A Novel Protein Elicitor PevL1, from Verticillium Lecanii 2, Induces Systemic Resistance against Bean Aphid (Megoura Japonica Matsumura) in Phaseolus Vulgaris L
Trinh Duy Nam1,2, Yusuf Ali Abdulle1, Khadija Javed1, Tum Sokea1 and Dewen Qiu1*
1State Key Laboratory of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, 100193, China
2Plant Protection Research Institute, Hanoi, Vietnam
*Corresponding Author (s): Dewen Qiu, State Key Laboratory of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, 100193, China
Received: 09 March 2020; Accepted: 17 March 2020; Published: 06 April 2020
Article Information
Citation: Trinh Duy Nam, Yusuf Ali Abdulle, Khadija Javed, Tum Sokea and Dewen Qiu. A Novel Protein Elicitor PevL1, from Verticillium Lecanii 2, Induces Systemic Resistance against Bean Aphid (Megoura Japonica Matsumura) in Phaseolus Vulgaris L. International Journal of Plant, Animal and Environmental Sciences 10 (2020): 81-94.
View / Download Pdf Share at FacebookAbstract
Abstract
In this paper, we reported a novel protein elicitor from Verticillium lecanii (PevL1), which can induce systemic resistance against M. Japonica) in Phaseolus Vulgaris L. The protein was purified and evaluated using de novo sequencing. The PevL1 gene consists of 402 bp with a theoretical molecular weight of 14.46 kDa. The PevL1 sequence was matched to a genomic sequence (GenBank accession no. OAR00290.1). The encoded PevL1 protein was expressed in Escherichia coli, and the recombinant protein was characterized by inducing systemic resistance against M. japonica. The effects of three PevL1 concentrations (24.0, 31.0, and 50.0 µg mL−1) at different temperatures (15, 20, and 25°C) were tested under laboratory conditions. The PevL1 protein was applied to three-leaved Phaseolus Vulgaris L., and newly emerged adult bean aphids (0 to 6 h old) were released on the treated leaves. The relative expression levels of key genes of the JA and SA plant defense-related pathways were also determined by RT-qPCR. The treated plants with PevL1 exhibited significant resistance to the bean aphid compared with the control plants.
Keywords
<p>Verticillium lecanii; Megoura japonica Matsumura; Resistance; PevL1; Protein elicitor; Phaseolus vulgaris L</p>
Article Details
Introduction
Phaseolus vulgaris L. is an annual legume crop with trifoliate broad leaves. It has a massive protein content and is a rich source of energy that provides dietary fiber, folic acid, and carbohydrates (filella et al. 1994). However, Phaseolus vulgaris L. is profoundly affected by many biotic stresses. Among them is attacked by the bean aphid (Megoura japonica Matsumura), which results in huge P. vulgaris L. yield losses worldwide (Munyasa et al. 2013). Thus, biological control results in the improved resistance of P. vulgaris L. against bean aphid. Verticillium lecanii is an important entomopathogenic fungus, which has been commercialized as a biological control agent against insect pests such as aphid and whitefly (Milner RJ 1997). The use of entomopathogenic fungi can manage several insect pests; however, they do not meet commercialization norms owing to the resulting slow target mortality rate (St Leger et al. 2009). It is necessary to explore new methods using advanced research and techniques to address plant pest-related issues. One new development is the use of herbivore management practices mediated by the use of different enzymes and proteins (Chen et al. 2007).
Plants are damaged by a wide range of pests, including insects, fungi, oomycetes, viruses, bacteria, nematodes, and mites, which have different infection forms and life cycles (Mingjia Chen et al. 2012). Plants can protect themselves against pest attacks through their varying resistance mechanisms (Díez-Navajas 2008 and Pieterse 2009). For instances, the salicylic acid (SA)-, ethylene-, and jasmonic acid (JA)-signaling pathways form defensive responses that protect plants from multiple pests by producing and combining various phytochemicals and defense-related proteins, such as steroid glycoalkaloids, polyphenol oxidase, and proteinase inhibitor proteins, as well as pathogenesis-related (PR) proteins and phytoalexins, which target physiological processes in the organisms (van Loon et al. 1998; Riera et al. 2018).
Additionally, cumulative defense-signaling pathways are activated in plants in response to herbivore attacks, but most jasmonate responses are related to chewing herbivores (Diezel 2009; Wu 2009), while salicylate responses have been linked to phloem-sucking insects, such as whitefly and aphid (Moran 2002; Zhang 2012). The interaction between SA and JA pathways might play a significant role in fine-tuning the defense response (Robert-Seilaniantz et al. 2011).
All the signals identifying plant cells, which trigger defensive reactions, are considered elicitors. Different organisms, such as bacteria, fungi, oomycetes, and viruses, can produce protein elicitors, including different peptides, oligosaccharides, glycoproteins, lipids, and proteins (Ellis et al. 2009; Islam et al. 2018; Nürnberger 1999). Gram-positive plant-pathogenic bacteria produce a multifunctional protein elicitor called hairpin (Wang et al. 2015; Wei et al. 1992). In addition, some fungal protein elicitors, such as Hrip1 of Alternaria tenuissima, PevD1 of Verticillium dahlia, PeBC1 of Botrytis cinerea, and MoHrip1 of Magnaporthe oryzae have been shown to activate the defense responses of plants (Kulye et al. 2012; Zhang et al. 2018).
In this paper, we isolated and purified a protein elicitor PevL1 which triggered defense responses and the effects of the enhanced basal defense of P. vulgaris L. against bean aphid infections. The results elucidated the interactions between bean aphid and P. vulgaris L. and provide a potential novel strategy for controlling the bean aphid.
Materials and methods
Fungal strains
- lecanii was collected from the Institute of Plant Protection, Beijing, China. The fungal strain was grown on potato dextrose agar medium in the dark at 25 ± 1°C for 20 days.
Plant cultures and insect rearing
- vulgaris L. was grown in sterile soil in plastic pots and were kept at 25 – 30°C with 60% – 70% relative humidity in a growth chamber. A colony of M. japonica was collected from the Institute of Plant Protection, Beijing, China, and then, bean aphids were reared on P. vulgaris L. in the cages (60 cm × 60 cm × 60 cm) in the growth chamber. The bean aphids were maintained at 21 ± 2°C with 60% – 70% relative humidity and a 16-h:8-h light:dark photoperiod.
Protein elicitor purification
The fungus V. lecanii was grown on potato dextrose agar dishes at 25°C for 20 days, and then the spores were harvested from the plates. The suspension was filtered through sterile cheesecloth. The conidial concentration was estimated as 1.0 × 108 conidia mL−1). The broth culture was prepared, and 5 mL of conidial suspension was added into 1 L of potato dextrose medium. The cultures were incubated at 160 rpm and 25°C for 6–8 days. The culture mycelia were removed by centrifugation at 4°C and 12,000 rpm for 20 min. The supernatants were passed through 0.45-µm pore-sized filters to produce the filtrates. The filtrates were added to ammonium sulfate at 80% saturation overnight at 4°C (in the refrigerator). After centrifugation at 4°C and 12,000 rpm for 20 min, the precipitate was re-suspended in a buffer (50 mM Tris HCI, pH 8.0) and was then dialyzed in 5 L of buffer (50 mM Tris-HCl, pH 8.0) at 4ºC (in the refrigerator) for 48h. The concentrates were collected, passed through a 0.45-μm filter after centrifugation at 4°C and 12,000 rpm for 20 min. The AKTA Explorer 10 System (Amersham Biosciences) was used to purify the protein. The crude protein was loaded onto a HiTrap™ Q HP column (5 mL; GE Healthcare) and pre-equilibrated with an elution buffer (50 mM Tris-HCl, pH 8.0). The bound proteins were eluted with a linear ion gradient of 0–1 M NaCl in elution buffer at a flow rate of 2 mL min−1. Protein peaks were collected and applied to a desalting column (GE Healthcare) for eliciting activity. The fraction having a higher biological activity was further analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE). Protein samples (25 µL protein + 5 µL protein loading buffer) for SDS–PAGE were reduced by boiling for 10 min. Protein samples were loaded on a vertical glass slab containing a 10% resolving gel and a 5% stacking gel fixed in SDS running buffer and run at 80 V and 500 mA for 15 min and 180 V and 500 mA for 45 min, respectively. The gel was stained in Coomassie brilliant blue for 1 h and finally disdained for 1 h in a buffer of 30% methanol and 10% acetic acid in distilled water.
Gene identification
The total DNA was extracted using a fungal DNA KIT (D3390-01; OMEGA Biotech). The combined results of the de novo sequencing and peptide sequence and BLAST searches in the NCBI (https://www.ncbi.nlm.nih.gov) database were used to amplify the entire coding sequence of the elicitor-encoding gene and were also used to design a primer pair. The primer sequences were designed as follows: forward primer, 5′-ATGAGCTCGTCCGTCGCCACAG-3′ and reverse primer, 5′-GCGCTC-CCGCGG GCTGAC-3′. The encoding gene was amplified from V. lecanii using PCR.
Expression and purification of the recombinant protein elicitor
The specific primers used to amplify the gene were as follows, forward primer (5'CGCGGATCCATGAGCTCGTCCGTCGCCACAG3' (BamdI is underlined site) and reverse primer (5'CCCAAGCTTGCGCTCCCGCGGGCTGAC3' (HindIII is underlined site). The PevL1 gene was inserted into the pET-30a vector using BamHI and HindIII restriction enzymes. The ligated plasmid was transformed into Escherichia coli Trans5a (TransGen Biotech, China). The recombinant vector was transferred into BL21 (DE3) (E. coli) for protein expression and 1.0mM isopropyl-β-D-1-thiopyran-galactopyranoside (Sigma, USA) was added to induce the recombinant protein’s expression. Bacterial cells were collected and centrifuged at 4°C and 5000 rpm for 15 min. The collected cells were re-suspended in a buffer (50 mM Tris-HCl 200 mM NaCl, pH 8.0), and an ultrasonic disruptor was used to interrupt the cells. The supernatant was harvested and centrifuged at 4°C and 12,000 rpm for 15 min. Protein was examined by SDS-PAGE. By using HisTrap HP, (GE Healthcare, USA) column was further purified by affinity chromatography a protein PevL1, with buffer (50 mM Tris HCl 200 mM NaCl 500 mM imidazole, pH 8.0) then eluted the bounded protein. The eluted protein was desalted with buffer (50 mM Tris HCl, pH 8.0) by using a desalting column (GE Healthcare, USA). The molecular weight of the purified recombinant elicitor was estimated using a protein marker (Thermo Scientific, USA).
Aphid choice test
Leaves of P. vulgaris L. were sprayed with PevL1 (50.0 µg mL−1) or buffer (50 mM Tris-HCl, pH 8.0) for control treatments, and after 24 h, the leaves were placed onto Petri plates containing 1% agar. In total, 30 bean aphids were maintained in a 2.0 mL plastic tube and were then released between PevL1-treated and control leaves (buffer treated). The numbers of bean aphids per leaf were recorded at 6, 24, and 48 h after their release. This experiment was repeated 15 times under environmentally controlled conditions of 21 ± 1°C and 60% – 70% relative humidity with a 16-h:8-h light:dark photoperiod.
Effects of the PevL1 protein on the reproductive biology of M. japonica
The PevL1 concentration was determined using a Bradford assay. Three different PevL1 concentrations (24.0, 31.0, and 50.0 µg mL−1) and buffer (50 mM Tris-HCl, pH 8.0) (control), the buffer was used to determine the effects of PevL1 against M. japonica. Bioassays were designed at the three-leaf stage of P. vulgaris L. For the treatment, 4–5 mL of elicitor solution per plant was sprayed until the plant was covered. The control plants were sprayed only with a buffer solution. Three freshly molted (0 – 6-h-old) apterous adult bean aphids per leaf were released 24 h after the application of PevL1 and the buffer (for control). The fertility was determined as the total number of offspring produced by careful observations at regular 3 hours intervals through the duration of the bioassay. This test was replicated randomly 10 times, and the bioassays were replicated randomly 3 times at 3 different temperatures (15, 20, and 25°C).
Plant RNA isolation and cDNA synthesis
The total RNA was collected from the treated leaf (50.0 µg mL−1 of PevL1) and the control leaf (treated buffer) using a plant RNA KIT (TransGen Biotech, China). first-strand cDNA was synthesized using the TransScript All-in-One SuperMix for RT-qPCR KIT (TransGen Biotech, China).
Reverse transcription-quantitative PCR (RT-qPCR)
To analyze the mechanisms that induce defense responses by PevL1 in P. vulgaris L., the expression levels of P. vulgaris L. SA- and JA-pathway genes were assessed. Real-time quantitative PCR was performed on an ABI 7500 RT PCR System (AppliedBiosystems, USA). Actin was used as a control. Relative expression levels were calculated using the 2–ΔΔCT method (Livak and Schmittgen 2001. Table 1 shows all the primers used for RT-qPCR amplification. For each sample, the reaction mixture contained 20 µL of the PCR mix, comprising 10 µL of 2× SYBR® Premix Ex Taq (TaKaRa Biomedical Technology, China), 0.5 µL of reverse and forward primers, 2 µL of cDNA template and 7 µL of dd H2O).
|
Gene ID |
Forward primer |
Reverse primer |
|
PHAVU_001G017800g |
GGGAGAAGCTGCTGAAACAC |
CCGACCTGAATATCGAAGGA |
|
PHAVU_003G111500g |
GAATTTCCCTGCTGCTCTTG |
CTGGCTTAGCCTCAGGAATG |
|
PHAVU_001G000800g |
AGCCGCATGCTGTTCTCTAT |
TTTTCATGAACAGCGCTCAC |
|
PHAVU_003G011600g |
TAGTGATGGTGCAGGAGCTG |
GATGCAAAGGCCTCATTGAT |
|
PHAVU_006G048600g |
CAGGATGCTTGGGATGATCT |
CAAGGGCCTTTCCTACTTCC |
|
PHAVU_008G057700g |
TGCTTCACATGAATGGTGGT |
CAACCCAAGTCTGCCACTTT |
|
PHAVU_008G272800g |
TCCTTGTTGATGCCCACATA |
CAAAGAAAAAGGGGGAGAGG |
|
PHAVU_009G022200g |
CCCATGCACAGTGTACCAAG |
ACCAATTAACCCCCAAGGAG |
|
β-actin |
GGAAAATCAGTCTCGGTTCAG |
TCATACAGCAGCAAGCAC |
Table 1: Primers used for RT-qPCR amplification and control genes
Experiment design and data analyses
All the experiments were designed with three independents repetition times. All the data were analyzed using Excel (Microsoft, USA) and Statistix 8.1 (Tallahassee, FL, USA) software. Significant differences among treatment factors (PevL1 concentrations and temperature regimes) were determined using a two-way ANOVA followed by an LSD test at α = 0.05. The aphid test choice data and RT-qPCR expression levels were analyzed using Student’s t-test at a probability level of α = 0.05.
Results
Purification of protein elicitor
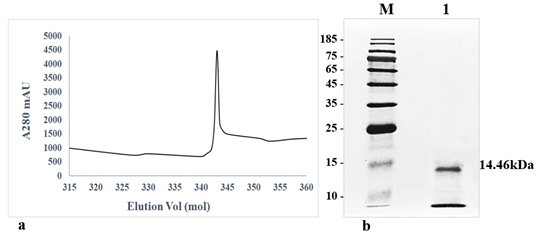
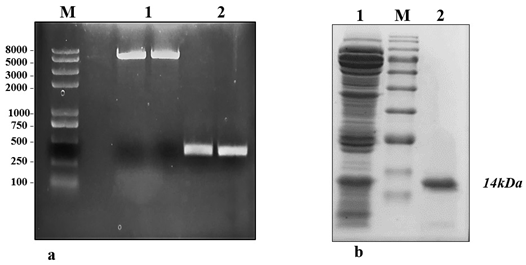
The crude protein from V. lecanii was dialyzed, and anion exchange chromatography was carried out and, finally, the protein was eluted with a buffer (50 mM Tris-HCl and 200 mM NaCl, pH 8.0). A protein peak was collected (fig. 1a) and examined using an SDS–PAGE gel, which displayed a single band with a molecular weight of 14.46 kDa (fig. 1b). The protein band was excised from the SDS–PAGE gel and identified by liquid chromatography-mass spectrometry. The outcomes were investigated using Mascot, and a similar protein (GenBank: OAR00290.1) was identified. This protein elicitor was named PevL1. The protein PevL1 has 402 bp and encodes 134 amino acids. The protein was cloned and transformed into the pET-30a (+) vector (fig. 2a), and then, it was expressed in E. coli BL21 (DE3). Using column chromatography, the recombinant protein was purified and desalted. The purified recombinant protein was checked using an SDS–PAGE gel, and a single protein band with a molecular weight of 14 kDa was observed (fig. 2b).
figure 2: Expression and purification of protein PevL1 in Escherichia coli. (a) M: Marker 2kplus II (pb); 1: PCR was used to amplify the full-length DNA sequence encoding PevL1. The length of the PevL1 gene is 402 bp; 2: The PevL1 gene was cloned and then ligated into pET30a (+) vector; (b): Purification of recombinant PevL1. M: Protein marker (kDa), 1: Total Escherichia coli expressed proteins, 2: Purified His-tagged PevL1.
Aphid choice test
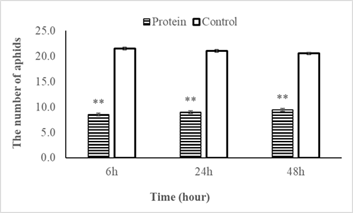
In total, 30 bean aphids were released between PevL1 treated and buffer treated leaves (control) for each repetition. The number of bean aphids on the treated leaves (50.0 µg mL−1 PevL1) was significantly lower than on the control at the three recording times of 6, 24, and 48 h after the release of the bean aphids (t28 = 2.04; P < 0.001). The numbers of aphids on the fluctuations were not significant at the three different data collection times. At the first data collection time (after 6 h), the average number of bean aphids selected on PevL1-treated leaves was 8.5 ± 2 bean aphids (occupying 28.4%), while the number of bean aphids on control leaves was 21.5 bean aphids (occupying 71.6%). The numbers of bean aphids after 24 and 48 h on protein-treated leaves increased but the changes were not significant. Specifically, the numbers of bean aphids recorded after 24 and 48 h of bean aphid transmission on PevL1-treated leaves were 9 ± 0.85 (occupying 30%) and 9.5 ± 0.92 (occupying 31.6%), respectively, while the numbers of bean aphids on buffer treated leaves (control) was recorded 21.0 ± 0.85 bean aphids (occupying 70%) and 20.5 ± 0.92 bean aphids (occupying 68.4%) respectively (fig. 3). Thus, the effect of the protein on the bean aphids was clear and stable after 48 h.
figure 3: Shows the number of bean aphids (mean±SE) that chose to infiltrate the leaves of Phaseolus vulgaris L. with protein PevL1 (50.0µg mL-1 concentration) treatment and with the untreated leaves (control) after 6, 24 and 48 h. The black bars display the data of treated leaves while the white bars showing the data of control leaves (untreated). The asterisk (**) display significant differences between treated leaves and untreated leaves (control) at P < 0.01 (n = 15; Student’s t- test).
Effects of the PevL1 protein on the reproductive biology of M. japonica
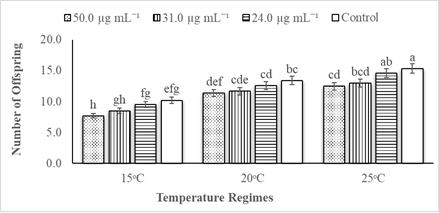
The statistical data analysis revealed that the impacts of different PevL1 concentrations on the reproductive capability of M. japonica correlated with different temperatures. The factors of temperature and protein concentrations both affected the reproductive capability of M. japonica (Table 2). The reproductive rate of M. japonica decreases as the PevL1 protein concentration increased (24.0, 31.0, and 50.0 µg mL−1) at the three different temperatures (15, 20, and 25°C). However, only the rate at the 50.0 µg mL−1 concentration was statistically significantly lower than the other rates at each of the three temperatures, but the numbers of offspring on 31.0 µg mL−1 PevL1-treated leaves decreased significantly at 15°C and 25°C. The M. japonica feeding on untreated (control) leaves reproduced at a greater rate during their life cycle compared with those feeding on PevL1-treated leaves. The maximum fecundity was recorded at 25°C, while the minimum fecundity was recorded at 15°C (fig. 4).
figure 4: Displays the mean fecundity (mean ± SE) of Megoura japonica on Phaseolus vulgaris L. treated protein PevL1 of 24.0, 31.0 and 50.0 µg mL-1 concentrations and on the control (untreated) at 15, 20 and 25oC regimes (n = 10). The different letters express significant differences between different concentrations of protein PevL1 and control (untreated) at each temperature regime (two-way ANOVA; LSD test at α = 0.05).
|
Source of Variation |
DF |
Sums of Squares |
Mean Squares |
F Ratio |
Prob |
|
Conc. |
3 |
34.0589 |
11.3530 |
9.40 |
0.000 |
|
Temp. |
2 |
147.662 |
73.8311 |
61.10 |
0.000 |
|
Conc. * Temp. |
6 |
1.00444 |
0.167407 |
0.14 |
0.988 |
|
Residual (Error) |
22 |
26.5828 |
1.20831 |
||
|
Total |
35 |
210.699 |
6.01997 |
||
|
CV%/Grand Mean |
9.2/11.6 |
Table 2: Analysis of variance (ANOVA) for the effect of different concentrations of PevL1 and temperature on the fecundity of Megoura japonica
Expression levels of SA- and JA-pathway genes in P. vulgaris L.
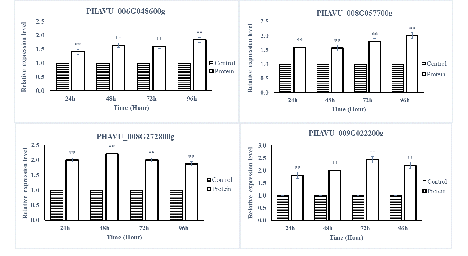
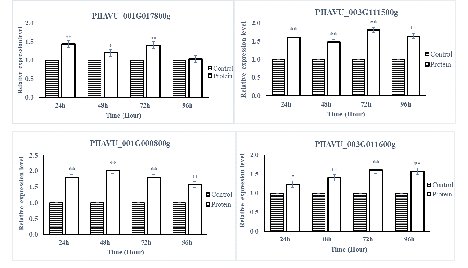
The SA- and JA-pathway marker genes in P. vulgaris L. after being sprayed with protein PevL1 were differentially expressed. There were significant differences in the expression levels of all the SA- and JA-pathway genes at different time intervals. The RT-qPCR results showed that all the SA-pathway genes were significantly up-regulated after 24, 48, 72, and 96h (fig. 5), while almost all the marker genes associated with the JA pathway were up-regulated, except one gene PHAVU_001G017800g, which displayed no difference between protein and buffer treatments after 96 h (fig. 6). The maximum levels of expression for all genes were detected at 48 and 72 h in plants treated with the partially purified protein compared with buffer.
figure 5: Displays the expression level analyses of SA pathway genes after the application of PevL1 elicitor protein in Phaseolus vulgaris L. at different time intervals. The white bars showing the effect of PevL1 elicitor treatments, black bars showing the effect of control treatments (treated buffer of 50mM Tris HCl, pH 8.0). The symbols (**) showing the significant difference between the PevL1 elicitor treatments and control treatments at P < 0.01 (Student’s t-test).
figure 6: Displays the expression level analyses of JA pathway genes after the application of PevL1 elicitor protein in Phaseolus vulgaris L. at different time intervals. The white bars showing the effect of PevL1 elicitor treatments while the black bars display the effect of control treatments (treated buffer of 50mM Tris HCl, pH 8.0). The symbols * and ** express the significant difference between the PevL1 elicitor and the control treatments at P < 0.05 and P < 0.01 (Student’s t-test).
Discussion
Protein elicitors play important roles in the signaling of the plant defense system and are vital tools for the control of pests. Pathogenic fungi, including those that are biotrophic and necrotrophic, comprise the main source of microbial elicitors (PAMPs or MAMPs) (Maffei et al. 2012). Protein elicitors, like PeaT1, PebC1, and PemG1 (Zhang et al. 2011; Mao et al. 2010; Qiu et al. 2009) from plant pathogenic fungi enhance the mechanisms of plant disease resistance. Moreover, some elicitors of M. oryzae might induce the defense responses against pathogen infection at the plant seedling stage (Qiu et al. 2009; Koga et al. 1998; Kanoh et al. 1993).
The fungal elicitors might reduce pathogen infection-related damage to plants. Likewise, a pre-treatment with PevL1 considerably improved responses to M. japonica infection compared with untreated control plants, suggesting that PevL1 boosts P. vulgaris L. resistance to M. japonica. PR genes of some plants, like PR-1a, PR-10a, and PR- 1b, may be expressed in plant resistance systems when the plant is subjected to pathogen infection (Liu et al. 2010; Sels et al. 2008). In response to the pathogen invasion, the plant defense system is regulated by a variety of signaling pathway networks, including SA, ethylene, and JA.
These studies focused on the evaluation of the elicitor PevL1, which was purified from V. lecanii, and evaluated its effects against M. japonica. The M. japonica population on P. vulgaris L. was treated with elicitor PevL1, which resulted in a slower reproduction rate compared with untreated control plants. These results were similar to those of previous studies. They demonstrated the negative impacts of applying exogenous protein elicitors, like benzothiadiazole, methyl jasmonate, and JA on the fitness traits, as well as population growth, of aphids (Boughton et al. 2006; Cooper and Goggin 2005). Additionally, the results were similar protein elicitors inducing resistance against sucking insect pests, such as against Tuta absoluta, through the co-expression of different proteinase inhibitors (Bostock et al. 2001; Hamza et al. 2018; Maffei et al. 2012). The outcomes are also in agreement with a study (Mallinger et al. 2011) in which a methyl salicylate elicitor played a major role in reducing a soybean aphid population by up to 40%. Similarly, the application of plant defense-related proteins and protease inhibitors can significantly reduce herbivorous pests on tomatoes (Boughton et al. 2006; Thaler et al. 1996).
Our results were in accordance with those of an earlier study in which M. japonica fed on SA strongly induces PR-1 expression, thereby, producing local resistance in Arabidopsis plants (Moran and Thompson 2001). Furthermore, our research indicated that PevL1 induced a significant strong up-regulation of the expression levels of all the JA and SA pathway-associated genes. These results demonstrate that herbivores, such as aphids, more strongly activate genes related to the SA-defense pathway than the JA-defense pathway (Ali and Agrawal 2012; Coppola et al. 2013).
Conclusions
A novel elicitor protein PevL1 was extracted from Verticillium lecanii exhibiting induced system resistance in Phaseolus vulgaris L. against M. japonica. This 402 bp elicitor protein encodes a polypeptide of 134 amino acids with a molecular weight of 14 kDa. In-vitro bioassays were performed with three different concentrations of this PevL1 protein (i.e. 24.0, 31.0 and 50 μm mL-1) against M. japonica on Phaseolus vulgaris L. the results showed significant (p < 0.05) and pronounced sub-lethal effects of exogenous application of PevL1 elicitor on M. japonica. By the application of PevL1 elicitor protein, aphids’ fecundity was significantly reduced as compared to control plants. Gene expression analysis done with RT-qPCR revealed that many plants defence associated genes, i.e. SA-responsive and JA-responsive genes, were up-regulated in the PevL1 treated plants. Overall, this study isolated a novel protein elicitor that could trigger plant systemic resistance against M. japonica.
Acknowledgments
We are especially thankful to the China Scholarship Council (CSC) for providing a Ph.D. scholarship.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ali JG, Agrawal AA. Specialist versus generalist insect herbivores and plant defense. Trends in plant science 17 (2012): 293-302.
- Bostock RM, Karban R, Thaler JS, et al. Signal interactions in induced resistance to pathogens and insect herbivores. European Journal of Plant Pathology 107 (2001): 103-111.
- Boughton AJ, Hoover K, Felton GW. Impact of chemical elicitor applications on greenhouse tomato plants and population growth of the green peach aphid, Myzus persicae. Entomologia Experimentalis et Applicata 120 (2006): 175-188.
- Chen J, Hua G, Jurat-Fuentes JL, et al. Synergism of Bacillus thuringiensis toxins by a fragment of a toxin-binding cadherin. Proceedings of the National Academy of Sciences 104 (2007): 13901-13906.
- Cooper WR, Goggin FL. Effects of jasmonate-induced defenses in tomato on the potato aphid, Macrosiphum euphorbiae. Entomologia experimentalis et applicata 115 (2005): 107-115.
- Coppola V, Coppola M, Rocco M, et al. Transcriptomic and proteomic analysis of a compatible tomato-aphid interaction reveals a predominant salicylic acid-dependent plant response. BMC genomics 14 (2013): 515.
- Diezel C, von Dahl CC, Gaquerel E, et al. Different lepidopteran elicitors account for cross-talk in herbivory-induced phytohormone signaling. Plant Physiology 150 (2009): 1576-1586.
- Diez-Navajas AM, Wiedemann-Merdinoglu S, Greif C, et al. Nonhost versus host resistance to the grapevine downy mildew, Plasmopara viticola, studied at the tissue level. Phytopathology 98 (2008): 776-780.
- Ellis C, Karafyllidis I, Wasternack C, et al. The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. The Plant Cell 14 (2002): 1557-1566.
- Ellis JG, Rafiqi M, Gan P, et al. Recent progress in discovery and functional analysis of effector proteins of fungal and oomycete plant pathogens. Current opinion in plant biology 12 (2009): 399-405.
- filella I, Penuelas J. The red edge position and shape as indicators of plant chlorophyll content, biomass and hydric status. International Journal of Remote Sensing 15 (1994): 1459-1470.
- Hamza R, Pérez-Hedo M, Urbaneja A, et al. Expression of two barley proteinase inhibitors in tomato promotes endogenous defensive response and enhances resistance to Tuta absoluta. BMC plant biology 18 (2018): 24.
- Islam W, Noman A, Qasim M, et al. Plant responses to pathogen attack: small RNAs in focus. International journal of molecular sciences 19 (2018): 515.
- Kanoh H, Haga M, Sekizawa Y. Transmembrane signalling operated at rice blade cells stimulated by blast fungus elicitor 2. Participation of calcium modulated protein. Journal of Pesticide Science-Pesticide Science Society of Japan-Japanese Edition 18 (1993): 325.
- Koga J, Yamauchi T, Shimura M, et al. Cerebrosides A and C, sphingolipid elicitors of hypersensitive cell death and phytoalexin accumulation in rice plants. Journal of Biological Chemistry 273 (1998): 31985-31991.
- Kulye M, Liu HU, Zhang Y, et al. Hrip1, a novel protein elicitor from necrotrophic fungus, Alternaria tenuissima, elicits cell death, expression of defence-related genes and systemic acquired resistance in tobacco. Plant, cell & environment 35 (2012): 2104-2120.
- Liu J, Wang X, Mitchell T, et al. Recent progress and understanding of the molecular mechanisms of the rice–Magnaporthe oryzae interaction. Molecular plant pathology 11 (2010): 419-427.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25 (2001): 402-408.
- Maffei ME, Arimura GI, Mithöfer A. Natural elicitors, effectors and modulators of plant responses. Natural Product Reports 29 (2012): 1288-1303.
- Mallinger RE, Hogg DB, Gratton C. Methyl salicylate attracts natural enemies and reduces populations of soybean aphids (Hemiptera: Aphididae) in soybean agroecosystems. Journal of economic entomology 104 (2011): 115-124.
- Mao J, Liu Q, Yang X, et al. Purification and expression of a protein elicitor from Alternaria tenuissima and elicitor-mediated defence responses in tobacco. Annals of Applied Biology 156 (2010): 411-420.
- Chen M, Zeng H, Qiu D, et al. Purification and characterization of a novel hypersensitive response-inducing elicitor from Magnaporthe oryzae that triggers defense response in rice. PLoS One 7 (2012).
- Milner RJ. Prospects for biopesticides for aphid control. Entomophaga 42 (1997): 227-239.
- Moran PJ, Thompson GA. Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant physiology 125 (2001): 1074-1085.
- Munyasa AJ, Chemining’wa GN, Kimani PM, et al. Evaluation of drought tolerance mechanisms in Mesoamerican dry bean genotypes. University of Nairobi, Nairobi (2013).
- Nürnberger T. Signal perception in plant pathogen defense. Cellular and Molecular Life Sciences CMLS 55 (1999): 167-182.
- Pieterse CM, Leon-Reyes A, Van der Ent S, et al. Networking by small-molecule hormones in plant immunity. Nature chemical biology 5 (2009): 308-316.
- Qiu D, Mao J, Yang X, et al. Expression of an elicitor-encoding gene from Magnaporthe grisea enhances resistance against blast disease in transgenic rice. Plant cell reports 28 (2009): 925-933.
- Riera N, Wang H, Li Y, et al. Induced systemic resistance against citrus canker disease by rhizobacteria. Phytopathology 108 (2018): 1038-1045.
- Robert-Seilaniantz A, Grant M, Jones JD. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annual review of phytopathology 49 (2011): 317-343.
- Sels J, Mathys J, De Coninck BM, et al. Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiology and Biochemistry 46 (2008): 941-950.
- St Leger RJ, Wang C, Stock S, et al. Entomopathonic fungi and the genomics era. Insect Pathogens: Molecular Approaches and Techniques, CABI (2009): 365-400.
- Thaler JS, Humphrey PT, Whiteman NK. Evolution of jasmonate and salicylate signal crosstalk. Trends in plant science 17 (2012): 260-270.
- Van Loon LC, Bakker PA, Pieterse CM. Systemic resistance induced by rhizosphere bacteria. Annual review of phytopathology 36 (1998): 453-483.
- Veit S, Wörle JM, Nürnberger T, et al. A novel protein elicitor (PaNie) from Pythium aphanidermatum induces multiple defense responses in carrot, Arabidopsis, and tobacco. Plant Physiology 127 (2001): 832-841.
- Wang H, Yang X, Guo L, et al. PeBL1, a novel protein elicitor from Brevibacillus laterosporus strain A60, activates defense responses and systemic resistance in Nicotiana benthamiana. Appl. Environ. Microbiol 81 (2015): 2706-2716.
- Wei ZM, Laby RJ, Zumoff CH, et al. Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science 257 (1992): 85-88.
- Wu J, Baldwin IT. Herbivory-induced signalling in plants: perception and action. Plant, cell & environment 32 (2009): 1161-1174.
- Zhang H, Yu P, Zhao J, et al. Expression of tomato prosystemin gene in Arabidopsis reveals systemic translocation of its mRNA and confers necrotrophic fungal resistance. new phytologist 217 (2018): 799-812.
- Zhang T, LUAN JB, QI JF, et al. Begomovirus–whitefly mutualism is achieved through repression of plant defences by a virus pathogenicity factor. Molecular Ecology 21 (2012): 1294-1304.
- Zhang W, Yang X, Qiu D, et al. PeaT1-induced systemic acquired resistance in tobacco follows salicylic acid-dependent pathway. Molecular biology reports 38 (2011): 2549-2556.


 Impact Factor: * 4.1
Impact Factor: * 4.1 Acceptance Rate: 75.32%
Acceptance Rate: 75.32%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 