Genetic Diversity of Eristalis Tenax (Linnaeus, 1758) as Insect Pollinator of Prunus Persica (L.) Stokes Flowers Based on Mtcoâ… Gene
Poonam Dhiman and Mahender Singh Thakur*
Department of Biosciences, Himachal Pradesh University, Shimla, Himachal Pradesh, India
*Corresponding Author: Mahender Singh Thakur, Department of Biosciences, Himachal Pradesh University, Shimla, Himachal Pradesh, India.
Received: 23 June 2023; Accepted: 10 July 2023; Published: 17 July 2023
Article Information
Citation: Poonam Dhiman, Mahender Singh Thakur. Genetic Diversity of Eristalis Tenax (Linnaeus, 1758) as Insect Pollinator of Prunus Persica (L.) Stokes Flowers Based on Mtcoâ… Gene. International Journal of Plant, Animal and Environmental Sciences 13 (2023): 37-45.
View / Download Pdf Share at FacebookAbstract
Eristalis tenax is an important insect pollinator of Prunus persica plant and samples were collected from seven localities of Himachal Pradesh i.e. Bathra (503m), Jaladi (508m), Hamirpur (786m), Rajgarh (1555m), Naldera (1887m), Summer Hill (2100m) and Mashobra (2146m). Phylogenetic relationship and multiple sequence alignment of all the sampled sequences of Eristalis tenax were analyzed by using mtCOI gene. Nucleotide composition analysis showed that percentage of A+T (69.03%) was higher than the percentage of C+G (30.97%) which showed that all the sequences were AT biased. Multiple sequence alignment showed three variable sites in the sequences of Eristalis tenax and the comparable values of transitions and transversions in the current study suggest the possible occurrence of genetic divergence over evolutionary time scales in Eristalis tenax of Himachal Pradesh.
Keywords
<p><em>Eristalis tenax</em>; <em>Prunus persica</em>; mtCOâ… ; Nucleotide composition; Multiple sequence alignment</p>
Article Details
1. Introduction
Pollinators are an essential part of the world's biodiversity because of their crucial ecological services to plants and crops. Pollinators enhance the genetic diversity in plants by cross pollination, that’s why they considered essential for the survival and maintenance of diversity of plants [1]. But at present time, pollinator populations have been declining due to habitat degradation, climate change, pollution and over-exploitation. Due to these reasons, many insect pollinators population have been reduced to small isolated fragmented groups and are under high risk of extinction [2]. The fundamental objective to study the genetic diversity is to use genetics knowledge to reduce the risk of pollinators extinction. Genetic diversity helps a species to adjust according to changing environment and maintain higher level of biodiversity at population and species level [3].
2. Material and Methods
In the present study, samples of Eristalis tenax were collected from seven localities of Himachal Pradesh from Prunus persica flowers (Table 1). DNA was extracted from the thorax or upper abdominal region of the collected insect specimens by using DNeasy blood and tissue Qiagen Kit method by following standardized protocol of the manufacturers. Extracted DNA was preserved in the -20°C for further use. Target DNA from mitochondrial gene, i.e. Cytochrome Oxidase subunit I was amplified using a pair of forward primers LCO1490 5’- GGTCAACAAATCATAAAGATATTGG-3’ and reverse primer HCO2198 5’- TAAACTTCAGGGTGACCAAAAAATCA-3’ [4]. Total volume of PCR reaction was 20 µl and reaction run in PCR as per standardized protocol.
|
S. No. |
Taxon |
Sample Location |
Geographical Location |
Genbank Accession No. |
||
|
Locality |
Longitude |
Latitude |
Altitude |
COI |
||
|
1 |
Eristalis tenax |
Bathra |
76°-21´46 |
31°-88´18 |
503 m |
OK444106 |
|
2 |
Eristalis tenax |
Jaladi |
76°-34´44 |
31°-77´85 |
508 m |
OK465102 |
|
3 |
Eristalis tenax |
Hamirpur |
76°-52´13 |
31°-68´62 |
786 m |
OK559908 |
|
4 |
Eristalis tenax |
Rajgarh |
77°-29´94 |
30°-85´00 |
1555 m |
OL589625 |
|
5 |
Eristalis tenax |
Naldera |
77°-18´69 |
31°-18´39 |
1887 m |
OQ359958 |
|
6 |
Eristalis tenax |
Summer Hill |
77°-13´99 |
31°-11´46 |
2100 m |
OK655835 |
|
7 |
Eristalis tenax |
Mashobra |
77°-22´83 |
31°-12´96 |
2146 m |
OL589638 |
Table 1: Localities of sample collection of Eristalis tenax with geographical location and the Genbank accession numbers of COI gene.
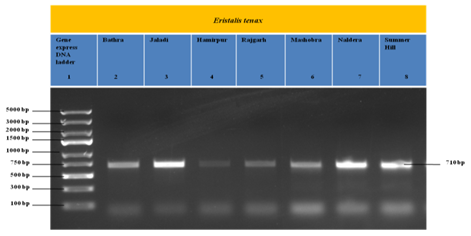
The amplified PCR product was analyzed on a 1.2% agarose gel electrophoresis and checked under UV light and documented. The amplified DNA fragments were extracted from agarose gel and purified using DNA/RNA purification Qiagen Kit. The primers used were the same primers used in PCR amplification and sequencing was done in “Big dye terminator version 3.1” cycle sequencing kit with a sequencing machine-ABI 3500xL Genetic analyzer. After completion of sequencing, all fasta format sequences obtained by Sanger sequencing was used for BLAST search to check the sequence homology at NCBI. All the sequences were edited and aligned using bioedit sequence alignment editor software. The edited sequences were submitted in the gene bank for accession Number (Table 1). The nucleotide content (A, T, G, C) of all the samples and the total C+G and A+T at first, second and third codon position were calculated using MEGA X software. DNADIST with the Kimura two parameter distance options were used to estimate divergence between sequences with a transition/ transversion ratio in the MEGA X software. Evolutionary analysis of obtained 7 sequences of sampled Eristalis tenax were conducted using Neighbor-Joining method and Kimura-2 parameter in MEGA X. Sequences were aligned by using the MEGA X software [5]. Analyses were performed on 1000 bootstrapped data sets generated by the program [6].
3. Results and Discussion
In the present study, DNA of Eristalis tenax collected from different altitudes of Himachal Pradesh was extracted and amplified using forward and reverse primers (Figure 1). The amplified products were sequenced by using Sanger sequencing and obtained fasta files were used to BLAST search in NCBI. The fasta format sequences were edited and mismatches were removed by using Bioedit sequence alignment editor software. The edited sequences were submitted in Genbank for accession number. Each sequence was accessed with accession number (Table 1). Multiple sequence alignment of seven sequences were performed by CLUSTAL Omega (1.2.4), which revealed three variable sites in COI sequences of Eristalis tenax (Figure 2).
3.1 Nucleotide content analysis of COI gene
The estimated transition/transversion bias (R) is 2.56 and the sites showing transition (75.42%) was higher than the sites showing transversion (24.6%) (Table 2). The total nucleotide content was 31.78% (A), 37.43% (T/U), 15.70% (C) and 15.01% (G). DNADIST with the Kimura two parameter distance option was used to estimate divergence between sequences with transition/ transversion ratio in the MEGA X software.
Table 2: Frequency percentage (%) of transitions and transversions and transition/transversion ratio (Ts/Tv) of COI gene.
3.2 Base composition at each codon positions
The percentage of A+T (69.03%) was higher than the percentage of C+G (30.97%) which showed that all the sequences were AT biased (Table 3). Nucleotide content of all the samples and total A+T and C+G were calculated by MEGA X software. Transition/transversion (Ts/Tv) ratio helpful in determining the degree and direction of natural selection. Study showed that overall transition/transversion bias (R) was 2.56 which indicated the positive or Darwinian selection in Eristalis tenax sequences of Himachal Pradesh. The comparable values of transitions and transversions in the current study suggest the possible occurrence of genetic divergence over evolutionary time scales.
Table 3: Mean frequencies (%) for base compositions at different codon positions for COI region.
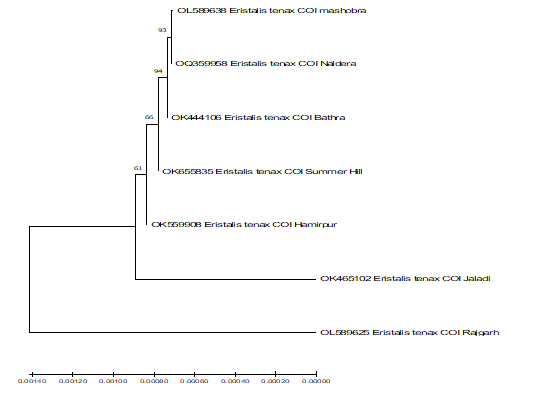
3.3 Phylogenetic analysis of mitochondrial Eristalis tenax COI sequences
Phylogenetic analysis of mitochondrial COI sequences was studied by constructing phylogenetic tree by Neighbor-Joining (NJ) method to study similarity and differences among different sequences (Figure 3). The phylogenetic relationship between seven COI sequences showed that the sequences of Mashobra and Naldera shared homology with each other and were also similar to the sequence of the Bathra. Summer Hill sequence found similar with Hamirpur. The sequence of Jaladi and Rajgarh were distinct from the other sequences. Distance matrix clearly signifies the very less difference among the sampled sequences shows that there is very less genetic diversity between them (Figure 3).
Present study showed that, molecular phylogenetics and genetic diversity among species can be useful in study the taxonomic and evolutionary relationship in different insect species. The mtCOI gene is used to study the intra-population genetic variation and also helps to study that how geographical variation changed the behavior and biology of insects [7]. Molecular markers are very helpful in determining the gene flow and genetic differences within and between insect species [8-10]. Many researchers use mtCOI gene for insect identification. Bouga et al. [11] classified the honeybee subspecies by using different molecular methods. Oldroyd et al. [12] used mtCOI gene for study the genetic divergence within species of Apis cerana in South India. Gaikwad et al. [13] studied the phylogenetic variation in Apis cerana of North Western Ghats of Maharashtra.
The present observations revealed that the percentage of A+T (69.03%) was higher than the percentage of C+G (30.97%), which showed that sequences were AT biased which were similar to the findings of Chalpathy et al. [14] who stated that honeybees sequences were AT biased and population showed natural selection from 12 localities in Karnataka. Similarly, Insuan et al. [15] studied the genetic diversity of Apis dorsata in Thailand and found limited genetic diversity in Apis dorsata samples. Present results are also accordance with the findings of Tanaka et al. [16], who studied the genetic variation of Apis dorsata dorsata from three different locations in Borneo and observed genetic variability among sequences of Apis dorsata dorsata of Borneo. In a similar study, Rukhsana et al. [17] studied the phylogenetic relationship of Apis cerana from Kerala using cytochrome oxidase subunit I gene (COI) and found significant variation in Apis species.
Significance of Study
Global climate change altered the intraspecific genetic diversity which is responsible for evolutionary changes and helps the species to adjust according to the new changing environmental conditions [18]. Sometimes, these variations will limit genetic diversity in populations and species, leading to population viability and extinction in extreme cases. To reduce the risk of extinction of species and ecosystems there is need of further characterization of species at species and subspecies level [19,20].
Acknowledgement
We are very grateful to Dr. N.K. Pandey, Director, ICAR-Central Potato Research Institute Shimla, for his cooperation and providing lab facility during the molecular work. We are very thankful to Scientist, Dr. Sundaresha Sidappa, Dr. Kailash Naga and Chander Mohan Singh Bist, ICAR-Central Potato Research Institute for their help during entire lab work. We are also thankful to UGC for funding facility.
Conflicts of Interest
The author declares no conflict of interest in the publication of this work.
References
- Wilcock C, Neiland R. Pollination failure in plants: why it happens and when it matters. Trends in Plant Science 7 (2002): 270-277.
- Frankham R, Ballou SEJD, Briscoe DA, et al. Introduction to conservation genetics. Cambridge University press (2002).
- Geffen E, Luikart G, Waples RS. Impacts of modern molecular genetic techniques on conservation biology. Key topics in conservation biology (2007): 46.
- Folmer O, Black M, Hoeh W, et al. DNA primers for amplification of mitochondrial cytochrome C oxidase subunit from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3 (1994): 294-299.
- Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35 (2018): 1547.
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39 (1985): 783-791.
- Hu J, Chen YD, Jiang ZL, et al. Global haplotype analysis of the whitefly Bemisia tabaci cryptic species in Asia. Mitochondrial DNA 26 (2015): 232-241.
- Cervera MT, Cabeza, JA, Simon B, et al. Genetic relationships among biotypes of Bemisiatabaci (Hemiptera: Aleyrodidae) based on AFLP analysis. Bulletin of Entomological Research 90 (2000): 391-396.
- Wong A, Forbes MR, Smith ML. Characterization of AFLP markers in damselflies: prevalence of codominant markers and implications for population genetic applications. Genome 44(2001): 677-684.
- Takami Y, Koshio C, Ishii M, et al. Genetic diversity and structure of urban populations of Pieris butterflies assessed using amplified fragment length polymorphism. Molecular Ecology 13 (2004): 245-258.
- Bouga A, Alaux C, Bienkowska M, et al. A review of methods for discrimination of honey bee populations as applied to European beekeeping. Journal of Apiculture Research 50 (2011): 51-84.
- Oldroyd BP, Reddy MS, Chapman NC, et al. Evidence for reproductive isolation between two colour morphs of cavity nesting honeybees (Apis) in South India. Insect Sociux 53 (2006): 428-434.
- Gaikwad GA, Gaikwad S, Shouche Y, et al. Phylogenetic variations found in Indian honeybees species, Apis cerana Fabr. of North Western Ghats of Maharashtra, India. Indian Journal of Experimental Biology 57 (2019): 55-58.
- Chalpathy CV, Puttaraju HP, Sivaram, V. A pilot study on genetic diversity in Indian honeybees – Apis cerana of Kernataka populations. Journal of Entomology and Zoological Studies 2 (2014): 07-13.
- Insuan S, Deowanish S, Klinbunga S, et al. Genetic differentiation of Giant Honeybee (Apis dorsata) in Thailand analysed by mitochondrial genes and microsatellites. Biochemical Genetics 45 (2006): 345-360.
- Tanaka H, Suka T, Kahono S, et al. Mitochondrial variation and genetic differentiation in honeybee (Apis cerana, Apis koschevnikovi and Apis dorsata) of Borneo. Tropics 13 (2003): 107-117.
- Rukhsana K, Akhilesh VP, Sebastian CD. Deciphering the molecular phylogenetics of the Asian honeybee, Apis cerana and inferring the phylogeographical relationships using DNA barcoding. Journal of Entomology and Zoology Studies 2 (2014): 218-220.
- Hoffmann AA, Sgro CM. Climate change and evolutionary adaptation. Nature 470 (2011): 479-485.
- Francuski L, Djurakic M, Ludoski J, et al. Landscape genetics and spatial pattern of phenotypic variation of Eristalis tenax across Europe. Journal of Zoological Systematics and Evolutionary Research 51 (2013): 227-238.
- Hallmann CA, Sorg M, Jongejans E, et al. More than 75 percent decline over 27 years in total flying insect biomass in protected areas. PloS one 12 (2017): e0185809.



 Impact Factor: * 4.1
Impact Factor: * 4.1 Acceptance Rate: 75.32%
Acceptance Rate: 75.32%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 