Evaluating Genetic Diversity of mtCOI Bombus haemorrhoidalis from Different Regions of Western Himalaya
Poonam Kumari and Mahender Singh Thakur*
Department of Biosciences, Himachal Pradesh University, Summerhill, Shimla- 171005, India
*Corresponding Author: Mahender Singh Thakur, Department of Biosciences, Himachal Pradesh University, Summerhill, Shimla- 171005, India
Received: 01 October 2023; Accepted: 09 October 2023; Published: 16 October 2023
Article Information
Citation: Kumari P, Thakur MS. Evaluating Genetic Diversity of mtCOI Bombus haemorrhoidalis from Different Regions of Western Himalaya. International Journal of Plant, Animal and Environmental Sciences.13 (2023): 60-67.
View / Download Pdf Share at FacebookAbstract
Bumblebees are a diverse group of crucial pollinators for agricultural food production and natural ecosystems worldwide. They are interesting insect pollinators to explore social evolution, behavior and ecology because they have both eusocial and solitary life-cycle stages as well as some social parasite species. Numerous reports of species declines cite the interrelated causes of pathogen spread, habitat loss, pesticide use and global temperature change. Our reliance on a small number of thoroughly researched species for agricultural pollination is particularly hazardous due to these threats to bumblebee diversity. Sanger sequencing was used to analyze the genetic diversity of mtCOI Bombus haemorrhoidalis, which was collected on Punica granatum (wild pomegranate) from seven different Western Himalayan regions. Omega revealed a total of 6 variable sites in the alignment between all the mtCO1 sequences of Bombus haemorrhoidalis. The estimated transition/transversion (Ts/Tv) bias of COI (R) is 1.67. The variation in nucleotide content almost completely exist in the third codon position due to AT rich region, as compared to the first and second position of codons. The transition/transversion (Ts/ Tv) ratio is significant in deducing the magnitude and direction of natural selection. Our study reveals clearly signifies the vast difference among the sampled species proves that there is genetic diversity between the samples collected from different areas of Western Himalaya.
Keywords
Genetic diversity of mtCOI Bombus haemorrhoidalis; Western Himalaya
Genetic diversity of mtCOI Bombus haemorrhoidalis articles; Western Himalaya articles
Article Details
1. Introduction
Wild pomegranate (Punica granatum L.) is one of the most important medicinal wild fruit crop and it is highly pollinated by wild insect pollinators viz., bees, bumblebees, beetles, flies etc. Bumblebees (Hymenoptera: Apidae) are pollinators of many wild as well as agricultural plants. They have high thermoregulatory behavior. They are the most reliable and effective pollinators due to their quick pollination, ability to burst the pollen sac with wing vibrations and capacity to forage in low ambient temperatures and light. Therefore, they act as important pollinators, primarily in alpine environments. Different species bumblebees like Bombus terrestris, B. impatiens, B. occidentalis and a number of other are used for commercial pollination of various crops within the world. These species are expensive to import and they compete for nesting locations, food and other resources with native pollinator species [1,2].
They are exceptionally fascinating social insects [3] but some species ranges and abundances have decreased as a result of local and global environmental changes, whereas those of other species have remained stable or even increased. Concerns have been raised for bumblebees, the plant species they pollinate, food security and ecosystem stability as a result of the decline in bumblebee abundance and distribution brought on by habitat loss, pathogen transmission, climate change and agrochemical exposure.
There are 250 species of bumblebees that are still alive today, which are divided into 15 subgenera [4-6]. In terms of shape, color patterning, food preference, disease incidence and life histories and ecologies, bumblebees exhibit significant interspecific variation [7-11].
There is still a lot to learn about bumblebees. For instance, little is known about the underlying genetic and chromosomal variety that results in these many phenotypes, particularly how differently they react to shifting circumstances. China is a hotspot of bumblebee species richness, home to around half of the 250 extant species, representing 14 of the 15 Bombus subgenera [4,6,12].
The amount of genetic diversity present within a species' populations is a critical component for species survival, as low levels of intraspecific genetic diversity will limit a species' capacity to adapt to present and future environmental changes [13-16].
However, the loss of allelic richness owing to drift can be made up for by high levels of gene flow and better dispersion abilities. Gene flow may be more restricted in smaller, less-connected populations, which reduces its buffering effect and raises the likelihood of brother-sister mating, which increases the likelihood of inbreeding and inbreeding depression [13-15]. The latter dynamics have the potential to further reduce genetic within small populations, potentially resulting in the extinction vortex, a vicious cycle that could eventually result in extinction [13-15].
2. Material and Methods
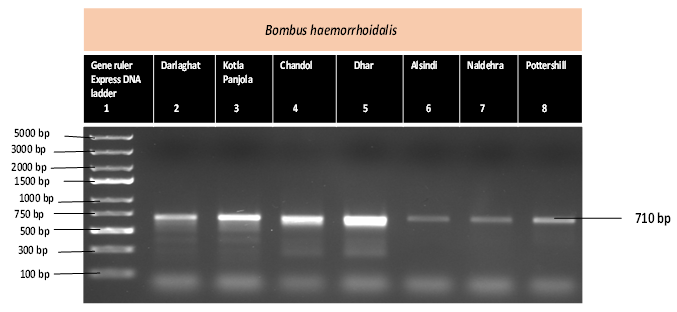
Genetic diversity of Bombus haemorrhoidalis, an insect pollinator on Punica granatum (wild pomegranate), based on mitochondrial COI gene were studied by collecting specimens from seven different areas of Western Himalaya and sequenced by Sanger sequencing. Gaps and mismatch were eliminated from the sequences and submit to NCBI genbank for accession numbers. All the seven sequences of Bombus haemorrhoidalis were accessed with accession numbers (Table 1, Figure 1). Alignment of DNA sequences were done by using multiple sequence alignment program CLUSTAL O. MEGA X was used for estimating evolutionary distances and phylogenetic analysis was constructed using Neighbor-Joining method [17]. Analyses were performed on 1000 bootstrapped data sets generated by the program [18]. This analysis involved 7 nucleotide sequences. All the data were calculated by MEGA X software. The nucleotide content of all the samples and the total C+G and A+T were calculated using MEGA software X. DNADIST with the Kimura two parameter distance option was used to estimate divergence between sequences with a transition/transversion ratio.
|
S. No. |
Species name |
Sample Location |
Geographical Location |
Genbank Accession No. |
||
|
Locality |
Longitude |
Latitude |
Altitude |
COI |
||
|
1 |
Bombus haemorrhoidalis |
Darlaghat |
76°-56´50 |
31°-13´14 |
1563 m |
OK483358 |
|
2 |
Bombus haemorrhoidalis |
Kotla Panjola |
77°-08´51 |
30°-51´09 |
1190 m |
OL347869 |
|
3 |
Bombus haemorrhoidalis |
Chandol |
77°-34´37 |
30°-93´60 |
1418 m |
OL658828 |
|
4 |
Bombus haemorrhoidalis |
Alsindi |
77°-12´31 |
31°-29´33 |
1132 m |
ON303732 |
|
5 |
Bombus haemorrhoidalis |
Dhar |
76°-82´85 |
31°-62´42 |
1360 m |
OL304916 |
|
6 |
Bombus haemorrhoidalis |
Pottershill |
77°-13´41 |
31°-12´13 |
2050 m |
OM432006 |
|
7 |
Bombus haemorrhoidalis |
Naldehra |
77°-18´69 |
31°-18´39 |
1887 m |
OL658831 |
Table 1: Places of sample collection of Bombus haemorrhoidalis with geographical location and the Genbank accession numbers of COI gene.
3. Results
Current study investigated the genetic diversity of Bombus haemorrhoidalis from seven localities of Western Himalaya by using NJ clustering based on COI gene. All the insect pollinators sampled specimens were collected on Punica granatum (wild pomegranate) and results are summarized as follow:
Multiple sequence alignment by CLUSTAL Omega
Multiple sequence alignment of seven COI sequences of Bombus haemorrhoidalis performed by CLUSTAL Omega revealed a total of 6 variable sites in the alignment between all the mtCO1 sequences of Bombus haemorrhoidalis (Figure 2).
Nucleotide content analysis of COI gene: According to the neutral theory, nucleotide polymorphism levels correlate to evolutionary rate, and the transition and transversion ratio within populations should be related to long-term evolutionary rate.
Comparative significance of Transitions and Transversions: The frequencies of transitions and transversions are mentioned in Table 2. The estimated transition/transversion (Ts/Tv) bias of COI (R) is 1.67. The percentage of sites showing transitions (75.42%) is higher than the number of sites showing transversions (24.58%). The nucleotide frequencies are 34.58% (A), 41.84% (T/U), 13.19% (C) and 10.39% (G).
Base composition at each Codon positions
In the present study, the nucleotide content (A,T,G,C) and the total C+G and A+T at first, second and third codon position of all the samples revealed the high numbers of polymorphic sites in the COI gene, evenly distributed among the 3 codon positions. Average A+T percentage (76.34%) were higher than C+T (23.66%) (Table 3). The variation in nucleotide content almost completely exist in the third codon position due to AT rich region, as compared to the first and second position of codons. The A+T bias was pronounced in general for this region for all codon positions.
The transition/transversion (Ts/Tv) ratio is significant in deducing the magnitude and direction of natural selection. The transitions (75.42%) and transversions (24.58%) depicts ratio of 3:1 (Table 2) and transition/transversion (Ts/Tv) bias of COI (R) is 1.67. The ratio greater than one indicates the positive or Darwinian selection and less than 1 implies purifying selection and neutral selection indicated by ratio equal to one. The current transitions/transversions value of 1.67 signifies the presence of positive genetic divergence in Bombus haemorrhoidalis of Western Himalaya. The current results suggested divergence between the Bombus haemorrhoidalis of Western Himalaya.
|
Transitions (%) |
Transversions (%) |
Ts/Tv ratio |
|||||||||||
|
COI Gene |
G/A |
C/T |
T/C |
A/G |
A/T |
A/C |
T/A |
T/G |
C/A |
C/G |
G/T |
G/C |
1.67 |
|
26.38 |
31.26 |
9.86 |
7.92 |
5.14 |
1.62 |
4.25 |
1.28 |
4.25 |
1.28 |
5.14 |
1.62 |
||
Table 2: Frequency percentage (%) of transitions and transversions and transition/transversion ratio (Ts/Tv) of mtCOI gene of Bombus haemorrhoidalis.
Table 3: Mean frequencies (%) for base compositions at different codon positions for COI region of Bombus haemorrhoidalis.
Phylogenetic analysis of mitochondrial CO1 of Bombus haemorrhoidalis samples
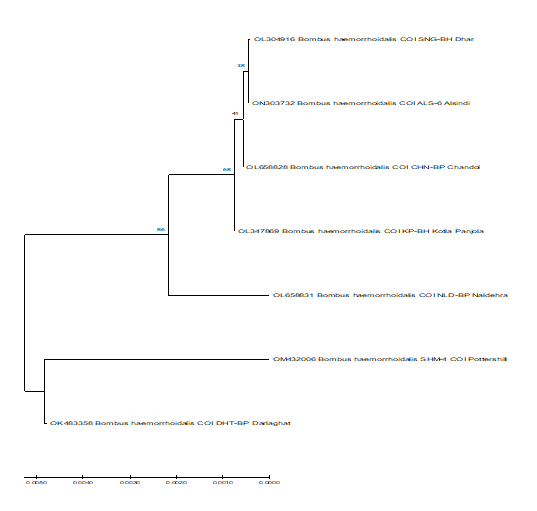
The phylogenetic relationship among Bombus haemorrhoidalis of the present study obtained through Neighbor-Joining (NJ), were shown in Figure 3. Phylogenetic relationship showed that among the seven samples sequences obtained from the present study area, the sample of Darlaghat and Potterhill were found to be closely related to each other. Sample of Alsindi and Dhar show homology with each other and close to the sample of Kotla Panjola and Chandol. Naldehra sample found to be distinct from other samples. Distance matrix clearly signifies the vast difference among the sampled species proves that there is genetic diversity between them (Figure 3).
4. Discussion
Form this study, we were able to estimate genetic diversity and to compare this with each other species collected from different areas on Punica granatum (wild pomegranate) in the Western Himalaya. This approach allowed us to conclude that phylogenetic relationship among the seven samples sequences obtained from the present study areas, the sample of Darlaghat and Potterhill were found to be closely related to each other. Sample of Alsindi and Dhar show homology with each other and close to the sample of Kotla Panjola and Chandol. Naldehra sample found to be distinct from other samples. Distance matrix clearly signifies the vast difference among the sampled species proves that there is genetic diversity between them.
According to numerous research [19-23], bumblebee species with declining population typically having lower genetic diversity level than stable bumblebee species. This phenomenon clearly indicate the decrease in genetic variety through time [21,22] as a result of one or more potential drivers of bee decline such as intensifying agriculture, emerging diseases and changing climates that result in smaller population sizes [24-27].
Studies using historical populations, however, discovered a comparable difference in genetic variation between declining and stable bumblebee species [28-31]. These studies also detected no significant decline in genetic diversity over a century in Belgium, possibly because dispersal counteracted drift effects. However, studies using historical populations found a similar difference in genetic variation between declining and stable bumblebee species [28-31], detecting no major drop in genetic diversity over one century in Belgium, possibly due to dispersal countering drift effects [31].
The latter finding suggests that there is no correlation between species abundance and genetic diversity, at least among populations in Belgium. This runs counter to the generally accepted idea that suggests that when a species is locally plentiful, its huge population should exhibit greater genetic diversity than that seen in areas where the species is less common [13,14,32]. This may not always be the case, though, for social insects like bumblebees [33].
As a result, our findings showed that determining a population's health state locally, particularly over a longer period of time, cannot be done just by counting the number of bumblebees present. Additionally, because bumblebee species with lower genetic diversity are less fit [23,34,35]. It appears that evaluating genetic diversity factors can provide an accurate estimate of how vulnerable a particular population or bumblebee species is to future population decrease. Genetic diversity has been proved to be important for the fitness of species because harmful mutations can be counterbalanced by high levels of heterozygozity [36-39]. Our study suggested that mitochondrial COI genes are well suited for determining genetic difference within species and there is need of extensive sampling and further characterization of genetic diversity with different mitochondrial genes to lessen the risk of extinction of species and ecosystems.
References
- Ghimire KC, Pandey A, Roka I, et al. Community dynamics of bumblebee across elevation gradients and habitat mosaics in Chitwan Annapurna Landscape, Nepal Heliyon 9 (2023): 1-13.
- Couvillon MJ, Jandt JM, Duong NHI, et al. Ontogeny of worker body size distribution in bumble bee (Bombus impatiens) colonies. Ecological Entomology 35 (2010): 424-435.
- Alem S, Perry CJ, Zhu X, et al. Associative mechanisms allow for social learning and cultural transmission of string pulling in an insect. PLoS Biology 14 (2016): e1002564.
- Williams PH. An annotated checklist of bumble bees with an analysis of patterns of description (Hymenoptera: Apidae, Bombini). Bulletin of the British Museum (Natural History) Entomology 67 (1998): 79-152.
- Cameron SA, Hines HM, Williams PH. A comprehensive phylogeny of the bumble bees, Biological Journal of Linnean Society 91 (2007): 161-188.
- Williams PH, Lobo JM, Meseguer AS. Bumblebees take the high road: climatically integrative biogeography shows that escape from Tibet, not Tibetan uplift, is associated with divergences of present day Mendacibombus. Ecography 41 (2018): 461-477.
- Williams PH. Phylogenetic relationships among bumble bees (Bombus Latr.): a reappraisal of morphological evidence. Systematic Entomology 19 (1994): 327-344.
- Sikora A, Kelm M. Flower preferences of the Wroclaw Botanical Garden Bumblebees (Bombus spp.). Journal of Apicultural Science 56 (2012): 27-36.
- Persson AS, Rundlof M, Clough Y, et al. Bumble bees show trait-dependent vulnerability to landscape simplification. Biodiversity Conservation 24 (2015): 3469-3489.
- Arbetman MP, Gleiser G, Morales CL, et al. Global decline of bumblebees is phylogenetically structured and inversely related to species range size and pathogen incidence. Proceedings of Biological Sciences 284 (2017): 20170204.
- Cameron SA, Sadd BM. Global trends in bumble bee health. Annual Review of .Entomology 65 (2020): 209- 232.
- Cheng Sun C, Huang J, Wang Y, et al. Genus-Wide Characterization of Bumblebee Genomes Provides Insights into Their Evolution and Variation in Ecological and Behavioral Traits. Molecular Biology Evolution 38 (2020): 486-501.
- Frankham R. Genetics and extinction. Biology Conservation, 126 (2005): 131-140.
- Zayed A. Bee genetics and conservation. Apidologie 40 (2009): 237-262.
- Habel JC, Husemann M, Finger A, et al. The relevance of time series in molecular ecology and conservation biology. Biological Review 89 (2014): 484-492.
- Koch JB, Looney C, Sheppard S, et al. Patterns of population genetic diversity and structure across bumblebee communities in the Pacific Northwest. Conservation Genetics 18 (2017): 507-520.
- Saitou N, Nei M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Molecular Biology and Evolution 4 (1987): 406-425.
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39 (1985): 783-791.
- Darvill B, Ellis JS, Goulson D. Population structure and inbreeding in a rare and declining bumblebee Bombus muscorum, (Hymenoptera: Apidae). Molecular Ecology 15 (2006): 601-611.
- Ellis JS, Knight ME, Darvill B, et al. Extremely low effective population sizes, genetic structuring and reduced genetic diversity in a threatened bumble bee species Bombus sylvarum, (Hymenoptera: Apidae). Molecular Ecology 15 (2006): 4375-4386.
- Goulson D, Lye GC, Darvill B. Decline and conservation of bumble bees. Annual Review of Entomology, 53 (2008): 191-208.
- Charman TG, Sears J, Green RE, et al. Conservation genetics, foraging distance and nest density of the scarce Great Yellow Bumblebee (Bombus distinguendus). Molecular Ecology 19 (2010): 2661-2674.
- Cameron SA, Lozier JD, Strange JP, et al. Patterns of widespread decline in North American bumblebees. Proceedings of National Academy of Sciences USA 108 (2011): 662-667.
- Potts SG, Biesmeijer JC, Kremen C, et al. Global pollinator declines: trends, impacts and drivers. Trends Ecology and Evolution 25 (2010): 345-353.
- Meeus I, Brown MJF, De Graaf DC, et al. Effects of invasive parasites on bumblebee declines. Conservation Biology 25 (2011): 662-671.
- Vanbergen AJ. The Insect Pollinators Initiative. Threats to an ecosystem service: pressures on pollinators. Frontiers in Ecology and Environment 11 (2013): 251-259.
- Rasmont P, Franzen M, Lecocq T, et al. Climatic risk and distribution atlas of European bumblebees. BioRisk 10 (2015): 1-236.
- Lozier JD, Cameron SA. Comparative genetic analyses of historical and contemporary collections highlight contrasting demographic histories for the bumblebees Bombus pensylvanicus and B. impatiens in Illinois. Molecular Ecology 18 (2009): 1875-1886.
- Lozier JD, Strange JP, Stewart IJ, et al. Patterns of range-wide genetic variation in six North American bumblebee (Apidae: Bombus) species. Molecular Ecololgy 20 (2011): 4870-4888.
- Maebe K, Meeus I, Ganne M, et al. Microsatellite analysis of museum specimens reveals historical differences in genetic diversity between declining and more stable Bombus species. PloS One 10 (2015): e0127870.
- Maebe K, Meeus I, Vray S, et al. A century of temporal stability of genetic diversity in wild bumblebees. Scientific Reports 6 (2016): 38289.
- Reed DH, Frankham R. Correlation between fitness and genetic diversity. Conservation Biology 17 (2003): 230-237.
- Maebe K, Karise R, Meeus I, et al. Level of genetic diversity in European bumblebees is not determined by local species abundance. Frontiers in Genetics 10 (2019): 1-9.
- Whitehorn PR, Tinsley MC, Brown MJF, et al. Genetic diversity, parasite prevalence and immunity in wild bumblebees. Proceedings of the Royal Society of London. Series B, Biological Science 278 (2011): 1195-1202.
- Whitehorn PR, Tinsley MC, et al. Genetic diversity and parasite prevalence in two species of bumblebee. Journal of Insect Conservation 18 (2014): 667-673.
- Goulson D. Bumblebees, behavior. Ecology and Conservation (Oxford: Oxford University Press) (2010): 336.
- Highlight contrasting demographic histories for the bumblebees Bombus pensylvanicus and B. impatiens in Illinois. Molecular Ecology 18 (2009): 1875-1886.
- Maebe K, Karise R, Meeus I, et al. Pattern of Population structuring between Belgium and Estonian bumblebees. Scientific Reports 9 (2019): 9651.
- Sheikh UAA, Ahmad, M, Imran M, et al. Distribution of Bumblebee, Bombus haemorrhoidalis Smith and its association with Flora In Lower Northern Pakistan. Pakistan Journal of Zoology 46 (2014): 1045-1051.



 Impact Factor: * 4.1
Impact Factor: * 4.1 Acceptance Rate: 75.32%
Acceptance Rate: 75.32%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 