Biological Evaluation and Anticancer Activity of Novel Binary Transition Metal Complexes Derived from Ortho Amino Benzo Hydroxamic Acid
D. Anitha Ranga Sree
Department of Chemistry, University College of Science, Osmania University, Hyderabad, Telangana, India
*Corresponding author: D. Anitha Ranga Sree, Department of Chemistry, University College of Science, Osmania University, Hyderabad, Telangana, India.
Received: 18 August 2025; Accepted: 22 August 2025; Published: 25 September 2025
Article Information
Citation: D. Anitha Ranga Sree. Biological Evaluation and Anticancer Activity of Novel Binary Transition Metal Complexes Derived from Ortho Amino Benzo Hydroxamic Acid. Journal of Pharmacy and Pharmacology Research. 9 (2025): 114-121.
View / Download Pdf Share at FacebookAbstract
Cobalt, Nickel, Copper, Zinc acetate salts are reacted with Ortho-amino benzo hydroxamic acid (OABH) in ethanol generate tetra co-ordinate complexes with the formula [M (II) (OABH) (H2O)] called as binary metal complex. Characterization of binary metal complexes are further calculated by IR, UV-VIS, ESR, microanalysis, molar conductance, magnetic susceptibility, mass spectra, and thermogravimetric analysis (TGA).M (II) being co-ordinate by tri- dentate OABH ligand and complexes are found to be mononuclear where it is checked by experimental data through enolic oxygen, amino, and the azomethine nitrogen atoms. Spectral data of UV-Vis and magnetic susceptibility have recommended the novel metal complexes show square planar and tetrahedral geometry. Complexes are tested for their abilities in biological activity. Antimicrobial tests of the ligand and novel binary complexes have been carried out. The outcome was evaluated with some well-known antibiotics. Synthesized metal complexes are appraised for the anticancer activity in cancer cells of A549 (lung cancer, CCL-185), SKNSH (Human neuroblastoma cancer, CCL-131), and Hela (cervical cancer, CCL-2) gives much more promising candidates to aim further structure activity-based studies.
Keywords
<p>Binary metal complexes, spectral, ortho-amino benzo hydroxamic acid, antimicrobial, anticancer activity</p>
Article Details
Introduction
Hydroxamic acids, a group of naturally occurring and synthetic weak organic acids, are widespread in the plant tissues, in metabolites of bacteria and fungi. The derivatives of hydroxamic acids have a wide variety of major important roles in biology and medicine. The chemistry and biochemistry of hydroxamic acids and their derivatives comprise considerable attention, due to their pharmacological, toxicological, and pathological properties [1]. Hydroxamic acids generally have low toxicities and have a range of activity in different types of biological systems as they act as growth factors, food additives, tumor inhibitors, anti-inflammatory, anti-asthmatic anti-leukemic, antituberculosis, antimicrobial agents [2], the pharmacophore of the moiety play important chemotherapeutic agents, advanced in human clinical trials, pharmaceutical drugs, pigments, and cell division factors. Hydroxamic acids have been found to react with both proteins and nucleic acids [3]. Hydroxamic acids are strong metal ion chelates [4] having one or more –CONHOH groups show a great number of applications in analytical chemistry [5,6]. Solid- phase synthesis has become an important tool, and there have been several reports describing syntheses of hydroxamic acid derivatives [7]. Hydroxamic acids are good ligands for metal ions and are used as indicators for detecting metal ions [8]. Hydroxamic acids have potentially able to form Chelated complexes. Complexes show better antimicrobial activities than the normal ligand [9, 10]. In therapeutic Chemistry, cancer is considered as one of the health concerns aspects to provide confronting the human race. To discover drugs incurring cancer is seems to be a most developing area in the pharmaceutical research field. The popularly known metal-based drug Cisplatin is used to cure cancer. Due to its resistant nature and prone to side effects such as failure of kidney, liver, and nausea is the main cause of toxicity with heavy metals. So much potential, fewer side effects, and low toxicity have to develop to cure cancer [37, 38, 39, and 40]. To improve activity promoted us to synthesis a new series of metal-containing hydroxamate ligand. The present study reports the preparation, characterization, antimicrobial and anticancer studies of binary complexes obtained from ortho-amino benzo hydroxamic acid and transition metal ions of Co (II), Ni (II), Cu (II), and Zn (II). The components present in the complexes are elucidated using elemental analysis, magnetic moment, mass, IR, 1H NMR, ESR, and molar conductance. The synthesized binary complexes were monitor further - antibacterial, minimum bacterial concentration, anti-fungal, with minimum fungicidal concentration. Compounds were screened for in-vitro against 4 cell lines using MTT assay. All compounds showed activity against four cell lines at micromolar concentration. The biological and anticancer activity of the ligand ortho-amino benzo hydroxamic acid and their metal complexes are reported.
Experimental
Materials
Anthranilic acid, 2-aminophenol, NaOH, ethyl alcohol, Co(OAc)24H20, Ni(OAc)24H20, Cu(OAc)2H20, Zn(OAc)22H20, 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyl-tetrazolium bromide, phosphate Buffer Saline (PBS) were supplied from Aldrich, and Sigma Chemicals. All analytical grade (AR)solvents are used.
Instruments
- • Elemental Analyses were performed on a Perkin-Elmer 240C (USA) elemental analyzer. Molar conductance of the metal complexes was measured in DMSO solution using a Digisun meter.
- • Magnetic susceptibilities of the complexes were measured on Guoy balance (Model 7550) using Hg [Co (NCS)4] as standard. The diamagnetic corrections of the complexes were made using Pascal’s constant.
- • The melting point of the binary complexes was determined on the Polmon instrument (model MP-96).
- • Electronic spectra were recorded on an Elico SL159 UV Visible spectrophotomer from 200-1100nm.
- • IR spectra were recorded in KBr discs on an FTIR spectrophotometer from 400 to 4000cm-1.
- • The mass spectra were recorded by ESI technique positive mode on BRUKER MAXIS High-Resolution Mass Spectrometer.
- • Thermogravimetric analysis (TGA) was carried out using a Shimadzu DT-50 thermal analyzer under a nitrogen atmosphere with a heating rate of 10oC/min.
- • Electron spin resonance (ESR) measurements of solid complexes were recorded at room temperature on Bruker EPR spectrometer at 9.706GHz(X-band), the microwave power was (1.0mW) with 4.0 G modulation amplitude, using 2, 2-diphenyl pyridyl hydrazone (DPPH) as standard (g=2.0037).
General procedure for the synthesis of Ortho-amino hydroxamic acid, binary metal complexes:
Synthesis of ligand (OABH)
Ortho-amino benzo hydroxamic acid (OABH) was prepared as described to a solution containing 5gm (0.03mole) of sodium hydroxide in 150ml of water, a solution of 7gm (0.1mole) of o-amino phenol in 100ml of absolute methanol was added and then 15.1gm (0.1mole) of anthranilic acid dissolved in methanol was added to the filtrate. The resulting solution was heated on a steam bath for 20min upon cooling the ppt of sodium –O- amino benzo hydroxamic acid was obtained which was filtered dissolved in a minimum quantity of water and the solution acidified to pH4 with acetic acid or HCl. The precipitate formed was recrystallized from ethanol and with TLC the purity of the compound is verified.
Preparation of binary Complexes
Ethanolic solution (20ml) of the ortho amino benzo hydroxamic acid 0.4gm (1mmol) and the metal salt of (1mmol) in ethanol (20ml) both solutions have to combine in a beaker heat the solution on a hot plate at 60oC for 6-12h with stirring. Quantity of the solution has to come down to one-half by evaporating, after 1 day the solid leftover was filtered and washed with hot petroleum ether(40-60oC), cold ethanol, and dried over anhydrous CaCl2 (under vacuum). The below-mentioned metal complexes were synthesized the binary complexes are formed with a ratio of 1:1(metal: ligand).
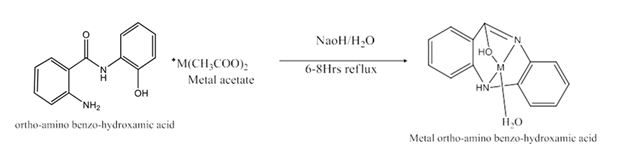
Synthesized route of Ortho-amino benzo hydroxamic acid (OABH) of metal complexes
Determination of metal content of the binary complexes
The metal content of the complexes was determined by the analytical methods described [11, 12, 13].
Screening for antimicrobial activity
In vitro studies: antimicrobial activity of Ortho-amino benzo hydroxamic acid (OABH) ligand, binary metal complexes towards the bacteria: Bacillus subtilis and Staphylococcus aureus, (gram +ve) Escherichia coli and Pseudomonas aeruginosa (gram-ve) in Agar well diffusion technique (Linday,1962) and fungi: Colletotricum capsici, Alternaria solani, in Agar well diffusion technique (Linday,1962). The antibacterial and antifungal activities are performed at 1mg/1ml concentrations in DMSO solvent. Tested samples are evaluated for Antimicrobial activity. As a general procedure,100µl of the test bacteria /fungi were grown in 10ml of fresh media until they reached an account of approximately 108 cells/ml and 105cells/ml for bacteria and fungi, respectively [14]. The microbial suspension was spread onto agar plates corresponding to the broth in which they were maintained. The bacterial strains: B.subtilis, S.aureus, E.coli, and P. aeruginosa were incubated for 24h at 37oC, and Fungi Strains: C. capsici, A. sonali, was incubated at 25oC to 30oC for 48h respectively then the diameters of the inhibition zones were measured in millimeters. Blank paper discs (Schleicher. and Schuell, Spain) with a diameter of 8.0mm were impregnated 10 µl of tested concentration of the stock solutions. Standard antibacterial drugs (Tetracycline) and an antifungal drug (Amphotericin B) were used as references to evaluate the potency of the tested compounds under the same conditions. The activity was determined by measuring the diameter of the zone showing complete inhibition (mm).10 µl of solvent (DMSO) was used as a negative control. Finally, the activity results are calculated as a mean of triplicates.
In vitro cytotoxicity evaluation
The cytotoxicity of the prepared binary metal complexes and the free ligand was based on in vitro growth of tumor cell lines in 96 well-cultured plates was kept for 24hr later 10µl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl- tetrazolium bromide (MTT,5mg/ml) was added to the wells. Again plates were incubated at 37oC for 4h. Culture media was discarded and wells were washed with phosphate Buffer Saline. To each well, 150 µl of DMSO was added and incubated for 30min. The cell-mediated reduction of tetrazolium salt to water-insoluble formazan crystals using doxorubicin as a standard control. Colour intensity was measured at 540nm in ELX800 Universal Microplate Reader [35] the cytotoxicity was assessed against a panel of four different cell lines.A549 derived from human lung cancer cell line (ATCC No.CCL-185), SKNSH derived from human neuroblastoma cancer cell line (ATCC No.CCL-131), Hela- derived from human cervical cancer line (ATCC No.CCL-2) and HEK – embryonic kidney (normal cell line) using MTT assay [36].
Results and Discussion
Ortho-amino benzo hydroxamic acid (OABH) and its binary metal complexes were synthesized and characterized by elemental analysis, molar conductance, magnetic susceptibility, IR, UV-Vis, ESR, mass spectra, and thermogravimetric analysis (TGA). All the metal complexes are colored, stable towards the air, insoluble in water- soluble in DMSO. The melting points of the complexes were higher than the ligand, revealing that metal complexes are much more stable than the ligand. The molar conductivity (Λm) of 1*104M solutions of the complexes in DMSO falls in the range 0.0-4.0 Ω-1mol-1cm2.these low values indicated that all of the complexes have non –electrolytic nature [15] Elemental data and some physical properties of (OABH) and its reported complexes are listed in table 1.
Table 1: Elementary analysis and physical properties of Ligand, and binary complexes.
|
Compound |
Color |
MP (°C) |
M.Wt |
Yield (%) |
C (%) |
N (%) |
H (%) |
M (%) |
Λm (Ω-1mol-1cm2) |
|
(OABH) (L1) |
Violet/ |
140 |
228 |
97 |
68.41 |
12.27 |
5.3 |
– |
– |
|
[CoL1(H2O)] |
Brown |
>300 |
287.18 |
90 |
54.38 |
9.75 |
4.21 |
20.5 |
1 |
|
[NiL1(H2O)] |
Brown |
>300 |
288.25 |
77 |
54.42 |
9.89 |
4.28 |
20.45 |
0.9 |
|
[CuL1(H2O)] |
Brown |
>300 |
291.79 |
86 |
53.51 |
9.6 |
4.15 |
21.63 |
2 |
|
[ZnL1(H2O)] |
Brown |
>300 |
293.66 |
84 |
53.2 |
9.54 |
4.12 |
22.27 |
0 |
FT-IR spectra
The coordination mode and sites of the ligand to the metal ions have been investigated by comparing the infrared spectra of the free ligands with binary metal complexes.
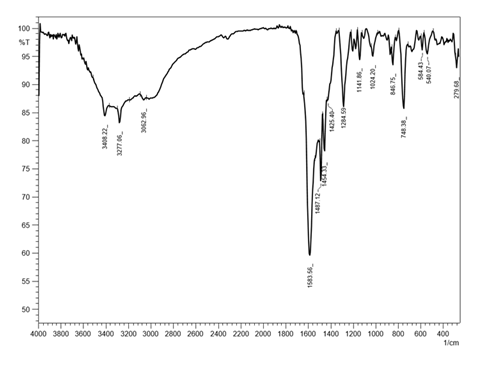
The characteristic IR data of the Ortho-amino benzo hydroxamic acid (OABH) and their metal complexes are listed below in Table 2. Infrared (cm-1. KBr of L1, binary complexes)
Table2: The characteristic IR data of the Ortho-amino benzo hydroxamic acid (OABH) and their metal complexes
|
Compound |
ν(OH) (cm-1) |
ν(OH) in-plane bending (cm-1) |
ν(C=N) (cm-1) |
ν(C–O) (cm-1) |
ν(M–N) (cm-1) |
ν(M–O) (cm-1) |
|
L1 |
3315 (w, br) |
1364 (m) |
1589 (s) |
1232 (m) |
– |
– |
|
[CoL1(H2O)] |
3408 (s, br) |
1452 (m) |
1583 (s) |
1284 (m) |
540 (w) |
420 (w) |
|
[NiL1(H2O)] |
3410 (s, br) |
1425 (m) |
1581 (s) |
1271 (m) |
538 (w) |
422 (w) |
|
[CuL1(H2O)] |
3410 (m, br) |
1423 (m) |
1585 (s) |
1292 (m) |
536 (w) |
426 (w) |
|
[ZnL1(H2O)] |
3414 (s, br) |
1429 (m) |
1579 (s) |
1290 (m) |
530 (w) |
427 (w) |
Note: (cm-1. KBr of L1, binary complexes)
In IR data binding nature of the ligand OABH to metal interaction in complex formation and its studies are conferred. The nature of bonding in the complexes and the structural characteristics are discussed. Metal ions are coordinated with OABH through azomethine and phenolic groups. The free ligand at L1 3315cm-1 was assigned to the ν (OH). The broadness of the band was attributed to the presence of the internal hydrogen bond OH…N=C [16]. IR data of binary, OABH showed a broadband 3408- 3414cm-1 assign to ν (OH) of the coordinated water molecule connected with the complexes. An appearance of a narrow intense band in binary complexes was seen at 748-758cm-1 is specific for the wagging mode of the co-ordinate water molecule [17]. The band at the 1589cm-1 characteristic of ν(C=N) in the free ligand, was shifted to the lower frequency region 1581cm-1 after co- ordination between OABH ligand and metal complexes [18]. The band at 1232cm-1 is ascribed to phenol C-O stretching vibration in the case of OABH [19] more signals were observed for metal complexes than that of the ligand. The nonligand bands at 530-540cm-1 and 420-427cm-1 were observed due to M-N and M-O, which gave evidence for bonding with azomethine nitrogen and phenol oxygen of the OABH metal ion [20]. From the above data with elemental analysis, it is concluded the binary complex formed with OABH ligand as tridentate ligand with ONO donor sites.
The IR spectra of the OABH ligand and binary Co (II) complex FTIR [CoL1(H2o)]
UV-VIS spectra and magnetic susceptibility are shown in Table.3
The geometry of the metal complexes is obtained from electronic spectra and magnetic measurements. The spectra of Ortho-amino benzo hydroxamic acid (OABH) binary metal complexes were recorded in 1*10-4M DMSO solution in the region 200-800nm. Ortho-amino benzo hydroxamic acid (OABH) L1 spectrum is at high-intensity absorption band 36,363 and 23,255 cm-1were assigned to intra ligand π-π*, n-π* transitions [21]. The electronic spectra of the binary metal complex data are attributed as in Co(II)band showed 22,779cm-1infers π-π* a ligand to metal transfer 26,666 shows n-π* 38,759cm-1 assigned to the intra ligand and 18691 cm-1 1A1g→1B1g transitions being characteristic as square-planar geometry [23]. Magnetic susceptibility is (3.87BM). Ni(II) displaying the strong absorption band at 22,883cm-1ascribed as a ligand to metal charge transfer (LMCT) π-π*, n-π* 38,910,27,027cm-1 showed high intense absorption band which is assignable to the intra ligand and 18,450 cm-1 shows spin allowed transitions3T1 (F)→3T1(P) characteristic as tetrahedral geometry [24]. Magnetic susceptibility is (2.97BM) [22]. Cu (II) the ligand exhibit the band at 38,759cm-1 can be assigned to intra ligand π-π*transition and exhibit redshift, the electronic spectral band at 22935cm-1 conveyable as a ligand to metal charge band and19230 cm-1 attributable to 2B1g→2A1g transition. Magnetic susceptibility of Cu (II) complex is (1.94BM) indicates an unpaired electron proposed to be as square-planar geometry. In Zn (II) complex is d10 configuration present intense band at 41, 666, 34, 480, 26,737 cm-1showed π-π*, n-π*, transitions ascribed as a ligand to metal charge (LMCT) transitions consign geometry is tetrahedral. Magnetic susceptibility is (0.0BM) diamagnetic property of zinc complex [25].
Table 3: UV-Visible spectra and magnetic susceptibility of (OABH) and its metal complexes
|
Compound |
UV-Vis (cm-1, DMSO) |
Assignments |
µeff (BM) |
|
L1 |
36,363; 23,255 |
π–π*, n–π* |
– |
|
[CoL1(H2O)] |
38,759; 26,666; 22,779; 18,691 |
π–π*, n–π*, ¹A1g → ¹B1g |
3.87 |
|
[NiL1(H2O)] |
38,910; 27,027; 22,883; 18,450 |
π–π*, n–π*, ³T1(F) → ³T1(P) |
2.97 |
|
[CuL1(H2O)] |
38,759; 22,935; 19,230 |
π–π*, n–π*, ²B1g → ²A1g |
1.94 |
|
[ZnL1(H2O)] |
41,666; 34,482; 26,737 |
π–π*, n–π*, CT |
0 |
ESR spectra
The ESR spectrum of binary Cu (II) complex recorded in the solid-state was assigned as paramagnetic possess square planar geometry was bound Cu (II) metal in the center of the complex [26]. The “g” tensor values of the Cu (II) complexes could be used to find the ground state [27]. Unpaired electron occupies the dx2-y2 orbital with 2B1g ground state resulting in g ║ >g┴. The values for g ║ is 2.10 and g┴ is 2.02 and g aver is 1/3(g┴ + 2 g ║) = 2.07. The observed ‘g’ values g ║ > g┴ > ge (2.0023) predicted that unpaired electron lies in the dx2-y2 orbital. A covalent environment g ║ is less than 2.3 but an ionic environment g ║ greater than and equal to 2.3 [28]. The Cu (II) complex value reported is less than 2.3 confirmed the ligand metal bond is covalent.
Thermogravimetric and mass spectra analysis
Table 4: Molecular weight of the metal complexes
|
Complex |
Molar Mass |
m/z values (relative intensity) |
|
[CoL1(H2O)] |
287.18 |
287a (80%), 269b (62%), 95c (71%) |
|
[NiL1(H2O)] |
288.25 |
288a (92%), 270b (65%), 96c (67%) |
|
[CuL1(H2O)] |
291.79 |
292a (84%), 274b (64%), 100c (63%) |
|
[ZnL1(H2O)] |
293.66 |
293a (88%), 275b (74%), 101c (81%) |
a = molecular ion peak (K)
b = (K – H2O)
c = (K – C13H10N2O + H2O)
The thermal behavior of metal complexes was investigated and characterized using thermogravimetric analysis (TGA) and differential scanning calorimetric (DSC). The sample was analyzed in a platinum pan under N2 and the temperature was linearly increased at 10o Cmin-1 over a temperature range of 20-900oC. The presence of water molecules is confirmed in synthesized binary metal complexes by thermal analysis. From the elemental analysis of each metal complex, proposed structures were found in agreement with formula weight to the decomposition of mass losses. The decomposition patterns were further confirmed from the mass spectral peaks in Table.4. Metal complexes started decomposing below 120oC indicating the presence of water molecule endothermic peak is observed in the range 340-420oC corresponding to the elimination of co-ordinate water molecule and organic part of the chelate leaving metal oxide at the final temperature. The residual metal oxide is suggested to be impregnated in the thermal analyzed values. The synthesized novel binary metal complexes are thermally stable [29].
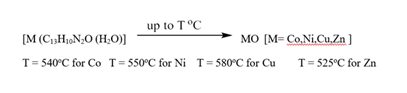
Table 5: Decomposition of metal complexes Thermo Gravimetrically
|
Complex |
Temperature (°C) |
Assignment |
Residue |
|
[CoL1(H2O)] |
540 |
C13H10N2 + H2O |
CoO |
|
[NiL1(H2O)] |
550 |
C13H10N2 + H2O |
NiO |
|
[CuL1(H2O)] |
580 |
C13H10N2 + H2O |
CuO |
|
[ZnL1(H2O)] |
525 |
C13H10N2 + H2O |
ZnO |
Conductivity
The molar conductivity (Λm) of 1*10-4M solutions of the binary metal complexes in DMSO falls in the range 0.0-
4.0 Ω-1mol-1cm2 these low values indicated that all of the complexes have non –electrolytic nature [30].
Antimicrobial activity
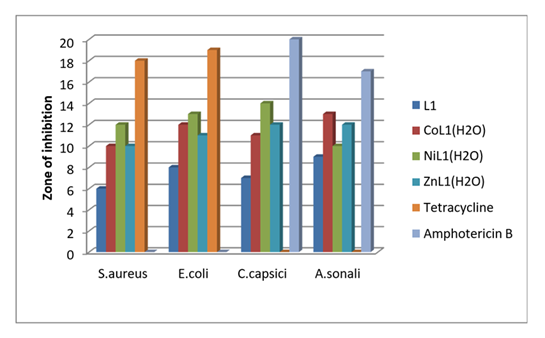
Table 6: Antimicrobial activity of OABH ligand and binary metal complexes
|
Compounds |
Anti-bacterial activity |
Anti-fungal activity |
||
|
S. aureus |
E. coli |
C. capsici |
A. sonali |
|
|
L1 |
6 |
8 |
7 |
9 |
|
[CoL1(H2O)] |
10 |
12 |
11 |
13 |
|
[NiL1(H2O)] |
12 |
13 |
14 |
10 |
|
[CuL1(H2O)] |
14 |
12 |
13 |
11 |
|
[ZnL1(H2O)] |
10 |
11 |
12 |
12 |
|
DMSO |
0 |
0 |
0 |
0 |
|
Tetracycline |
18 |
19 |
– |
– |
|
Amphotericin B |
– |
– |
20 |
17 |
The OABH, binary metal complexes were tested for their inhibitory effects on the growth of bacteria: S.aureus(gram +ve), E. coli (gram –ve), and fungi: C.capsici and A.sonali because such organisms can attain resistance to antibiotics through biochemical and morphological modification [31]. The susceptibility of the ligand and their metal complexes were determined by measuring the size of the inhibition diameter. The concentration for these samples was maintained as 1mg ml-1 in DMSO. The results thus obtained (table 6) and fig .2 are explained by chelation theory [32]. The mode of action of the compounds may involve the formation of a hydrogen bond through the azomethine group with the active centers of cell constituents, resulting in interference with the normal cell. [33]. The variation in the activity of different complexes against different organisms depends either on the impermeability of the cells of the microbes or the difference in the ribosome of microbial cells. A comparison of the biological activity of the synthesized compounds with known antibiotics, Tetracycline and Amphotericin B. The results have been indicated that S.aureus(gram +ve), E.coli (gram –ve) C.capsici and A.sonali, OABH (L1) show less activity whereas in binary complexes[CuL1(H2O)] shown good activity [CoL1(H2O)] [NiL1(H2O)] [ZnL1(H2O)] are shown moderate activity towards bacterial and fungal strains when compared with standard Tetracycline and Amphotericin B. According to chelation theory, the polar nature of the metal ion got decreased because overlapping of ligand orbital and contributing the charge of a metal ion with donor groups upon then increase π- electrons delocalization over complete chelate ring increases which makes the way to enter the ternary metal compounds penetrate in the membrane of lipid which stops the binding sites of metal to the enzyme of microbes. These complexes interrupt the process of respiration of the cell and drop the synthesis of proteins leads to the arrest of further growth of the organisms [34].
Table 7: Cytotoxicity results shown for binary metal complexes and OABH
|
IC50 values in (µg/ml) |
||||
|
A549 |
SKNSH |
Hela |
HEK |
|
|
L1 |
25.6 |
20.2 |
19.4 |
68.9 |
|
[CoL1(H2O)] |
12.1 |
12.7 |
10.8 |
NA |
|
[NiL1(H2O)] |
NA |
85 |
NA |
120 |
|
[CuL1(H2O)] |
18.2 |
67 |
20.5 |
143 |
|
[ZnL1(H2O)] |
NA |
NA |
NA |
190 |
Doxorubicin (standard control)
NA-not active
A549 derived from human lung cancer cell line (ATCC No.CCL-185), SKNSH derived from human neuroblastoma cancer cell line (ATCC No.CCL-131), Hela- derived from human cervical cancer line (ATCC No.CCL-2), and HEK – embryonic kidney (normal cell line).
The IC50 values (50% inhibitory concentration) were calculated from the plotted absorbance data for the dose-response curves. IC50 values in (µg/ml) are expressed as the average of two independent experiments. Doxorubicin – IC50 value (µg/ml) is 0.47.
Conclusions
Binary metal (II) complexes of transition series have been synthesized by reaction of metal acetate with tridentate ONO of OABH by reflux. Binary metal complexes of OABH and its metal complexes show square planar and tetrahedral geometry. After subjecting the ligand and metal complexes inhibition zone in mm of both bacteria and fungi is monitored. The binary metal complexes show more reactivity than ligand OABH which suggests that new complexes have potent antimicrobial activity. The results are compared with standard antibiotics. The synthesized binary complexes exhibit anti-cancer activities towards the cytotoxicity was assessed against a panel of four different cell lines.A549 derived from the human lung cancer cell line (ATCC No.CCL-185), SKNSH derived from the human neuroblastoma cancer cell line (ATCC No.CCL-131), Hela- derived from human cervical cancer line (ATCC No.CCL-2) and HEK – embryonic kidney (normal cell line). The results were encouraging to the synthesis of more efficient metal complexes.
Acknowledgment
We thank the Head, Department of Chemistry, for providing facilities to carry out the experiments at UGC Center in the University of Hyderabad, for providing spectral and analytical data. We are thankful to the Head, Department of Botany and Chemistry, University College for Women, Koti, Hyderabad for their kind support to perform biological studies.
References
- Paniago EB, Carvalho S Ciencia e Cultura 40 (1988): 629.
- weber G. Cancer Res 43 (1983): 3466. (b) Miller mJ. Acc Chem Res 19 (1986): 49. (c) Miller MJ. Chem Rev 89 (1989): 1563.
- Niemeyer HM, Pesel E, Copaja SV, et al. Changes in hydroxamicacid levels of wheat plants induced by aphid feeding. Phytochemistry 28 (1989): 447.
- (a) Kurzak B, Kozlowski H, Farkas E. Coord Chem Rev 114 (1992): 169. (b) Boukhris S, Souizi A, Robert A. Tetrahedron Lett 37 (1996): 179.
- Davidson DJJ. Chem Ed 17 (1940): 81.
- Agrawal YK. Rev. Anal. Chem 3 (1980).
- (a) Floyd CD, Lewis CN, Patel SR, Whittaker M. Tetrahedron Lett 37 (1993): 8045. (b) Zhang W, Zhang L, Li X, et al. J Comb Chem 3 (2001): 151.
- Yale H.L. Chem. Rev. 33 (1943): 209.
- Gumus F, Algul O, Eren G, Eroglu H, Diril N, Gur S, Ozkul A. Eur. J. Med. Chem. 38 (2003): 473.
- Kumar Sau D, Batcher RJ, Chandhuri S, Saha N. Mol. Cell. Biochem. 253 (2003): 21.
- Rishi AK, Garg BS, Singh RP. Fresenius J. Anal. Chem. 259 (1972): 125.
- Vogel AI. Textbook of Quantitative Chemical Analysis, 5th ed., Longmans, London, 1998.
- Macarovici CG. Inorganic Quantitative Chemical Analysis. Editura Academiei R.S.R., Bucuresti, 1979.
- Pfaller MA, Burmeister L, Bartlett MA, Rinaldi MG. J. Clin. 26 (1988): 1437.
- Dean JA. Lange’s Book of Chemistry, 4th ed., McGraw-Hill, New York, 1992.
- Nakamoto K. Infrared and Raman Spectra of Inorganic and Coordination Compounds, 5th ed., Wiley-Interscience, New York, 1997.
- Adams DM. Metal Ligand and Related Vibrations. Edward Arnold, London, 1967.
- Hundekar MA, Sen DN. Indian J. Chem. Sec. A 23 (1984): 477.
- Saxena A, Tandon JP. Polyhedron 3 (1984): 681.
- Thomas M, Nair MKM. Synth. Inorg. Met.-Org. Chem. 25 (1995): 471–479.
- Lever ABP. Inorganic Electronic Spectroscopy. Elsevier, New York, 1968.
- Dutta RL, Syamal A. Elements of Magnetochemistry. East West Press, New Delhi, 1992.
- Raman N, Raja SJ, Joseph J, Raja JD. J. Chil. Chem. Soc. 52 (2007): 1138.
- Soliman AA. Spectrochim. Acta A 65 (2006): 1180.
- El-Shahawi MS, Shoair AF. Spectrochim. Acta A 60 (2004): 121.
- Greenwood NN, Earnshaw A. Chemistry of the Elements. Pergamon Press, New York, 1984.
- Hathway BJ, Tomlinson AAG. Coord. Chem. Rev. 5 (1970): 1.
- Kivelson D, Neiman R. J. Chem. Phys. 35 (1961): 149.
- Katsuki T. Coord. Chem. Rev. 140 (1995): 189.
- Dean JA. Lange’s Handbook of Chemistry, 4th ed., McGraw-Hill, New York, 1992.
- Moohmed GG, Zayed MA, Abdallah SM. J. Mol. Struct. 979 (2010): 62–67.
- Lawrence PG, Harold PL, Francis OG. Antibiot. Chemother. 5 (1980): 1597.
- Colombo D, Franchini L. Eur. J. Med. Chem. 40 (2005): 69–74.
- Dharmaraj N, Viswanathamurthi P, Natarajan K. Trans. Met. Chem. 26 (2001): 105.
- Lee M, Rhodes AL, Wyatt MD, Forrow S. J. Am. Chem. Soc. 76 (1954): 3047.
- Mosmann T. Rapid colorimetric assay for cellular growth and survival; application to proliferation and cytotoxicity assays. J. Immunol. Methods 65 (1983): 55–63.
- Boulikas T, Vougiouka M. Oncol. Rep. 10 (2003): 1663.
- Angeles-Boza AM, Bradely PM, Fu PKL, Wicke SE, Bacsa J, Dumbar KM, Turro C. Inorg. 43 (2004): 8510.
- Galanski M, Arion VB, Jakupec MA, Keppler BK. Pharm. Des. 9 (2003): 2078.
- Wang D, Lippard SJ. Nat. Drug Discov. 4 (2005): 307.


 Impact Factor: * 3.3
Impact Factor: * 3.3 Acceptance Rate: 74.39%
Acceptance Rate: 74.39%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 