorough investigation [47].
3. The Genomic Arrangement of SARS-CoV-2
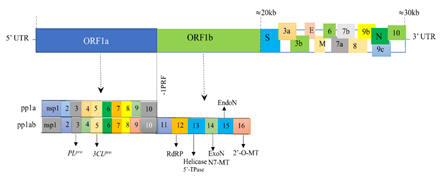
The SARS-CoV-2 is a single stranded positive sense RNA virus of nearly 30 kilobases (kb) size. The genome of SARS-CoV-2 shares 79-82% similarity with that of SARS-CoV and MERS-CoV, which generally have five nucleotide variations [48]. The genome of SARS-CoV-2 contains a 5′ cap head structure at one end and a 3′ poly(A) tail at another end with 38% of the genome has Guanine-Cytosine (GC) content [49]. Every SARS-CoV-2 virus possess 14 open reading frames (ORFs) encoding 7096 amino acid residues long polyprotein in 27 distinct proteins essential for viral replication, nucleocapsid and spike formation [4,30,48]. The longest ORF, called ORF1, contains replicase gene particularly in the upstream region of the ORF1a/b that make up two-thirds (nearly 20kb) of the genome while the remaining third (about 10kb) of the viral genome taken by 13 ORFs composed of the structural and accessory genes (Figure 2) [50]. The 5′ terminus of the genome contains a leader sequence and untranslated region (UTR) that contains ORF1 expressing many nsps required for viral replication, transcription and possibly immune evasion [51]. In particular, the ORF1a gene encodes for polyprotein pp1a comprising 10 nsps while the ORF1b gene, next to ORF1a, encodes polyprotein pp1ab possessing up to 16 nsps [48]. The pp1a and pp1ab polyproteins undergo autoproteolytic cleavage to form the viral replicase-transcriptase complex (RTC) encoding proteins having several unique or unusual enzymatic activities [50]. Besides, there are transcriptional regulatory sequences (TRS) at 5’ end at the beginning of each structural or accessory genes necessary for the expression of each of these genes [48,49]. The 3′end of SARS-CoV-2 genome on the other side contains four structural genes and eight accessory genes as well as other unidentified genes required for viral RNA synthesis [30]. Structural genes are responsible for expressing four viral structural proteins: S, E, M, and N proteins [51]. The accessory genes are distributed within the structural genes and encode nine putative accessory proteins, 3a, 3b, 6, 7a, 7b, 8, 9b, 9c, and 10 [48]. Interestingly, SARS-CoV-2 uniquely possesses ORF3b and ORF10 encoding genes that are not found in SARS-CoV. Accessory proteins generally have no known function and are almost non-essential for replication except some that have been indicated from a recent genetic analysis to play important roles in viral pathogenesis [50,52]. The recently identified new variants of SARS-CoV-2 are reported to result from the gene mutations typically occurring in five genes, namely, S, N, ORF8, ORF3a, and ORF1ab [53]. Collectively, the genome of SARS-CoV-2 is typically organized in the order of 5′-leader-UTR-replicase-S–E–M–N–3′UTR–poly (A) tail with accessory genes interspersed within the structural genes at the 3′ end of the genome [30].
Figure 2: Schematic illustration of the genomic arrangement of SRAS-CoV-2. The single-stranded RNA genome of SARS-CoV-2 (30kb) consisting of two large genes, the ORF1a and ORF1b genes, which encode pp1a and pp1ab comprising 10 and16 nsps respectively. The nsps assemble into RTC which is a multienzyme complex involving PLpro (nsp3); 3CLpro (nsp5); RdRP (nsp12); helicase and 5′-TPase (nsp13); ExoN(nsp14); N7-MT (nsp14); EndoN (nsp15); 2’-O-MT (nsp16). The structural genes encode four common structural proteins (S, E, M, and N proteins). The ORFs also contains genes the so-called accessory genes encoding accessory proteins (3a, 3b, 6, 7a, 7b,8, 9b, 9c, and 10).
Abbreviations: 3CLpro- 3 Chymotrypsin Like Protease; E- Envelope Protein; EndoN- Endoribonuclease; ExoN- Exoribonuclease; M- Membrane Protein; N- Nucleocapsid Protein; nsps- Non-Structural Proteins; N7-MT- N7-Methyl Transferase; ORF- Open Reading Frame; pp- Polyprotein, 2’-O-MT, 2’-O-methyl transferases; PLpro- Papain-Like Protease; -1PRF- -1 Programmed Ribosomal Frameshifting Nucleocapsid Proteins; RdRP- RNA Dependent RNA Polymerase; RTC- Replicase-Transcriptase Complex; S- Spike Protein; 5’-TPase- 5′-Triphosphatase.
4. The Life Cycle of SARS-CoV-2
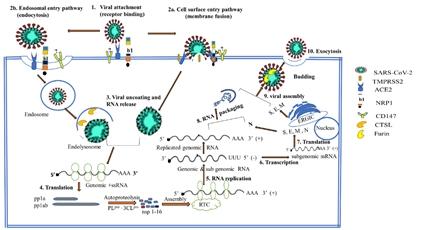
Generally, CoV entry into host cells is a significant determinant of viral infectivity and pathogenesis [54]. SARS-CoV-2 is mainly transmitted through person-to-person transmission, primarily by spraying droplets from the nose, eyes or mouth of infected individuals. This is followed by the entrance of viral particles during breathing into the lungs through the respiratory tract [6]. SARS-CoV-2 is generally suggested to use a different-step mechanism to infect host cells, especially lung alveolar cells. The virus first binds to the cell surface receptor for viral attachment, then membrane fusion occurs to directly release viral RNA into the cytoplasm or the virus reaches endosomes via receptor mediated endocytosis to fuse viral and lysosomal membranes and release the viral genome. This is then followed by translation, RNA synthesis and packaging, viral assembly, and virion release [4,54]. In this review, the life cycle of SARS-CoV-2 is discussed under the following headings: viral attachment, viral entry, viral uncoating and RNA release, translation of replicase proteins, replication and transcription, RNA packaging and viral assembly, and finally, exocytosis and virion release.
4.1 Viral Attachment
It is the virus–host cell interactions that determine the cellular entry and spread of SARS-CoV-2 across the tissues, suggesting its crucial role for viral invasion and viral tissue tropism. To infect human, the attachment between the virus and the host cell is initiated by the interaction between the S proteins of SARS-CoV-2 and its surface receptor on human cells in a lock and key fashion [6,10]. The process of viral attachment to and invasion of human cells occur using different cellular receptors. The S1 subunit of the S protein of SARS-CoV-2 recognizes host surface receptor (ACE2) to enable the virus to enter and infect the cell. Like SARS-CoV, the ACE2 receptor on alveolar epithelial cells of the lower respiratory tract has been reported to be a key cell-surface receptor to harbor SARS-CoV-2 [55-57]. Besides, recent PCR analyses have revealed that ACE2 receptors are also expressed in vascular endothelial cells, renal tubular epithelium, and gastrointestinal tract, and to a lesser extent, in the heart, adipose, and reproductive tissues to bind with viruses [58-62]. In particular, the host ACE2 receptor bind to the RBD of S1 subunit in SARS-CoV-2 with affinity (dissociation constant, Kd=15nM) 10–20 fold higher than to that of SARS-CoV (Kd=325.8 nM) [63]. This is due to the super binding affinity of glutamine (Gln) 493 and asparagine (Asn) 501 residues in RBM with ACE2 [63,64]. This possibly explain the more sensitivity of the S protein of SARS-CoV-2 to ACE2 than the S protein of SARS-CoV, and hence the higher capacity of SARS-CoV-2 for human cell infection. In addition to ACE2, the CD147 (also known as Basigin or EMMPRIN) has been identified as an alternative cell-surface receptor for human cell mediating SARS-CoV-2 invasion and spread to cause COVID 19 [65]. Furthermore, more recent publications have demonstrated a novel host factor for SARS-CoV-2 called neuropilin 1(NRP1). NRP1 is a host factor that abundantly expressed in the respiratory, olfactory epithelial cells and endothelial cells. This host factor serves as a co factor for viral entry into cells, especially in those cells with low level of ACE2 receptors. Thus, NRP1 may serve as an alternative or independent gateway for SARS-CoV-2 entry and invasion of the human cells [66,67].
4.2 Viral Entry
Following receptor binding, the SARS-CoV-2 entry requires S protein priming and activation through proteolytic cleavage at S1/S2 and S2’ sites respectively to facilitate viral entry into a host cell and to activate viral and target cell membrane fusion [68-70]. Proteolysis at S1/S2 site can occur by host protease enzymes, including extracellular proteases such as TM protease serine 2 (TMPRSS2) and lysosomal cysteine proteases such as cathepsin L (CTSL) that have essential roles in priming S protein of SARS-CoV-2 for entry [23,51]. Based on the cleavage sites of S protein and the use of TMPRSS2 for entry, accumulated evidence indicated that the SARS-CoV-2 entry mechanism can be cell surface or endosomal entry pathway (Figure 3) [18,19,71].
4.3 Cell Surface Entry Pathway
The cell surface entry pathway, also termed as the membrane-TMPRSS2 or early pathway, involves the trimming of S protein at S1/S2 and S2’ cleavage sites on the cell surface by host proteases such as TMPRSS2, trypsin, and human airway trypsin-like protease (HAT) at neutral pH [14,72]. In SARS-CoV, the S1/S2 cleavage site of the S protein is located at R667, whereas the S2’ site is situated at R797. These sites are highly conserved in all CoVs, suggesting its functionally relevance to cell surface or early SARS-CoV-2 entry mechanism using TMPRSS2 [14]. Unlike SARS-CoV, SARS-CoV-2 possess additional unique cleavage site that potentially preactivated (or pre-primed) by proprotein convertase (PPC) enzyme family (furin) at the S1/S2 boundary to facilitate viral entry, particularly in TMPRSS2-mediated cell surface entry pathway [54]. This specific furin cleavage site, also referred to as PPC motif (or polybasic site), incorporates four distinct amino acids (RRAR) located at the interface between the S1 RBD and the S2 FP within the S protein[69,73]. Thus, furin cuts the full-length S protein at S1/S2 polybasic cleavage site into S1 and S2 functional subunits that possibly provide additional host cell surface receptor binding sites, indicating the highly contagious nature of SARS-CoV-2 compared to SAR-CoV [74-76]. Furin mediated cleavage at the S1/S2 priming site is followed by the activation of SARS-CoV 2 via TMPRSS-2 [77]. Furthermore, the presence of multiple furin cleavage sites within S protein of SARS-CoV-2 increases the likelihood S protein cleavage by furin-like proteases and thereby enhances its infectivity and pathogenicity [78,79].This is confirmed by another study demonstrating that SARSCoV-2 virus with a natural deletion of the S1/S2 furin cleavage site is associated to attenuated pathogenicity in hamster models [80]. Overall, a receptor binding, specifically with protease-cleaved S1 protein potentiates SARS-CoV-2 entry and host cell infectivity [54,81]. Furin-mediated cleavage at S1/S2 junction of the S protein facilitates S1 binding to host ACE2 receptor, while this pre-priming exposes the C-end rule peptide on S1 of SARS-CoV-2 and enables binding to the b1 subdomain of NRP1 [66]. After the first cleavage of S protein in concert with the binding of S1 to target cell surface receptor, the prefusion trimer destabilized and resulting in shedding of the S1 subunit and insertion of the N terminus of the newly released S2 subunit into the cell membrane [19,82]. This promotes further proteolytic cleavage of the S2 subunit at the S2’ site into FP and S2’ domains by host extracellular proteases such as TMPRSS2 and HAT, allowing membrane fusion via the combined actions of FP and HR domains [54,81]. In native S protein, the FP and HR segments of the S2 remain stably sequestered but following S2’ site cleavage, they undergo a series of conformational changes [83]. This conformational change exposes the FP and allow its insertion into the target cell membrane, and hence triggers membrane fusion by providing attachment points for drawing the viral and cellular membranes together [14]. This reduces the distance between the viral and host cell membrane, and now the HR1 domain of the S2 protein becomes in close apposition to the target cell membrane, while the HR2 domain is in close proximity to the side of viral membrane. Then, HR2 folds back to HR1 to form a stable hexameric bundle in an antiparallel manner in the fusion core, allow the pulling of the viral membrane toward cellular membrane and tightly binds to it, thereby enhance the fusion of two membranes and subsequent viral entry [22,84].
Figure 3: The schematic diagram of the life cycles of SARS-CoV-2. S protein of SARS-CoV-2 binds to ACE2 (or NRP1 and CD147) (1) and the virus enter to host cell either via cell surface entry pathway involving priming of S protein by TMPRSS2 (or other serine proteases) followed by viral and host membrane fusion (2a) or through endosomal entry pathway involving receptor mediated endocytosis into endosomes, which then fuse with lysosome to form endolysosome where S protein cleaved by lysosomal enzyme such as CTSL (2b). Then viral uncoating and RNA release into the host cell cytoplasm occur (3) and translation of RTC directly from RNA genome followed (4). Genomic RNA undergoes replication to produce replicated viral genome (5); transcription (6) and translation to synthesize structural proteins (S, M, E, and N protein) required to form new virions (7). Viral RNA packaging of the replicated genomic RNA with N protein (8) and viral assembly with structural proteins (9) occur. Finally, after furin-mediated processing of S protein, exocytosis to release new viruses out of the host cell environment takes place. ACE2- Angiotensin Converting Enzyme 2; CTSL- Cathepsin L; 3CLpro- 3 Chymotrypsin Like Protease; ERGIC- Endoplasmic Reticulum–Golgi Intermediate Compartment; NRP1- Neuropilin 1; PLpro- Papain-Like Protease; RTC- Replicase-Transcriptase Complex; +ssRNA- Positive Sense Single Stand RNA.
4.4 Endosomal Entry Pathway
The cell surface or early entry mechanism is generally preferably used by a certain TMPRSS2 expressing cells during SARS-CoV-2 entry and infection [14]. In absence of TMPRSS2 or other exogenous proteases, the endosomal pathway has been observed to serve as an alternative SARS-CoV-2 entry mechanism in which the internalization of the virus occurs either through clathrin-dependent or clathrin-independent receptor mediated endocytosis of viral particles enclosed within the vesicle [10]. This pathway is also called the endosome-cathepsin or late pathway of viral entry since the cleavage of S protein occurs in late phase of infection within the endosome where the S protein is processed (activated) by CTSL at a low pH, triggering the fusion between the viral and host endosomal membrane [85-88]. The CTSL is one of the eleven human cathepsins that cleaves S protein at T678 which is localized at 11 amino acids downstream of the S1/S2 cleavage site and 120 amino acids upstream of S2’site [14]. Prior studies proposed the possibility of another protease to cleave at S2’ site in the low pH of endosomes to fully activate the fusogenic potential of the S protein [82]. Generally, the predominantly utilized SARS-CoV-2 entry mechanisms may vary with the types of host cells. The recent studies using immortalized kidney epithelial cell lines such as Vero E6 (from African green monkey) and 293T (from embryonic human) demonstrated that these cells do not express the cell surface protease TMPRSS2 and hence SARS-CoV-2 infection is dependent on the endosomal entry pathway and cathepsin inhibitors were effectively blocked the viral entry [64,75,76,89]. In contrast, TMPRSS2 expressing cells, such as the lung and intestinal epithelial cells are vulnerable to viral entry preferably via the cell surface or early entry pathway and hence TMPRSS2 inhibitors become more effective while impairs the efficacy of cathepsin inhibitors according to the different studies [64,75,76,89]. A recent study has suggested that SARS-CoV-2 entry inhibition may require blockade of both the cell surface and endocytic pathways [90]. However, further extensive studies are required to clearly explain the role of the two entry pathways of SARS-CoV-2 and whether individual inhibition of the cell surface or endocytic pathways will provide effective therapeutic benefit against COVID-19.
4.5 Viral Uncoating and RNA Release
In cell surface or TMPRSS2-mediated entry mechanism, the viral nucleocapsid directly released into the host cell cytoplasm, while in endosomal pathway, the endo-lysosomal compartment opens to release the SARS-CoV-2 virus into the cytoplasm via the action of lysosomal enzymes. This is followed by uncoating of viral nucleocapsid through the host cell proteasomal degradation of N protein to release a single stranded viral RNA genome into the cytoplasm [4,10,85].
4.6 Translation of Replicase Proteins
Once the viral genetic material is fully released into the cytoplasm, the direct translation of the replicase gene from the positive sense viral genomic RNA using the host cell machinery called ribosome to generate replicase proteins important for viral replication [4,30]. The replicase gene from ORF1a/b encodes two large ORFs, named rep1a and rep1b that are involved in expressing two co-terminal polyproteins (pp) called pp1a and pp1ab. During the translation of pp1ab there is a molecular mechanism that controls the expression of protein in SARS-CoV-2 called -1 programmed ribosomal frameshifting (-1PRF). Available evidence indicated that the -1PRF of SARS-CoV-2 differ from -1PRF of SARS-CoV only with a single nucleotide and this subtle difference does not have impact on the rate of translation [50]. The replicase pp1a and pp1ab in SARS-CoV-2 are subsequently autoproteolytically cleaved into the 10 and 16 individual nsps by papain-like protease (PLpro) encoded within nsp3 and by the main protease called chymotrypsin like protease (3CLpro) encoded within nsp5 [4]. The former enzyme is responsible for cleaving the nsp1/2, nsp2/3, and nsp3/4 boundaries, while the latter is involved in the cleavage of the rest eleven boundaries [10]. Then the nsps assemble into the RTC that involves multiple enzymes, including nsp7-nsp8 primase complex, RNA dependent RNA polymerase (RdRP) within nsp12, RNA helicase and 5′-triphosphatase (5’-TPase) within nsp13, RNA cap-modifying methyltransferases, such as N7-methyl transferase (N7-MT) within nsp14 and 2-O-methyl transferase (2-O-MT) within nsp16, an exoribonuclease (ExoN) within nsp14, and an endonuclease (EndoN) within nsp15 (Figure 2) [4,10]. These enzymes are important to mediate RNA replication and transcription using sub-genomic RNA as a template. Furthermore, the RTC creates a conducive environment for RNA synthesis by mediating the rearrangement of the rough ER-derived membranes to form double-membrane vesicles (DMV) in the cytoplasm of the infected cell, where viral replication and transcription take place [4,91].
4.7 Replication and Transcription
Besides as a template for translation of the replicase polyproteins, the positive sense viral RNA used as a template for replication. The RTC uses the existing positive sense viral genomic RNA as a template to generate full-length anti-sense RNAs which subsequently serve as templates for synthesizing various copies full-length positive sense RNA genomes called genomic RNA. The novel replicated positive sense genomic RNA is now used as a viral genome to be encapsulated with N protein during the packaging of the new virion [50]. However, the RTC does not always replicate the entire viral genome, sometimes it stops early and creates a shorter RNA strand, known as sub genomic RNA. Thus, discontinuous or fragmented transcription called nested sub genomic RNA transcription produces 3′ nested sub-genomic mRNAs from the canonical TRS and only the ORF closest to the 5′ end of sub genomic RNAs undergo translation though it has several ORFs [92,93]. The SARS-CoV-2 expresses nine sub genomic RNAs (S, 3a, E, M, 6, 7a, 7b, 8, and N) which serve as precursors (or as mRNAs) to produce structural proteins (S, E, M, and N proteins) and accessory proteins through translation using host ribosome attached to the ER membrane [30,50]. The viral genomic RNA synthesis involving replication and transcription is catalyzed by nsp12 RdRP with the help of nsp7 and nsp8 as cofactors [94]. The nsp14 exoribonuclease provide the RTC 3’-5’ proofreading feature [95].
4.8 RNA Packaging and Viral Assembly
In this step, the viral RNA genome is packaged and combined with the viral protein to form a new virus. The S, E, and M structural proteins are first inserted into the ER and then transported via the secretory pathway into the endoplasmic reticulum–Golgi intermediate compartment (ERGIC) [4,30].The M protein directs most protein–protein interactions required for viral assembly. When M protein is expressed along with E protein, viral progeny are formed after incorporating nucleocapsids, suggesting these two proteins function together to produce envelopes [48]. The nucleocapsids are formed from replicated and packaged viral genomes encapsulated by N protein within the cytoplasm, and as a result they fuse within the ERGIC membrane in order to enhances viral envelopment and hence creating mature virions enclosed within the Golgi vesicle by budding in the ERGIC [10]. In the meantime, the S proteins, although not required for assembly, able to traffic to the ERGIC and interact with the M protein incorporated into virions at this step. M protein interactions provide the impetus for envelope maturation [10,49]. Regardless of the viral entry mechanisms, furin mediated processing of S protein at the S1/S2 cleavage site is thought to occur following SARS-CoV-2 replication and viral assembly in ERGIC prior to the release of new viruses into the extracellular environment [15].
4.9 Exocytosis and Virion Release
Following the combination of viral proteins and genome into the mature virions, the novel virions are eventually exported from infected cells by fusing smooth walled vesicles enclosing virion with host cell membrane to release the virus outside the cell through exocytosis. Now the new virus is ready to infect another nearby cell or person [10,50].
5. SARS-CoV-2–Host Protein Interactions (interactome)
Besides the interaction between S protein of SARS-CoV-2 and host cell surface receptors (ACE2, CD147, and NRP1), recent studies using interactome maps revealed the intricate molecular interactions between the several viral proteins of SARS-CoV-2 with human proteome, known as virus-host interactome [96-98]. A study by Gordon et al identified 332 interactions between SARS-CoV-2 proteins and human proteins. These interacting proteins are predominantly found in the lung tissues compared to other tissues, supporting the idea that SARS-CoV-2 preferentially hijacks proteins that are expressed in lung tissue [96]. The interactions between SARS-CoV-2 proteins and human proteins have been recently demonstrated in many biological processes [96-99]. Interactome map studies indicated that viral nsp5, nsp8, nsp13, and E protein are associated with the epigenetic and gene-expression regulators such as histone deacetylase 2 (HDAC2) [96,98]. In particular, nsp5 has been identified to have high confidence interaction with HDAC2 and may inhibit the transport of HDAC2 into nucleus and could possibly hinder the ability of HDAC2 mediated inflammatory and interferon response of the host cell [100,101]. Besides, the vesicle trafficking of the host was found to be associated with the interacting proteins of SARS-CoV-2, accounted for nearly 40% of viral proteins. The study by Gordon and his coworkers were identified the host interactions with viral nsp2 (Wiskott–Aldrich syndrome protein and scar homology (WASH)), nsp6 (vacuolar ATPase and Sigma receptors), nsp7(Rab proteins), nsp8 (signal recognition particle (SRP)), nsp10 (AP2), nsp13(organization of the centrosome and Golgi), ORF3a (homotypic fusion and protein sorting (HOPS), E protein (AP3), M protein (morphology of the ER and vacuolar ATPase)), and ORF8 (protein quality control in the ER), ORF9c (Sigma receptors). The viral protein interaction with Sigma receptors have been implicated in lipid modification and the ER stress response [96]. In addition, 3CTpro (nsp5) of SARS-CoV-2 was reported to affect the trafficking into the ER and mitochondria [102]. SARS-CoV-2 interacts with the host ubiquitin ligases required for ubiquitination of proteins. Viruses usually hijack ubiquitination pathways of the host cell for replication and pathogenesis44. The ORF10 of SARS-CoV-2 interacts with ubiquitin ligases, particularly to members of a cullin-2 E3 ligase complex, to hijack ubiquitination and to degrade restriction factors [96,103]. Furthermore, SARS-CoV-2 interacts with host translation machinery via N protein which binds to the stress granule proteins and host mRNA-binding proteins such as the mTOR-regulated translational repressor, casein kinase 2 (CK2), and mRNA decay factors [96]. N protein also interacts with RNA processing and regulation of the host cell [104-106]. SARS-CoV-2 interacts with the host cytoskeleton via nsp1 and nsp13 and reorganizes it for efficient cell entry and controls host transcriptional processes to support viral protein translation [98]. The SARS-CoV-2 also interacts directly with innate immune signaling proteins using various viral proteins and hence dysregulates innate cellular defenses. The interactions between the viral nsp13, nsp15 and ORF9b with the interferon pathway while nsp13 and ORF9c interact with the NF-κβ pathway have recently been reported an interactome map study [96]. Other studies also reported that the two E3 ubiquitin ligases that regulate antiviral innate immune signaling targeted by viral ORF3a and nsp9 [107,108]. This is also supported with Messina et al. who demonstrated the viral-host interactome, particularly the S protein interactions with the components of the host innate immunity, such as toll Like receptors (TLR), cytokines, and chemokines, as well as with lipid metabolism [99]. In addition, the host interacts with nsp1 in DNA replication, nsp8 in ribonucleoprotein complex biogenesis and RNA processing and regulation, nsp7, nsp8, nsp13, N protein, and ORF9b in cell signaling, nsp7, nsp9, nsp15, and ORF6 in nuclear transport machinery, nsp4, nsp8, and ORF9c in mitochondria, and nsp9 the extracellular matrix [96]. Overall, deciphering the functional interaction between the host and SARS-CoV-2, can be used to shed light on the host–virus interaction in the dynamic process of SARS-CoV-2 infection and pathogenesis. This may pave the way to better understand how the host cellular signaling pathways hijacked during this viral infection to replicate and evade innate immunity. Besides, virus-host interactions may also provide key information to guide new antiviral therapeutic and prophylactic drug repurposing [96,97].Therapies targeting the host-virus interface, where the occurrence of mutational resistance is less likely, could potentially give promising long lasting, broad-spectrum therapeutic modalities [109]. A recent interactome map noted 69 existing drugs known to target host proteins or associated pathways that interact with SARS- CoV-2 [96].
6. Concluding Remarks
Conclusively, COVID 19 is a newly emerged global pandemic caused by novel CoV called SARS-CoV-2. SARS COV-2 is an enveloped, crown shaped beta CoV with a positive-sense single-stranded RNA genome possessing an overall amount of nearly 30kbs. The structure of the SARS-CoV-2 genome is organized into 14 ORFs that encodes four structural proteins, eight accessory proteins and several nsps. The structural proteins involve S, M, E, and N proteins. The S protein, composed of S1 and S2 subunits, is important for viral entry to the host cells. The viral entry involves binding of S1 protein (via its RBD) with the host cell surface receptors, such as ACE2 receptors, CD147, and NRP1 for viral attachment, subsequently it fuses with host cell membrane or internalizes to endosomes, and fuse viral and lysosomal membranes with the help of S2 protein (via FP and HR) to release its viral genome into the cytoplasm. Eventually, the viral replication, packaging, and release occur and the novel virus becomes ready to infect another cell or person. M protein has a crucial role in viral assembly while E protein has a profound role in viral release in addition to its involvement in viral assembly. On the other hand, N protein plays a key role in viral genome replication, transcription and packaging. Furthermore, SARS-CoV-2 proteins have been recently revealed to interact with several host proteins that play important roles in usurping cellular machinery, which may be helpful to target cellular signaling pathways and likely contribute to development of safe and effective anti-COVID-19 therapies or vaccines.
Acknowledgments
Not applicable.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
Not applicable.
Availability of Data and Materials
Not applicable.
Ethical Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors report no conflict of interests.
References
- Abebe EC, Dejenie TA, Shiferaw MY, et al. The newly emerged COVID-19 disease: a systemic review. Virology journal 17 (2020): 1-8.
- Zhu Z, Lian X, Su X, et al. From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respiratory research 21 (2020): 1-14.
- Bos R, Rutten L, van der Lubbe JE, et al. Ad26 vector-based COVID-19 vaccine encoding a prefusion-stabilized SARS-CoV-2 Spike immunogen induces potent humoral and cellular immune responses. npj Vaccines 5 (2020): 1-11.
- Boopathi S, Poma AB, Kolandaivel P. Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics 2020 (2020):1-10.
- Lazarus JV, Ratzan SC, Palayew A, et al. A global survey of potential acceptance of a COVID-19 vaccine. Nature medicine 2020 (2020): 1-4.
- Guihot A, Litvinova E, Autran B, et al. Cell-mediated immune responses to COVID-19 infection. Frontiers in immunology 11 (2020): 1662.
- Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. The lancet 395 (2020): 507-513.
- Singh RDM, Malim N, Khan MAR, et al. Life Cycle of Covid-19 Stages and Treatment.
- Siracusano G, Pastori C, Lopalco L. Humoral Immune Responses in COVID-19 Patients: A Window on the State of the Art. Frontiers in Immunology 11 (2020).
- Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Coronaviruses (2015): 1-23.
- Adedokun KA, Olarinmoye AO, Mustapha JO, et al. A close look at the biology of SARS-CoV-2, and the potential influence of weather conditions and seasons on COVID-19 case spread. Infectious Diseases of Poverty 9 (2020): 1-5.
- Neuman BW, Kiss G, Kunding AH, et al. A structural analysis of M protein in coronavirus assembly and morphology. Journal of structural biology 174 (2011): 11-22.
- Lai MM. Coronaviridae: the viruses and their replication (2001).
- Murgolo N, Therien AG, Howell B, et al. SARS-CoV-2 tropism, entry, replication, and propagation: Considerations for drug discovery and development. PLoS Pathogens 17 (2021): e1009225.
- Huang Y, Yang C, Xu X-f, et al. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacologica Sinica 41 (2020): 1141-1149.
- Wang Q, Zhang Y, Wu L, et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181 (2020): 894-904. e9.
- Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581 (2020): 215-220.
- Tang T, Bidon M, Jaimes JA, et al. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral research 178 (2020): 104792.
- Millet JK, Whittaker GR. Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology 517 (2018): 3-8.
- Chambers P, Pringle CR, Easton AJ. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. Journal of General Virology 71 (1990): 3075-3080.
- Xia S, Zhu Y, Liu M, et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cellular & molecular immunology 2020 (2020): 1-3.
- Deng Y, Liu J, Zheng Q, et al. Structures and polymorphic interactions of two heptad-repeat regions of the SARS virus S2 protein. Structure 14 (2006): 889-899.
- Kim CH. SARS-CoV-2 evolutionary adaptation toward host entry and recognition of receptor O-Acetyl sialylation in virus–host interaction. International journal of molecular sciences 21 (2020): 4549.
- Klausegger A, Strobl B, Regl G, et al. Identification of a coronavirus hemagglutinin-esterase with a substrate specificity different from those of influenza C virus and bovine coronavirus. Journal of virology 73 (1999): 3737-3743.
- Cornelissen L, Wierda C, Van Der Meer FJ, et al. Hemagglutinin-esterase, a novel structural protein of torovirus. Journal of virology 71 (1997): 5277-5286.
- Arndt AL, Larson BJ, Hogue BG. A conserved domain in the coronavirus membrane protein tail is important for virus assembly. Journal of virology 84 (2010): 11418-11428.
- Ahmed SF, Quadeer AA, McKay MR. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses 12 (2020): 254.
- Kelly JA, Olson AN, Neupane K, et al. Structural and functional conservation of the programmed− 1 ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2). Journal of Biological Chemistry 295 (2020): 10741-10748.
- Thomas S. The structure of the membrane protein of SARS-CoV-2 resembles the sugar transporter semiSWEET. Pathogens and Immunity 5 (2020): 342.
- Wang Y, Grunewald M, Perlman S. Coronaviruses: an updated overview of their replication and pathogenesis. Coronaviruses 2020 (2020): 1-29.
- Hung AY, Sheng M. PDZ domains: structural modules for protein complex assembly. Journal of Biological Chemistry 277 (2002): 5699-5702.
- Javier RT, Rice AP. Emerging theme: cellular PDZ proteins as common targets of pathogenic viruses. Journal of virology 85 (2011): 11544-11556.
- De Maio F, Cascio EL, Babini G, et al. Enhanced binding of SARS-CoV-2 Envelope protein to tight junction-associated PALS1 could play a key role in COVID-19 pathogenesis (2020).
- Liao Y, Yuan Q, Torres J, et al. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology 349 (2006): 264-275.
- Tseng YT, Wang SM, Huang KJ, et al. SARS-CoV envelope protein palmitoylation or nucleocapid association is not required for promoting virus-like particle production. Journal of biomedical science 21 (2014): 1-11.
- Gupta MK, Vemula S, Donde R, et al. In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics (2020): 1-11.
- Kamau A, Kulmanov M, Arold ST, et al. Functional pangenome analysis suggests inhibition of the protein E as a readily available therapy for COVID-2019. (2020).
- Rahman MS, Islam MR, Alam ARU, et al. Evolutionary dynamics of SARS-CoV-2 nucleocapsid protein and its consequences. Journal of medical virology 93 (2021): 2177-2195.
- Chang CK, Sue SC, Yu TH, et al. Modular organization of SARS coronavirus nucleocapsid protein. Journal of biomedical science 13 (2006): 59-72.
- Kumar A, Parveen A, Kumar N, et al. Characterization of nucleocapsid (N) protein from novel coronavirus SARS-CoV-2. Preprints (2020).
- Kuo L, Koetzner CA, Masters PS. A key role for the carboxy-terminal tail of the murine coronavirus nucleocapsid protein in coordination of genome packaging. Virology 494 (2016): 100-107.
- Grunewald ME, Fehr AR, Athmer J, et al. The coronavirus nucleocapsid protein is ADP-ribosylated. Virology 517 (2018): 62-68.
- Chen Y, Yu Z, Yi H, et al. The phosphorylation of the N protein could affect PRRSV virulence in vivo. Veterinary microbiology 231 (2019): 226-231.
- Fu J, Chen R, Hu J, et al. Identification of a novel linear B-cell epitope on the nucleocapsid protein of porcine deltacoronavirus. International journal of molecular sciences 21 (2020): 648.
- Ju B, Zhang Q, Ge J, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584 (2020): 115-119.
- Wajnberg A, Amanat F, Firpo A, et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science 370 (2020): 1227-1230.
- Ray M, Sarkar S, Rath SN. Druggability for COVID-19: in silico discovery of potential drug compounds against nucleocapsid (N) protein of SARS-CoV-2. Genomics & Informatics 18(2020).
- Wu A, Peng Y, Huang B, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell host & microbe 27 (2020): 325-328.
- Naqvi AAT, Fatima K, Mohammad T, et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease (2020): 165878.
- Rastogi M, Pandey N, Shukla A, et al. SARS coronavirus 2: from genome to infectome. Respiratory Research 21 (2020): 1-15.
- Abduljalil JM, Abduljalil BM. Epidemiology, genome, and clinical features of the pandemic SARS-CoV-2: a recent view. New microbes and new infections 35 (2020): 100672.
- Hu B, Zeng LP, Yang XL, et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS pathogens 13 (2017): e1006698.
- Wang C, Liu Z, Chen Z, et al. The establishment of reference sequence for SARS-CoV-2 and variation analysis. Journal of medical virology 92 (2020): 667-674.
- Shang J, Wan Y, Luo C, et al. Cell entry mechanisms of SARS-CoV-2. Proceedings of the National Academy of Sciences 117 (2020):11727-11734.
- Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature microbiology 5 (2020): 562-569.
- Jiang F, Yang J, Zhang Y, et al. Angiotensin-converting enzyme 2 and angiotensin 1–7: novel therapeutic targets. Nature Reviews Cardiology 11 (2014): 413.
- Lukassen S, Chua RL, Trefzer T, et al. SARS-CoV-2 receptor ACE 2 and TMPRSS 2 are primarily expressed in bronchial transient secretory cells. The EMBO journal 39 (2020): e105114.
- Subramanian A, Vernon K, Slyper M, et al. RAAS blockade, kidney disease, and expression of ACE2, the entry receptor for SARS-CoV-2, in kidney epithelial and endothelial cells. (2020).
- Jing Y, Run-Qian L, Hao-Ran W, et al. Potential influence of COVID-19/ACE2 on the female reproductive system. Molecular human reproduction 26 (2020): 367-373.
- Chen L, Li X, Chen M, et al. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovascular research 116 (2020): 1097-1100.
- Lamers MM, Beumer J, van der Vaart J, et al. SARS-CoV-2 productively infects human gut enterocytes. Science 369 (2020): 50-54.
- Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. The lancet 395 (2020): 1054-1062.
- Xia S, Lan Q, Su S, et al. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal transduction and targeted therapy 5 (2020): 1-3.
- Wan Y, Shang J, Graham R, et al. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. Journal of virology 94 (2020).
- Wang K, Chen W, Zhou YS, et al. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. (2020).
- Cantuti-Castelvetri L, Ojha R, Pedro LD, et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370 (2020): 856-860.
- Daly JL, Simonetti B, Klein K, et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 370 (2020): 861-865.
- Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of 2019-nCoV on virus entry and its immune cross-reactivity with spike glycoprotein of SARS-CoV. (2020).
- Andersen KG, Rambaut A, Lipkin WI, et al. The proximal origin of SARS-CoV-2. Nature medicine 26 (2020): 450-452.
- Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. cell 181 (2020): 271-280. e8.
- Yang N, Shen HM. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. International journal of biological sciences 16 (2020): 1724.
- Simmons G, Zmora P, Gierer S, et al. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral research 100 (2013): 605-614.
- Jaimes JA, Millet JK, Whittaker GR. Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 site. IScience 23 (2020): 101212.
- Hoffmann M, Kleine-Weber H, Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Molecular cell 78 (2020): 779-784. e5.
- Coutard B, Valle C, de Lamballerie X, et al. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral research 176 (2020): 104742.
- Rabaan AA, Al-Ahmed SH, Haque S, et al. SARS-CoV-2, SARS-CoV, and MERS-COV: a comparative overview. Infez Med 28 (2020): 174-184.
- Bestle D, Heindl MR, Limburg H, et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life science alliance 3 (2020).
- Hasan A, Paray BA, Hussain A, et al. A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics (2020): 1-9.
- Millet JK, Whittaker GR. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus research 202 (2015): 120-134.
- Lau SY, Wang P, Mok BWY, et al. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerging microbes & infections 9 (2020): 837-842.
- Belouzard S, Millet JK, Licitra BN, et al. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4 (2012): 1011-1033.
- Hoffmann M, Hofmann-Winkler H, Pöhlmann S. Priming time: how cellular proteases arm coronavirus spike proteins. Activation of Viruses by Host Proteases: Springer (2018): 71-98.
- Kawase M, Kataoka M, Shirato K, et al. Biochemical analysis of coronavirus spike glycoprotein conformational intermediates during membrane fusion. Journal of virology 93 (2019).
- Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annual review of biochemistry 70 (2001): 777-810.
- Hoffmann M, Kleine-Weber H, Krüger N, et al. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. BioRxiv (2020).
- Wedrowska E, Wandtke T, Senderek T, et al. Coronaviruses fusion with the membrane and entry to the host cell. Annals of Agricultural and Environmental Medicine 27 (2020): 175-183.
- Algarroba GN, Rekawek P, Vahanian SA, et al. Visualization of severe acute respiratory syndrome coronavirus 2 invading the human placenta using electron microscopy. American journal of obstetrics and gynecology 223 (2020): 275-278.
- Chen Y, Guo Y, Pan Y, et al. Structure analysis of the receptor binding of 2019-nCoV. Biochemical and biophysical research communications 525 (2020): 135-140.
- Tortorici MA, Walls AC, Lang Y, et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nature structural & molecular biology 26 (2019): 481-489.
- Tian X, Li C, Huang A, et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerging microbes & infections 9 (2020): 382-385.
- Van Hemert MJ, Van Den Worm SH, Knoops K, et al. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog 4 (2008): e1000054.
- Kim D, Lee JY, Yang JS, et al. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914-21. e10.
- Sawicki SG, Sawicki DL, Siddell SG. A contemporary view of coronavirus transcription. Journal of virology 81 (2007): 20-29.
- Gao Y, Yan L, Huang Y, et al. Structure of RNA-dependent RNA polymerase from 2019-nCoV, a major antiviral drug target. BioRxiv (2020).
- Asghari A, Naseri M, Safari H, et al. The Novel Insight of SARS-CoV-2 Molecular Biology and Pathogenesis and Therapeutic Options. DNA and Cell Biology 39 (2020): 1741-1753.
- Gordon DE, Jang GM, Bouhaddou M, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583 (2020): 459-468.
- Perrin-Cocon L, Diaz O, Jacquemin C, et al. The current landscape of coronavirus-host protein–protein interactions. Journal of translational medicine 18 (2020): 1-15.
- Suryawanshi RK, Koganti R, Agelidis A, et al. Dysregulation of cell signaling by SARS-CoV-2. Trends in Microbiology (2020).
- Messina F, Giombini E, Agrati C, et al. COVID-19: viral–host interactome analyzed by network based-approach model to study pathogenesis of SARS-CoV-2 infection. Journal of translational medicine 18 (2020): 1-10.
- Barnes PJ. Role of HDAC2 in the pathophysiology of COPD. Annual review of physiology 71 (2009): 451-464.
- Xu P, Ye S, Li K, et al. NOS1 inhibits the interferon response of cancer cells by S-nitrosylation of HDAC2. Journal of Experimental & Clinical Cancer Research 38 (2019): 1-16.
- Dewe JM, Fuller BL, Lentini JM, et al. TRMT1-catalyzed tRNA modifications are required for redox homeostasis to ensure proper cellular proliferation and oxidative stress survival. Molecular and cellular biology 37 (2017).
- Timms RT, Zhang Z, Rhee DY, et al. A glycine-specific N-degron pathway mediates the quality control of protein N-myristoylation. Science 365 (2019).
- Knoops K, Kikkert M, Van Den Worm SH, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol 6 (2008): e226.
- Shah PS, Link N, Jang GM, et al. Comparative flavivirus-host protein interaction mapping reveals mechanisms of dengue and Zika virus pathogenesis. Cell 175 (2018): 1931-45. e18.
- Heaton NS, Moshkina N, Fenouil R, et al. Targeting viral proteostasis limits influenza virus, HIV, and dengue virus infection. Immunity 44 (2016): 46-58.
- Kondo T, Watanabe M, Hatakeyama S. TRIM59 interacts with ECSIT and negatively regulates NF-κB and IRF-3/7-mediated signal pathways. Biochemical and biophysical research communications 422 (2012): 501-507.
- Li S, Wang L, Berman M, et al. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity 35 (2011): 426-440.
- Prussia A, Thepchatri P, Snyder JP, et al. Systematic approaches towards the development of host-directed antiviral therapeutics. International journal of molecular sciences 12 (2011): 4027-4052.


 Impact Factor: * 3.0
Impact Factor: * 3.0 Acceptance Rate: 76.32%
Acceptance Rate: 76.32%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
