Cell-Penetrating Peptides and DNA-Free Genome Editing in Plants
Han Wu1, Zhipeng Zhang1,Fulai Ke1, Pengxiang Jia2, Kai Zhu1#, Jianqiu Zou1#
1Sorghum Research Institute, Liaoning Academy of Agricultural Sciences, Shenyang, China
2Zhejiang Wanli University, Ningbo, China
*Corresponding Author: Jianqiu Zou, Kai Zhu, Sorghum Research Institute, Liaoning Academy of Agricultural Sciences, Shenyang, China
Received: 9 July 2021; Accepted: 20 July 2021; Published: 30 July 2021
Article Information
Citation: Han Wu, Zhipeng Zhang, Fulai Ke, Pengxiang Jia, Kai Zhu, Jianqiu Zou. Cell-Penetrating Peptides and DNA-Free Genome Editing in Plants. International Journal of Plant, Animal and Environmental Sciences 11 (2021): 474-484.
View / Download Pdf Share at FacebookAbstract
Genome editing technology based on the clustered regularly interspaced short palindromic repeats/Cas system is an important achievement of molecular biology. Nevertheless, public concerns regarding genetically modified organisms and regulatory restrictions have limited the use of this technology. Cell-penetrating peptides (CPPs) are protein transduction domains, usually less than dozens of amino acids, which have the ability to take cargo and penetrate the cell membrane of recipient cells. CPPs have only minor cytotoxicity and are ideal transfection tools for DNA-free transfection and genome editing in plants. In this review, we discuss the classifications and translocation mechanisms of CPPs, as well as the brief history of CPPs mediated DNA-free genome editing
Keywords
<p>Cell-penetrating peptide; Genome editing; DNA-free; Clustered regularly interspaced short palindromic repeats/Cas system; Genetically modified organism</p>
Article Details
1. Introduction
The third-generation genome editing system represented by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas is becoming the core technology for crop molecular breeding. This technology largely depends on genetic transformation. In theory, if the foreign gene is not eliminated after the editing event, the sgRNA and CRISPR/Cas9 gene in the recipient may continue to function, resulting in nonspecific double-strand breaks and repair. Therefore, off-target effects may be retained [1-7]. In Arabidopsis, the foreign genes can be eliminated, and the mutant genes can be retained in the T2 generation by fast-R tag-assisted screening [8]; however, public concerns regarding the safety of genetically modified organisms (GMOs) and regulatory restrictions have limited the use of CRISPR/Cas technology [9].
DNA-free transfection can be used to avoid the integration of foreign DNA into the genome of recipient cells. With the increased use of DNA-free transfection technology, represented by cell-penetrating peptides (CPPs) and biolistics, it is possible to overcome public concerns and achieve efficient DNA-free site-specific editing of target genes [10, 11].
In this review, we discuss the classifications and translocation mechanisms of CPPs, with an emphasis on their use in DNA-free genome editing technology.
2. CPPs
CPPs are usually less than dozens of amino acids in length and have the ability to carry biomolecules across the cell membrane of recipient cells. As early as 1988, CPPs were identified in the TAT protein from human immunodeficiency virus (HIV)-1 [12]. Subsequently, researchers have found that some short peptides also have the ability to penetrate the cell membrane.
2.1 Classifications
Based on physicochemical properties, CPPs can be categorized into three groups: cationic, hydrophobic, and amphipathic (Table 1). Most CPPs belong to the cationic group and typically contain polyarginine, which results in a positive charge. For example, TAT is a typical cationic CPP encoded in the genome of HIV-1, and its dimer TAT2 is successfully utilized in plant transformation [13]. Additionally, Bilichak et al. inserted R9 and mCherry genes into the same expression frame, and after prokaryotic expression, the fusion protein (R9-cys-mCherry) was successfully transfected into wheat microspores. The amphipathic group, including VT5 and MAP, shows amphipathicity owing to the presence of lysine residues in their sequences [14]. The amphipathic α-helical motif usually has hydrophobic groups on one side, whereas the other side contains cationic and anionic groups [15]. Hydrophobic CPPs from signal peptides contain only apolar residues, such as methionine, valine, or alanine [16]; fibroblast growth factor (FGF) and Pep-7 belong to this group.
|
Group |
Name |
Sequence |
Reference |
|
Cationic |
TAT |
RKKRRQRRR |
[13] |
|
R9 |
RRRRRRRRR |
[25] |
|
|
Amphipathic |
MAP |
KLALKALKALKAALKLA |
[50] |
|
VT5 |
DPKGDPKGVTVTVTVTVTGKGDPKPD |
[51] |
|
|
Hydrophobic |
FGF |
PIEVCMYREP |
[52] |
|
Pep-7 |
SDLWEMMMVSLACQY |
[53] |
2.2 Translocation mechanism
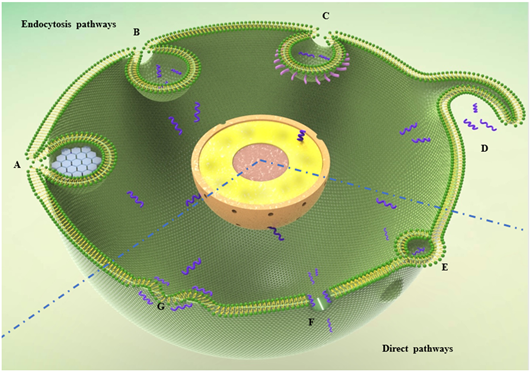
CPPs typically penetrate the cell membrane via direct translocation and endocytosis pathways (Figure 1). In the endocytosis pathway, once CPPs attach to the membrane, clathrin and caveolae are recruited. Finally, the coated CPPs and their cargo are translocated into the cytoplasm. In the clathrin-independent pathway, CPPs undergo macropinocytosis without clathrin or caveolae recruitment [17].
Direct translocation is an energy-independent pathway [18]. Positively charged amino acid residues interact with membrane phospholipids, interfere with membrane stability, and finally permit facilitate membrane penetration. There are three models for penetration of CPPs into the lipid bilayer: 1) inverted micelles [19], 2) the carpet model [20], and 3) pore formation [21]. The inverted micelle model was obtained from nuclear magnetic resonance imaging of penethrin internalization [19]. Cationic residues of CPPs were found to interact with negatively charged phospholipid groups, leading to the production of micelles enclosing the peptide and cargo. Through the hydrophobic motif in CPPs, micelles break apart in the cytoplasm and release their cargo. In the carpet model, cationic residues interact with phospholipids, leading to rotation of the peptide and interaction of the hydrophobic motif with the hydrophobic tails of phospholipids. CPP cargo then penetrates the membrane via breaks [22]. In the pore model, the hydrophobic motif of CPPs create bonds with the lipid, leading to the formation of transmembrane pores by hydrophilic residues [23]. The mechanism of translocation varies depending on the properties of CPPs. The nature and culture conditions of recipient cells can also affect the uptake mechanism. Several translocation modes may occur in a single experiment [15] (Figure 1).
Endocytosis pathways: A) Clathrin-dependent model.The hexagon represents clathrin. B) clathrin or caveolae independent pathway. C) caveolae-mediated model.The pink bar around the caveolae presents caveolin. D) macropinocytosis process; Direct pathways: E) inverted micelles model, whereby CPPs interact with lipid, leading to the formation of inverted micells for up take; F) pore model, where CPPs interact with polar groups of phospholipids and forming a pore for penetrating; G) carpet model, where CPPs transiently destabilize the lipid leading to the reorganization for translocation.The blue short wave lines in this figure represent cell penetrating peptides translocating into a target plant cell via different pathways.
2.3 Cytotoxicity
CPPs show weak cytotoxicity at lower concentrations [14, 16, 24, 25]. In basic research in mammalian cells, CPPs exhibit almost no toxic effects [26]. For example, even when used at a high concentration of 30 μM, antennapedia, TAT, and R9 show no obvious toxic effects [27]. In plant diploid cell transfection, trypan blue and FDA (fluorescein diacetate) staining results also demonstrated that CPPs had only minor effects on the viability of onion epidermal cells and wheat mesophyll cells, respectively [28, 29]. In wheat haploid transfection experiments, CPPs were shown to have weak effects on the viability of microspore cells [13]. Moreover, the toxicity of the fusion protein is related to the length and concentration of CPPs [24]. Taken together, these studies have shown that low concentrations of CPPs do not strongly inhibit recipient cell viability.
3. DNA-free transfection
Transfection refers to the process of introducing exogenous biomacromolecules (e.g., DNA, RNA, or protein) into recipient cells. By contrast, genetic transformation, also known as transgenic technology, refers to the transfer of exogenous DNA into recipient cells and integration into the genome for stable inheritance. However, the use of genetic transformation is limited by public concerns regarding GMOs as well as regulatory restrictions. Therefore, efficient DNA-free (nontransgenic) transfection systems are essential for promoting the application of genome editing in plant breeding.
3.1 Biolistics
Biolistics (also known as microprojectile bombardment) is a direct and physical method to transfer foreign biomacromolecules into recipient cells. This method was first invented by Sanford and colleagues at Cornell University [30]. In this method, microcarriers encapsulated with exogenous biomacromolecules have sufficient kinetic energy to penetrate the cell wall and membrane of recipient cells. Although biolistics is a traditional transgenic method, when exogenous macromolecules encapsulated on microcarriers are proteins or RNA, rather than DNA, biolistics can be considered an important DNA-free transfection method [10, 11]. Because there is no exogenous DNA involved in the entire process, this DNA-free transfection technology may have broad applications. Biolistics can be utilized in various recipient cells that cannot be infected by Agrobacterium. Nevertheless, the physical damage caused by microparticles may also decrease cell viability.
3.2 CPP-mediated DNA-free transfection
CPPs are ideal transfection tools that can be combined with biomacromolecules and carry them into recipient cells [14, 16, 25, 26, 31]. The DNA from CPPs is very short, which may facilitate insertion into expression constructs containing the target gene. Fusion proteins can then be obtained by prokaryotic expression. Bilichak et al. constructed a gene encoding R9 and red fluorescent protein in a single expression construct; after prokaryotic expression, the fusion protein (R9-cys-mCherry) was successfully transfected into wheat microspores. Moreover, the CPP fusion protein was found to be an effective alternative to CRISPR ribonucleoprotein particle (RNP) transfection. In another study, the genome of human HEK293T cells was successfully edited by CPP-mediated DNA-free transfection; researchers expressed a cationic CPP gene (nine consecutive tandem arginines, R9) and the Cas9 gene together and then incubated fusion protein with sgRNA containing R9 synthesized in vitro to form a complex. Finally, with the help of R9, the fusion protein was successfully transfected into human HEK293T cells for DNA-free genome editing. CPP-mediated DNA-free transfection is still being developed [32] and may become an alternative to other transfection technologies.
4. DNA-free genome editing
4.1 Genome editing
Site-specific genome editing is a technology used to modify the DNA sequence of a target organism at the genome level [33]. This approach has been shown to have important applications in gene function analysis [34], genetic improvement [35, 36], and target gene activation [37]. Specific endonucleases, including zinc finger nucleases [38], transcription activator-like effector nucleases [39], and CRISPR/Cas proteins [35, 36, 40], have been successfully used in crop genetic improvement. Genome editing system based on the CRISPR/Cas system is becoming a core technology for crop molecular breeding. Using sgRNA, specific endonucleases can generate DNA double-strand breaks at a designated sequence. The DNA repair mechanism can then facilitate gene base insertion, deletion, or DNA fragment replacement at the target site, leading to site-specific editing of the target gene.
4.2 RNA-guided engineered nuclease (RGEN) RNPs
Currently, the most commonly used DNA-free plant genome editing system is the RGEN RNP editing system. RGEN RNP technology is used to incubate CRISPR/Cas protein (typically Cas9) and gRNA in vitro, leading to the formation of a ribonucleoprotein complex for translocation and then cutting the target DNA in the nuclei. The CRISPR/Cas protein is ultimately degraded by endogenous proteases, thereby realizing DNA-free genome editing [41, 42]. RGEN RNPs were first applied to nematodes and human 293 cells [41, 42] and were subsequently applied to genome editing in Arabidopsis, lettuce, tobacco [43], and wheat microspores [44]. In 2017, Kim et al. successfully applied CRISPR/cpf1 RNPs to edit soybean FAD2 and tobacco AOC genes. The editing efficiency was as high as 11.7%, and no off-target effects were detected [45], representing a major milestone in DNA-free genome editing for crop genetic improvement.
Biolistics have also been used to mediate the DNA-free transfection of RGEN RNPs. Researchers attached RGEN RNPs to a gold carrier and transferred them into maize somatic cells, whereby site-directed mutagenesis was successfully achieved for the maize leafless tongue gene, male fertility gene, and ALS2 gene [10]. In 2017, Liang et al. also achieved biolistics-mediated DNA-free RGEN RNP transfection into bread wheat. Despite these promising results, biolistics may cause unexpected physiological damage to recipient cells, thereby reducing cell viability.
4.3 CPP-mediated DNA-free genome editing
CPPs may be used as an alternative approach for CRISPR RNP intracellular delivery, and recent studies have reported the application of CPP-mediated DNA-free genome editing [32, 44].
Nuclear localization is essential for DNA-free genome editing; indeed, genome editing can only be achieved via translocation of the CPP-cargo into the nucleus [29]. Arginine-rich peptides (e.g., TAT and R9) generally have a nuclear localization function and can interact with nuclear pore complexes to facilitate the transport of the complex into the nucleus. Ramakrishna et al. first obtained human HEK293T gene-edited cells. Prokaryotic expression of a cationic CPP gene (9 arginine, 9R) and Cas9 gene was performed, and the purified fusion protein was then incubated with 9R-sgRNA in vitro. Finally, HEK293T cells were successfully transfected and edited via CPPs. Bilichak et al. also attempted to transfect wheat microspores with a CPP-mCherry fusion protein and eventually edit the genome by delivering ZNF protein [25, 44]. Compared with other chemical transfection methods, CPPs have low cytotoxicity, making them suitable for use in plant experiments (16, 25, 26, 44, 46-48). Accordingly, CPP-mediated transfection may become an important approach for DNA-free genome editing [32].
5. Perspectives
CPP-mediated transformation and transfection have broad potential applications. First, in addition to DNA-free genome editing, CPPs and embryogenesis-related transcription factor fusion proteins, such as CPP-BBM and CPP-WUS, are expected to improve cell proliferation rates, overcoming the bottleneck of genotype dependency, and resulting in a generic regeneration system for genome editing. Moreover, the CPP-dCas9 fusion protein [49] can transiently increase target gene expression levels without causing double strand breaks. Overall, this novel approach for analyzing gene function at the RNA level can complement gene knockout and transformation experiments. Thus, the development of CPPs and DNA-free genome editing techniques are major milestones in molecular breeding and experimental botany.
References
- Fu Y, Foden JA, Khayter C, et al. High-frequency off-target mutagenesis induced by CRISPR/cas nucleases inhuman cells. Nature Biotechnology 31 (2013): 822-826.
- Pattanayak V, Lin S, Guilinger JP, et al. High-throughput profiling of off-target DNA cleavage revealsRNA-programmed Cas9 nuclease specificity. Nature Biotechnology 31 (2013): 839-843.
- Cradick TJ, Fine EJ, Antico CJ, et al. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Research 41 (2013): 9584-9592.
- Lin Y, Cradick TJ, Brown MT, et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNAsequences. Nucleic Acids Research, 42 (2014): 7473-7485.
- Cho SW, Kim S, Kim Y, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Research 24 (2014):132-141.
- Hsu PD, Lander ES, Zhang F. Development, and applications of CRISPR-Cas9 for genome engineering. Cell 157 (2014): 1262-1278.
- Tsai SQ, Zheng Z, Nguyen NT, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nature Biotechnology 33 (2015):187-197.
- Castel B, Tomlinson L, Locci F, et al. Optimization of T-DNA architecture for Cas9-mediated mutagenesis in Arabidopsis. PLOS ONE 14 (2019): e0204778.
- Waltz E. CRISPR-edited crops free to enter market, skip regulation. Nature Biotechnology 34 (2016): 582.
- Svitashev S, Schwartz C, Lenderts B, et al. Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nature Communications 7 (2016): 13274.
- Liang Z, Chen KL, Li TD, et al. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nature Communications 8 (2017): 14261.
- Frankel AD, Chen L, Cotter RJ. Cellular uptake of the TAT protein from human immunodeficiency virus. Proceedings of the National Academy of Sciences. PNAS 85 (1988): 6297-6300.
- Chugh A, Amundsen E, Eudes F. Translocation of cell penetrating peptides and delivery of their cargoes in triticale microspores. Plant Cell Report 28 (2009): 801-810.
- Huang YW, Lee HJ. Cell-penetrating peptides for medical theranostics and targeted drug delivery. Peptide Applications in Biomedicine, Biotechnology and Bioengineering (2018): 359-370.
- Guo Z, Peng H, Kang J, et al. Cell-penetrating peptides: possible transduction mechanisms and therapeutic applications (review). Biomed Rep 4 (2016): 528-534.
- Derakhshankhah H, Jafari S. Cell penetrating peptides: A concise review with emphasis on biomedical applications. Biomedicine & Pharmacotherapy 108 (2018): 1090-1096.
- Mäger I, Langel K, Lehto T, et al. The role of endocytosis on the uptake kinetics of luciferin-conjugated cell-penetrating peptides. BBA - Biomembranes 1818 (2012): 502-511.
- Bode SA, M Thévenin, Bechara C, et al. Self-assembling mini cell-penetrating peptides enter by both direct translocation and glycosaminogly can-dependent endocytosis. Chemical Communications 48 (2012): 7179-7181.
- Derossi D, Chassaing G, Prochiantz A. Trojan peptides: the penetratin system for intracellular delivery. Trends in Cell Biology 8 (1998): 84-87.
- Pouny Y, Rapaport D, Mor A, et al. Interaction of antimicrobial dermaseptin and its fluorescently labeled analogues with phospholipid membranes. Biochemistry 31 (1992): 12416-12423.
- Herce HD, Garcia AE: Molecular dynamics simulations suggest a mechanism for translocation of the HIV-1 TAT peptide across lipid membranes. Proc Natl Acad Sci USA 104 (2007): 20805-20810.
- Matsuzaki K, Sugishita K, Miyajima K. Interactions of an antimicrobial peptide, magainin 2, with lipopolysaccharide-containing liposomes as a model for outer membranes of gram-negative bacteria. Febs Letters 449 (1999): 221-224.
- Deshayes S, Plénat T, Aldrian-Herrada G, et al. Primary amphipathic cell-penetrating peptides: structural requirements and interactions with model membranes. Biochemistry (2004).
- Cardozo AK, Buchillier V, Mathieu M, et al. Cell-permeable peptides induce dose- and length-dependent cytotoxic effects. Biochimica et Biophysica Acta 1768 (2007): 2222-2234.
- Bilichak A, Luu J, Eudes F. Intracellular delivery of fluorescent protein into viable wheat microspores using cationic peptides. Frontiers in Plant Science 6 (2015): 666.
- Chugh A, Eudes F, Shim YS. Cell-penetrating peptides: nanocarrier for macromolecule delivery in living cells. IUBMB Life 62 (2010): 183-193.
- Jones SW, Christison R, Bundell K, et al. Characterization of cell-penetrating peptide-mediated peptide delivery. British Journal of Pharmacology 145 (2005): 1093-1102.
- Chang M, Chou JC, and Lee HJ. Cellular internalization of fluorescent proteins via arginine-rich intracellular delivery peptide in plant cells. Plant Cell Physiol 46 (2005): 482-488.
- Chugh A, Eudes F. Translocation and nuclear accumulation of monomer and dimmer of HIV-1 Tat basic domain in triticale mesophyll protoplasts. Biochimica et Biophysica Acta 1768 (2007): 419-426.
- Sanford JC. The development of the biolistic process. In Vitro Cellular and Developmental Biology-Plant 36 (2000): 303-308.
- Ziemienowicz A, Shim YS, Matsuoka A, et al. A novel method of transgene delivery into triticale plants using the Agrobacterium T-DNA-derived nano-complex. Plant Physiology 158 (2012): 1503-1513.
- Ran YD, Liang Z, Gao CX. Current and future editing reagent delivery systems for plant genome editing Science China Life Sciences 60 (2017): 490-505.
- Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339 (2013): 819-823.
- Miao CB, Xiao K, Hua CS, et al. Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proceedings of the National Academy of Sciences. PNAS 155 (2018): 6058-6063.
- Jiang W, Zhou H, Bi H, et al. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum, and rice. Nucleic Acids Research 41(2013): 1-12.
- Yuan M, Zhu J, Gong LM, et al. Mutagenesis of FAD2 genes in peanut with CRISPR/Cas9 based gene editing. BMC Biotechnology 19 (2019): 24.
- Konermann S, Brigham MD, Trevino AE, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517 (2015): 583-588.
- Bibikova M, Golic M, Golic KG, et al. Targeted chromosomal cleavage, and mutagenesis in Drosophila using zincfinger nucleases. Genetics 161 (2002): 1169-1175.
- Li T, Liu B, Spalding MH, et al. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nature Biotechnology 30 (2012): 390-392.
- Endo A, Masafumi M, Kaya H, et al. Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Scientific Reports 6 (2016): 38169.
- Cho SW, Lee J, Carroll D, et al. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics 195 (2013): 1177-1180.
- Kim S, Kim D, Cho SW, et al. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Research 24 (2014): 1012-1019.
- Woo JW, Kim J, Kwon SI, et al. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nature Biotechnology 33 (2015): 1162-1164.
- Bilichak A, Sastry-Dent L, Sriram S, et al. Genome editing in wheat microspores and haploid embryos mediated by delivery of ZFN proteins and cell-penetrating peptide complexes. Plant Biotechnology Journal 18 (2020).
- Kim S, Jeong H, Kim EY. Genomic and transcriptomic landscape of Escherichia coli BL21(DE3). Nucleic Acids Research 45 (2017): 5285-5293.
- Huang YW, Lee HJ, Tolliver LM, et al. Delivery of nucleic acids and nanomaterials by cell-penetrating peptides: opportunities and challenges. BioMed Research International (2015): 1-16.
- Alizadeh S, Irani S, Bolhassani A, et al. HR9: an important cell penetrating peptide for delivery of HCV NS3 DNA into HEK-293T cells. Avicenna Journal of Medical Biotechnology 12 (2020).
- Geng J, Guo X, Wang L, et al. Intracellular delivery of DNA and protein by a novel cell-permeable peptide derived from dot1l. Biomolecules 10 (2020).
- Cheng AW, Wang H, Yang H, et al. Multiplexed activation of endogenous genes by CRISPR-on an RNA-guided transcriptional activator system. Cell research 23 (2013): 1163-1171.
- Oehlke J, Scheller A, Wiesner B, et al. Cellular uptake of an alpha-helical amphipathic model peptide with the potential to deliver polar compounds into the cell interior nonendocytically. Biochim. Biophys Acta 1414 (1998): 127-139.
- Magzoub M, Sandgren S, Lundberg P, et al. N-terminal peptides from unprocessed prion proteins enter cells by macropinocytosis. Biochem Biophys Res Commun 348 (2006): 379-385.
- Nakayama F, Yasuda T, Umeda S, et al. Fibroblast growth factor 12 (FGF12) translocation into intestinal epithelial cells is dependent on a novel cell penetrating peptide domain: Involvement of internalization in the in vivo role of exogenous FGF12. J Biol Chem 286 (2011): 25823-25834.
- Gao C, Mao S, Ditzel HJ, et al. A cell penetrating peptide from a novel pVII-pIX Phage displayed random peptide library. Bioorg Med Chem 10 (2002): 4057-4065.


 Impact Factor: * 4.1
Impact Factor: * 4.1 Acceptance Rate: 75.32%
Acceptance Rate: 75.32%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 