ACEII Gene Analysis Exposes SARS-Cov-2 as A Potential Threat to Agricultural and National Security
Ruhl Michael W1*, Jenkins Tracie M2†
1*Grand Canyon University, College of Doctoral Studies, Phoenix, AZ 85017, USA
2University of Georgia, Department of Entomology, Athens, GA 30602, USA
†Professor Emerita
*Corresponding Author: Ruhl Michael W, Grand Canyon University, College of Doctoral Studies, Phoenix, AZ 85017, USA
Received: 8 July 2021; Accepted: 16 July 2021; Published: 29 July 2021
Article Information
Citation: Ruhl Michael W, Jenkins Tracie M. ACEII Gene Analysis Exposes SARS-Cov-2 as A Potential Threat to Agricultural and National Security. International Journal of Plant, Animal and Environmental Sciences 11 (2021): 443-455.
View / Download Pdf Share at FacebookAbstract
Coronavirus is now a significant human pathogen with the emergence of SARS-CoV-2. However, until now, there has been no data to support a threat to agricultural industries. Using a comparative genomic protein analysis, this study examined the angiotensin-converting enzyme II (ACEII) gene of 17 animal species of animals. The 20 known SARS-CoV-2 ribosomal binding domain (RBD)-ACEII gene interaction sites were compared to the 17 animal species to determine their potential susceptibility to the SARS-CoV-2 virus. Using the known bat host’s (XP_032963186) number of binding sites as a threshold, we note that all animal species examined in this study contained significant numbers (≥10) of SARS-CoV-2 binding sites and should be considered at serious risk for SARS-CoV-2 infection. The data from this study suggests SARS-CoV-2 imposes a grave threat to the safety and security of the agricultural industry. Urgent studies are needed to determine if infected animals can transmit SARS-CoV-2 before and after processing.
Keywords
<p>Agriculture</p>
Article Details
1. Introduction
Until recently, it was unknown whether animals could become infected by SARS-CoV-2. Despite the widespread suspicion that “SARS-CoV-2, originated from a bat (RaTG13| MN996532.1),” it remains unclear whether other animal species may be viable primary or secondary hosts. Preliminary data [1], suggest that various animal groups contain SARS-CoV-2 interaction sites between the SARS-CoV-2 spike protein ribosomal binding domain (RBD) and the angiotensin-converting enzyme II (ACEII) gene. This RBD has 16 amino acid residues capable of interacting with 20 ACEII amino acid sites [1, 2].
Since the onset of the SARS-CoV-2 pandemic, we have identified and verified that tigers [3, 4] and lions serve as SARS-CoV-2 hosts. To determine if SARS-CoV-2 poses a potential threat to agricultural security, this study examines the SARS-CoV-2 spike protein RBD sites capable of interacting with the ACEII gene of 17 animals identified as having agricultural significance.
2. Materials and Methods
Human and animal ACEII gene sequences (Table 1) were queried on the NCBI gene database [5]. Sequences were then sequestered into a Notepad++ [6] text file and saved using the fasta format suffix. The resulting fasta file of ACEII protein sequences was then imported into UGENE v34 [7] and aligned using Muscle [8, 9] default algorithm. A second alignment was performed using the Cobalt tool within the NCBI site [10]. The second alignment was necessary as UGENE lacks a nexus file format option when exporting alignments. The nexus file format was more easily imported to MS Word, where it was annotated.
Using UGENE, a distance matrix was generated using the following parameters: Distance algorithm = Similarity, Profile mode = Percentage, save profile to file= checked File = Comma-separated (.CSV). Generated CSV file was imported into MSExcel, annotated, and exported as a PDF. The human reference sequence must be in the first position when generating a distance matrix. The distance matrix generation process was repeated after removing all amino acid residues other than the 20 known ACEII interactive sites.
Using ASCII gene alignment, all sequences were examined for SARS-CoV-2/ACEII gene interaction sites using the 20 known human sites as the reference [1, 2]. Amino acid changes were annotated and represented graphically using MSWord and SnagIt Editor [11]. The number of matching sites was denoted into an MS Excel spreadsheet and exported as PDF.
Using human (NP_001358344) reference sequence, amino acids that differ from human SARS-CoV-2 interaction sites were queried manually and recorded in MS Word table format. The similarity value obtained from the lowest scoring known host was used as a threshold value to determine the possibility of COVID-19 infection.
3. Results
Amino acid alignment of the ACEII binding domain (Figure 1) reveals a highly homologous binding domain between species. Most SARS-CoV-2/ACE II interactive sites in this binding domain lie within the α-helices and β-sheets of the ACEII complex structure.
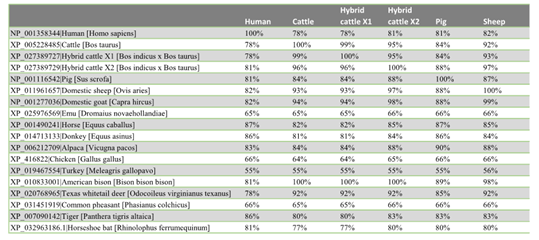
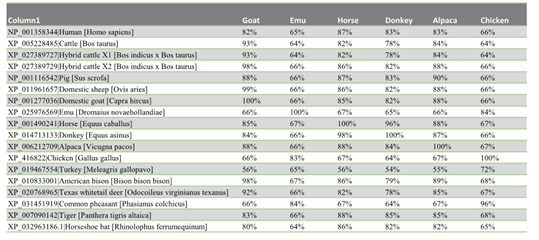
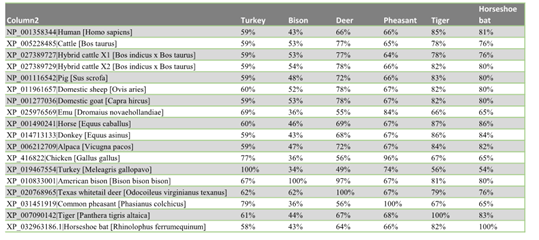
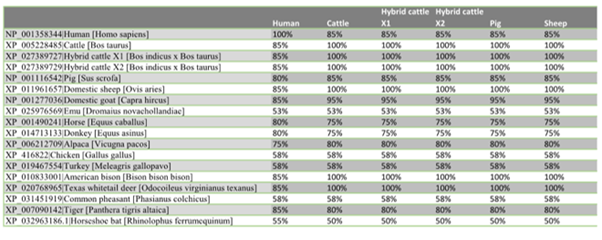
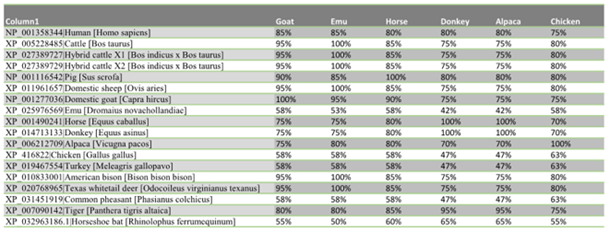
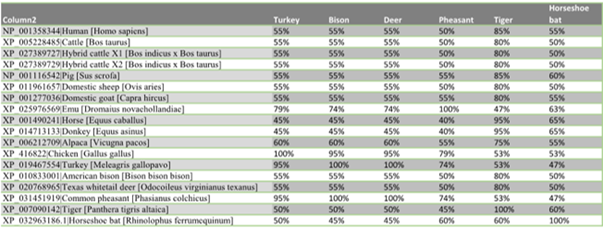
The similarity distance matrix demonstrates that the human ACEII gene shares ≥78% whole gene identity to all species in this study except the four birds: emu 65%, turkey 55%, chicken 66%, and pheasant 66% (Fig. 2a-c). When only the 20 ACEII interaction sites are analyzed, all species except the four birds, emu 53%, turkey 58%, chicken 58%, and pheasant 58%, in addition to bat (55%), demonstrate ≥75% similarity (Fig. 3a-c).
Our study noted the following AA residue differencesin the hydrophobic pockets created by F28, L79, Y83, and L97: Birds p.L79N, Pig p.L79I, and all others except donkey and horse p.L79M; birds and bat p.Y83F; and birds p.L97I in birds (Figure 1). F28 was conserved in all species.
The known host species with the least number of SARS-CoV-2/ACEII complex structure interaction sites were identified as being horseshoe bat (XP_032963186) with ten interaction sites. The known host tiger and cattle, sheep, goat, bison, and deer contained the most SARS-CoV-2 interaction sites outside humans (Table 2). Species with the greatest number of SARS-CoV-2 interaction sites contained 17, including cattle, sheep, goat, bison, deer, and tiger. Donkey, horse, and alpaca have 15 binding sites.
Figure 1a, 1b, 1c, 1d: ACEII protein sequence alignment. Only regions containing SARS-CoV-2 interaction sites are shown. Bold white letters with black background indicate the similarity of SARS-CoV-2 interaction sites to the human ACEII reference sequence. Clear boxes denote alpha-helices, shaded boxes denote β-sheets. Conserved interactive amino acid residues are seen at F28, E37, L45, N331, K354, D356, and R358.
Figure 2a, 2b, 2c: Distance matrix
Figure 3a, 3b, 3c: Matrix table using only 20 known SARS-CoV-2/ACEII interaction sites.
|
Accession number|common name [scientific name] |
|
NP_001358344|Human [Homo sapiens] |
|
XP_005228485|Cattle [Bos taurus] |
|
XP_027389727|Hybrid cattle X1 [Bos indicus x Bos taurus] |
|
XP_027389729|Hybrid cattle X2 [Bos indicus x Bos taurus] |
|
NP_001116542|Pig [Sus scrofa] |
|
XP_011961657|Domestic sheep [Ovis aries] |
|
NP_001277036|Domestic goat [Capra hircus] |
|
XP_025976569|Emu [Dromaius novaehollandiae] |
|
XP_001490241|Horse [Equus caballus] |
|
XP_014713133|Donkey [Equus asinus] |
|
XP_006212709|Alpaca [Vicugna pacos] |
|
XP_416822|Chicken [Gallus gallus] |
|
XP_019467554|Turkey [Meleagris gallopavo] |
|
XP_010833001|American bison [Bison bison bison] |
|
XP_020768965|Texas whitetail deer [Odocoileus virginianus texanus] |
|
XP_031451919|Common pheasant [Phasianus colchicus] |
|
XP_007090142|Tiger [Panthera tigris altaica] |
|
XP_032963186.1|Horseshoe bat [Rhinolophus ferrumequinum] |
Table 1: Accession list.
|
Accession number|Common name |
SARS-CoV-2 Binding Sites (20 possible) |
|
XP_005228485|Cattle |
17 |
|
XP_027389727|Hybrid cattle X1 |
17 |
|
XP_027389729|Hybrid Cattle X2 |
17 |
|
NP_001116542|Pig |
15 |
|
XP_011961657|Domestic sheep |
17 |
|
NP_001277036|Domestic goat |
17 |
|
XP_025976569|Emu |
10 |
|
XP_001490241|Horse |
15 |
|
XP_014713133|Donkey |
15 |
|
XP_006212709|Alpaca |
15 |
|
XP_416822|Chicken |
10 |
|
XP_019467554|Turkey |
10 |
|
XP_010833001|American bison |
17 |
|
XP_020768965|Texas whitetail deer |
17 |
|
XP_031451919|Common pheasant |
10 |
|
NP_001358344|Human |
20 |
|
XP_007090142|Tiger |
17 |
|
XP_032963186|Horseshoe bat |
10 |
Table 2: SARS-CoV-2/ACEII site interaction values.Accessions in the black background are known as SARS-CoV2 hosts.
4. Discussion
4.1 Structural analysis
It has been previously described that the unique SARS-CoV-2 F486 site can bind deep in the ACEII hydrophobic pockets created by F28, L79, Y83, and L97 [1]. The interspecies AA residue differences of those sites noted in this study conserve the hydrophobic pockets in the ACEII complex structure. It retains the hydrophobic pockets by the use of other hydrophobic residues. Of note is the p.Y83F substitution in birds and bats, which creates π-stacking of aromatic residues, an effect that can alter drug effectiveness/design. It also establishes a disulfide bond with F486 increasing virus binding affinity (k=6) [12]. This improved binding affinity may alter our AA interaction threshold estimate, 10 in known host bat, for establishing infection in a host. It is conceivable that these species with p.Y83F would not require as many sites to cause infection since they could theoretically acquire similar virus-host bonding strengths with fewer interacting residues.
Other than birds, all animals in this study contained at least 15 SARS-CoV-2/ACEII complex structure interaction sites. This number of interaction sites is well above our threshold value established by the know bat host at 10. The number of interaction sites may or may not cause concern, but it certainly leads to a high index of suspicion to their SARS-CoV-2 host viability.
4.2 Historical coronavirus evidence in agricultural species
It is essential to understand that ALL species in this study have been previously identified as coronavirus hosts; cattle [13] chicken, pheasant [14], turkey [15] pig [16], sheep [17], goat, bison [13], emu [18], donkey, horse, [19-21], alpaca, [22, 23] bison [24, 25] and deer [24]. While not SARS-CoV-2, they were of the same closely related viral coronavirus genus. Therefore, we cannot disregard the possibility of SARS-CoV-2 until proven otherwise.
It is still not known what the pathogenic effect, if any, will be in SARS-CoV-2 infected animals. However, looking at historical data from bovine coronavirus (BCoV) infections [26], it is conceivable that SARS-CoV-2 could overcome BCoV as the leading cause of morbidity and mortality in cattle < 2years old.
4.3 SARS-CoV-2 impact on agricultural security
“Agricultural animals are typically kept in large numbers and in close proximity” to each other. As a result, the possibility of one animal infecting another is exceptionally high. Even with free-range animals, their natural herd tendencies sustain a viral transmission risk. While numerous studies have shown that viruses can spread quickly with a high degree of population penetrance, no studies are found to document coronavirus transmission to humans or other animals through consumption or process handling. However, the absence of this data does not eliminate this prospect.
SARS-C0V-2 has demonstrated surface survivability of up to 9 days [27]. In excreted body fluids, it can survive >7 days [28]. Our understanding is that survivability in biological tissues has yet to be determined, but we can infer survivability of the virus is also >7 days from excreted fluids. Within biological tissues, the virus’ survivability could increase beyond 20 days when refrigerated [29], a vital process to advert food spoilage. A commonly used method to increase food preservation is gamma irradiation [30, 31]. However, this process may not inactivate enough of the virus to prevent viral transmission, as demonstrated from gamma irradiation studies with the Ebola virus [32]. In that study, it was determined that “no dosage could be considered to inactivate 100% of a sample” [32]. Principally, the viral transmission was possible after gamma irradiation.
Infected animals and humans may also contaminate a water supply if in direct contact. Studies have shown that coronavirus inactivation in water can take over 100 days at 4οC and approximately ten days at 23οC [33]. This study also indicates the possibility of human-to-animal transmission through an open water source.
4.4 Mitigation and control
Coronavirus is now a significant human pathogen with the emergence of SARS-CoV-2. This pathogen has caused severe illness and death in humans. However, until recently, no evidence existed to suggest the agricultural industry was in jeopardy until the recent confirmation of SARS-CoV-2 infections in several Bronx Zoo tigers and lions. To date, it is unknown if the tigers and lions contracted the disease through human interaction, another vector, or a contaminated food source.
Understanding the potential impact of SARS-CoV-2 on animals is paramount to agricultural and national security [34-37]. Until now, there has been no data to support a threat to the global food supply. Existing control measures are not adequate to mitigate SARS-CoV-2 propagation. Agricultural industries should begin preparing for a worst-case scenario before they are forced to respond to one.
Using the human ACEII gene sequence as a reference limit this study since actual SARS-CoV-2/ACEII animal complex structure interactions could differ in animals. Another limitation is that SARS-CoV-2 survivability in tissues and water must be inferred from other coronaviruses and may vary under identical environmental conditions. Finally, the lack of animal testing precludes a definitive viral susceptibility analysis. Nevertheless, our work establishes the framework for a SARS-CoV-2 infection risk amongst animals. Future studies should test living and processed animal samples to determine primary and secondary host viability.
5. Summary
Using a comparative genomic protein analysis, this study found that all animal species in this study contained significant numbers of SARS-CoV-2/ACEII complex structure interaction sites and should be considered at serious risk for SARS-CoV-2 infection. Data from this study suggest SARS-CoV-2 imposes a grave threat to agricultural and national security. Urgent studies are needed to determine if infected animals can transmit SARS-CoV-2 before and after processing.
Acknowledgments
We gratefully acknowledge the authors, originating and submitting laboratories of the sequences from the NCBI Database on which this research is based. I would like to thank the University of Georgia-Griffin faculty for their valuable guidance in molecular processes.
Funding
No outside funding sources.
Author contributions
Michael Ruhl was performed data query, protein analysis, literature review, developed the initial manuscript, and produced tables and graphics. Tracie Jenkins was the primary editor and contributed to genomic analysis.
Competing interests
Authors have no competing interests to declare.
Data and materials availability
All accessions are publicly available and listed in Table 1. All software used is publicly available, and citations are provided in body text.
References:
- Chen Y, Guo Y, Pan Y, et al. (2020). Structure analysis of the receptor binding of 2019-nCoV. Biochemical and Biophysical Research Communications 525 (2020): 135-140.
- Li F, Li W, Farzan M, et al. Structural biology: Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309 (2005): 1864-1868.
- Gollakner R, Capua I. Is COVID-19 the first pandemic that evolves into a panzootic? Veterinaria Italiana (2020).
- Mahdy M. An Overview of SARS-CoV-2 and Animal Infection (2020).
- Anjay A. National Center for Biotechnology Information (NCBI). Bethesda, Maryland, USA (2012).
- Ho D. Notepad++ (2020). In Retrived from: http://notepad-plus-plus. org (7.8.5).
- Okonechnikov K, Golosova O, Fursov M, et al. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28 (2012): 1166-1167.
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32 (2004a): 1792-1797.
- Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5 (2004b): 113.
- Papadopoulos JS, Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences 23 (2007): 1073-1079.
- Bragg DC. Snagit, Version 6.1 for Windows. Bulletin of the Ecological Society of America 83 (2002): 255-256.
- Chen K, Kurgan LA, Ruan J. Prediction of flexible/rigid regions from protein sequences using k-spaced amino acid pairs. BMC Structural Biology 7 (2007): 25.
- Weiyan YHDNX, Chengping L. Seroepizootiological Survey of Bovine Coronavirus in Cattle, Human and Other Animals [J]. Journal of Nanjing Agricultural University 2 (1990).
- Culver FA, Britton P, Cavanagh D. RT-PCR detection of avian coronaviruses of galliform birds (Chicken, Turkey, Pheasant) and in a parrot. Methods in Molecular Biology 454 (2008): 35-42.
- Guy JS, Barnes HJ, Smith LG, et al. Antigenic Characterization of a Turkey Coronavirus Identified in Poult Enteritis- and Mortality Syndrome-Affected Turkeys. Avian Diseases 41 (1997): 583.
- Woo PC, Lau SK, Tsang CC, et al. Coronavirus HKU15 in respiratory tract of pigs and first discovery of coronavirus quasispecies in 5’-untranslated regions. Emerging Microbes & Infections 6 (2017): e53.
- Tråvén M, Carlsson U, Lundén A, et al. Serum Antibodies to Bovine Coronavirus in Swedish Sheep. Acta Veterinaria Scandinavica 40 (1999): 69-74.
- Kummrow MS. Ratites or Struthioniformes. In Fowler’s Zoo and Wild Animal Medicine 8 (2015): 75-82.
- Fielding CL, Higgins JK, Higgins JC, et al. Disease Associated with Equine Coronavirus Infection and High Case Fatality Rate. Journal of Veterinary Internal Medicine 29 (2015): 307-310.
- Nagano S. Effect of a sterilization locker with ethylene oxide gas. Journal of the Japan Veterinary Medical Association 64 (1982): 535-539.
- Oue Y, Morita Y, Kondo T, et al. Epidemic of Equine Coronavirus at Obihiro Racecourse, Hokkaido, Japan in 2012. Journal of Veterinary Medical Science 75 (2013): 1261-1265.
- Crossley B, Mock R, Callison S, et al. Identification and Characterization of a Novel Alpaca Respiratory Coronavirus Most Closely Related to the Human Coronavirus 229E. Viruses 4 (2012): 3689-3700.
- Jin L, Cebra CK, Baker RJ, et al. (2007). Analysis of the genome sequence of an alpaca coronavirus. Virology 365 (2007): 198-203.
- Amer HM. Bovine-like coronaviruses in domestic and wild ruminants. Animal Health Research Reviews 19 (2019): 113-124.
- Harms NJ, Jung TS, Andrew CL, et al. Health status of reintroduced wood bison (Bison bison athabascae): Assessing the conservation value of an isolated population in northwestern Canada. Journal of Wildlife Diseases 55 (2019): 44-53.
- Boileau MJ, Kapil S. Bovine Coronavirus Associated Syndromes. In Veterinary Clinics of North America - Food Animal Practice 26 (2010): 123-146.
- Kampf G, Todt D, Pfaender S, et al. (2020). Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. In Journal of Hospital Infection 104 (2020): 246-251.
- Lai MYY, Cheng PKC, Lim WWL. Survival of Severe Acute Respiratory Syndrome Coronavirus. Clinical Infectious Diseases 41 (2005): e67-e71.
- Guionie O, Courtillon C, Allee C, et al. (2013). An experimental study of the survival of turkey coronavirus at room temperature and +4°C. Avian Pathology 42 (2013): 248-252.
- Diehl JF. Food irradiation - Past, present and future. Radiation Physics and Chemistry 63 (2002): 211-215.
- Piri I, Babayan M, Tavassoli A, et al. The use of gamma irradiation in agriculture. African Journal of Microbiology Research 5 (2011): 5806-5811.
- Hume AJ, Ames J, Rennick LJ, et al. Inactivation of RNA viruses by gamma irradiation: A study on mitigating factors. Viruses 8 (2016).
- Gundy PM, Gerba CP, Pepper IL. Survival of Coronaviruses in Water and Wastewater. Food and Environmental Virology 1 (2009): 10-14.
- Batie SS, Healy RG. Future of American agriculture as a strategic resource (1980).
- Casagrande R. Biological terrorism targeted at agriculture: The threat to US national security. Nonproliferation Review 7 (2000): 92-105.
- Horn FP, Breeze RG. Agriculture and Food Security. Annals of the New York Academy of Sciences 894 (1999): 9-17.
- Winters LA. Digging for Victory: Agricultural Policy and National Security. The World Economy 13 (1990): 170-191.






 Impact Factor: * 4.1
Impact Factor: * 4.1 Acceptance Rate: 75.32%
Acceptance Rate: 75.32%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 