Pharmacokinetics of KA2237, A Novel Selective Inhibitor of PI3K- β and PI3K-δ, in Patients: A First-in-Human Study Using PK Modelling to Predict Drug Concentrations During Dose Escalation
James Dow1, Graham Trevitt2, Elisabeth Bone1, Kemal Haque1, Loretta J. Nastoupil3
1Karus Therapeutics Ltd. Harwell Innovation Centre, Harwell Oxford, Oxfordshire OX11 0QG, UK
2Sygnature Discovery. BioCity, Pennyfoot Street, Nottingham NG1 1GR, UK
3Department of Lymphoma/Myeloma, Division of Cancer Medicine, The University of Texas MD Anderson Cancer Center, Houston, TX, USA
*Corresponding author: James Dow, JLClinDev Ltd. London W13 8BH, UK.
Received: 03 March 2023; Accepted: 10 March 2023; Published: 31 March 2023
Article Information
Citation: James Dow, Graham Trevitt, Elisabeth Bone, Kemal Haque, Loretta J. Nastoupil. Pharmacokinetics of KA2237, A Novel Selective Inhibitor of PI3K-β and PI3K-δ, in Patients: A Firstin- Human Study Using PK Modelling to Predict Drug Concentrations During Dose Escalation. Journal of Pharmacy and Pharmacology Research. 6 (2023): 28-38
View / Download Pdf Share at FacebookAbstract
Aims: KA2237, an oral, potent and selective, inhibitor of the PI3K β and δ isoforms, was evaluated for safety, tolerability and pharmacokinetics (PK) in patients with B-cell lymphoma. KA2237 is metabolised by CYP3A4/5 but also demonstrated mechanism-based inhibition (MBI) of CYP3A4/5. An MBI mechanistic dynamic model was used to predict drug accumulation after repeat dosing of KA2237. This model, along with clinical safety data, was used to guide safe dose escalation.
Methods: An open-label, single arm, dose escalation study was carried out in patients, dosed orally with KA2237 at 50, 100, 200 and 400 mg once daily. Complete plasma profiles were obtained on Day 1 and Day 14 of dosing and pre-dose (Cmin) samples were obtained on Days 2-7. The MBI model was validated and used to calculate drug levels and predict potential drug accumulation during dose escalation.
Results: KA2237 elimination half-life was around 20-30 h, compatible with once daily dosing regimens. The accumulation of KA2237 was around 4-fold after the highest dose of 400 mg and around 3-fold after administration of 200 mg, which is considered the maximum tolerated dose (MTD). The MBI model accurately predicted this accumulation.
Conclusions: Drugs that demonstrate MBI and potential auto-inhibition can be successfully developed, provided that models are developed to assess the extent of accumulation prior to the start of FIH clinical studies. This, along with the close monitoring of drug levels and clinical safety data can be used to guide dose escalation and lead to the safe conduct of clinical studies.
Keywords
<p>KA2237, CYP3A4, MBI, PK modelling, PI3K, FIH, B-cell lymphoma</p>
Article Details
1. Introduction
The phosphoinositide 3-kinases (PI3Ks) constitute a family of enzymes that phosphorylate the 3’- hydroxyl head group of phosphoinositides leading to the inhibition of a cellular signalling cascade which includes the downstream effector kinase AKT [1]. KA2237 (4-(1H-Indol-4-yl)-6-(morpholin-4-yl)-12-[(1S,4S)-2-oxa-5-azabicyclo [2.2.1] heptan-5-ylmethyl]-8-oxa-3,5,10-triazatricyclo [7.4.0.02,7] trideca-1(13),2(7),3,5,9,11-hexaene; succinic acid) is a potent inhibitor of two Class I PI3K isoforms, PI3K-p110δ and PI3K-p110β in in vitro assay systems. KA2237 inhibited anti-IgM stimulated AKT phosphorylation (PI3K- p110δ dependent) and basal levels of AKT phosphorylation in the PTEN mutant cell line, PC-3 (PI3K- p110β dependent), with IC50s in the low nM range. By contrast, IC50s for the inhibition of AKT phosphorylation driven by PI3K-p110α or PI3K-p110γ were greater than 500 nM, demonstrating the selectivity of KA2237.
As PI3K-p110β and PI3K-p110δ are known to have essential roles in the growth and maintenance of tumours, the effects of KA2237 on the growth of cancer cell lines were determined. As some of the most sensitive cell lines in this panel were derived from haematological cancers, the data supported the selection of this indication for evaluation in the early clinical development of KA2237. Despite therapeutic advances, there remains a considerable need for novel therapies for B-cell lymphomas. Although a high proportion of patients show response to initial therapy, many fail to achieve durable remissions and experience recurrent disease. Agents that target molecular pathways associated with the development and progression of lymphoma are likely to be highly effective and are desirable. The p110δ isoform of the PI3K enzyme is mainly expressed in lymphocytes and has been an attractive therapeutic target, with several PI3Kδ inhibitors demonstrating meaningful efficacy in B-cell lymphomas [2-4]. Targeting the p110β isoform may further overcome tumour growth and escape mechanisms by mitigating the upregulation of the PI3K/AKT pathway, particularly in PTEN-deficient lymphomas [5-7]. KA2237 is an oral, potent and selective inhibitor of the PI3K-p110- β and - δ isoforms.
CYP3A4 is the most abundant human cytochrome P450 enzyme and many drugs are metabolised by this enzyme. CYP3A4 can be inhibited reversibly or irreversibly and is involved in a number of clinically relevant drug-drug interactions. Irreversible inhibition, commonly referred to as mechanism-based inhibition (MBI), involves the metabolism of an inhibitor by CYP3A4 to a metabolite that inactivates the enzyme in a concentration and time dependent manner [8].
KA2237 is a substrate for CYP3A4 and CYP3A5, but not for the other major human CYPs, and demonstrated low, reversible inhibition of all the major human CYPs. However, when KA2237 was pre-incubated for 30 min with human liver microsomes, using midazolam as probe substrate, there was a shift in IC50 indicating the possibility of time-dependent inhibition of *CYP3A4/5. As the shift in IC50 was evident when samples pre-incubated without NADPH were compared to samples pre-incubated with NADPH, it was considered that KA2237 caused metabolism-dependent inhibition of CYP3A and that there was no evidence that KA2237 per se was a time-dependent inhibitor of CYP3A. This increase in inhibition upon pre-incubation was irreversible, demonstrating that KA2237 is a MBI of CYP3A. Thus, KA2237 has the potential for auto-inhibition of its metabolism.
Due to this potential for auto-inhibition, an MBI mechanistic dynamic model was developed and used to predict drug accumulation after repeat dose of KA2237. This model, along with clinical safety data, was used to guide safe dose escalation. The aim of the Phase I, single arm study was to investigate the safety, tolerability, pharmacokinetic properties and pharmacodynamic effects of KA2237, in order to determine the MTD based on dose limiting toxicity and to assess preliminary anti-tumour activity in patients with relapsed/refractory (R/R) B-cell lymphoma. The study was also designed to define a recommended Phase 2 (RP2) dose.
*Throughout the paper CYP3A will be used to designate CYP3A4/5
2. Methods
2.1 Study design and subjects
This Phase I clinical trial (NCT02679196) was approved by the Institutional Review Board of the MDAnderson Cancer Center, Houston, Texas and was conducted in accordance with Good Clinical Practice. The study was a first in human (FIH), open-label, single arm, non-randomised, study in patients ≥ 18 years, ECOG ≤ 2, with B-cell lymphoma (indolent or aggressive), R/R or intolerant of established therapies. Patients were enrolled using a 3+3 dose escalation, multiple ascending dose (MAD) design. KA2237 was given orally on a once daily continuous schedule until progression or unacceptable toxicity.
As in vitro studies suggested that KA2237 had the potential for MBI of CYP3A, concomitant use of drugs which were substrates of CYP3A and had narrow therapeutic margins were contraindicated. Strong inhibitors and inducers of CYP3A were also contraindicated in this study. Patients were advised to avoid eating grapefruit/drinking grapefruit juice while taking KA2237. The starting dose was derived using the methodology described in the FDA guidance document for anticancer pharmaceuticals [9] and was based on 1/6th of the highest non-severely toxic dose (HNSTD) in the non-human primate (NHP) and a conversion factor of 3.1 to obtain the human equivalent dose (HED). This dose level was anticipated to provide concentrations at steady state that were comparable to those effective in a mouse syngeneic tumour model. Four dose levels, 50, 100, 200 and 400 mg once daily KA2237 were investigated. There were N=6 patients in the 50 mg and 200 mg cohorts, N=3 patients in the 100 mg cohort and N=5 in the 400 mg cohort. Patients were instructed to eat a meal up to one hour prior to dosing, if a patient was unable to eat a meal, or a meal could not be provided, the meal was replaced by administration of a liquid food supplement. Water was available ad libitum except for 1 hour before and 1 hour after dosing. Approximately 240 mL water was given to each patient at the time of drug administration.
2.2 Determination of KA2237 in plasma
A validated quantitative analytical method for the determination of KA2237 in human K2-EDTA plasma samples by LC-MS/MS was developed. [Morpholine-D8]- KA2237 was used as internal standard. Results showed suitable linearity, selectivity and stability and demonstrated that the validated method was suitable for the determination of KA2237 in human plasma samples. The assay was linear from 1.0 ng/mL (LLOQ) to 1000 ng/mL (ULOQ) and long-term stability showed that KA2237 was stable for at least 5 months in plasma at -20°C. (PRA Health Sciences - Early Development Services, Bioanalytical Laboratory,10836 Strang Line Road, Lenexa, KS 66215, USA).
2.3 PK samples and analysis
Complete plasma concentration profiles were collected from each patient on Day 1 and Day 14 of the first cycle of treatment. Samples for PK were drawn pre-dose and at 0.25, 0.5, 1, 2, 4, 6, 8, 12, and 24 hours post dose on Day 1 and Day 14. On days 3 to 7 samples were drawn pre-dose (Cmin). The derivation of PK parameters for KA2237 from plasma concentration-time data was done using non compartmental analysis (Phoenix WinNonlin Version 6.4; Certara Princeton, NJ, USA). Actual elapsed time from dosing was used to estimate all individual pharmacokinetic parameters. The following PK parameters were calculated where possible: Tmax (h): time to reach maximum observed concentration; Cmax (ng/mL): maximum observed plasma concentration; Cmin (ng/mL): minimum observed plasma concentration (pre-dose sample); Half-life (h): apparent terminal elimination half-life = ln(2)/Lambda_z; where Lambda_z is the terminal phase rate constant estimated by linear regression analysis of the log transformed plasma concentration-time data; AUCtau (ng.h/mL): area under the plasma concentration-time curve over the dosing interval (24h) (AUC was estimated using the linear/log trapezoidal rule); R (AUCtau): measure of accumulation after repeat dosing (AUCtau Day 14/AUCtau Day 1).
2.4 Statistical Analysis
SAS software version 9.3 (SAS Institute Inc, Cary, NC, USA) was used for statistical analysis. All the individual data was summarised using descriptive statistics.
2.5 Mechanistic dynamic MBI model for KA2237
A mechanistic dynamic model was developed to guide dose selection and predict plasma concentrations during dose escalation for this FIH study. This model included the prediction of the potential effects of CYP3A MBI on the repeat dose PK of KA2237 in patients.
2.5.1 Initial Model Description
A single compartment PK model was proposed, which assumes first order absorption of dosed compound from a site, A, to the central compartment, C, at rate Ka. The model assumes first order elimination from the central compartment, C, at rate Ke. The dose available at the central compartment is corrected for oral bioavailability, F. The model breaks down total clearance into the contributions of extra-hepatic and biliary clearance, plus clearance due to metabolism by hepatic CYP3A4 and CYP3A5. For the MBI model, the fraction of hepatic metabolic clearance due to CYP3A4 is defined as Fm.
Clearance by CYP3A4, CL3A4 is a function of the relative abundance of active 3A4, 3A4active where CL3A4 = Clhep_metab*Fm*3A4active
If all other hepatic metabolism is by CYP3A5, then clearance due to CYP3A5, CL3A5 = Clhep_metab*(1-Fm)*3A5active
2.5.2 Modelling Mechanism Based Inhibition
Using a turnover model of enzyme inactivation, CYP3A4 and CYP3A5 abundance in the absence of MBI, is a function of the zero-order formation of enzyme with rate constant Ksyn and first order degradation with rate constant Kdeg of each enzyme. If the rate constants for these are equal, the relative abundance at steady state will be equal to 1 and this is assumed to be the case. A value of Kdeg = 0.0193 h-1 is reported to be optimal for CYP3A4 by Rowland Yeo et al [10], and has been assumed to be the same for CYP3A5. MBI is assumed to inactivate the enzyme under Michaelis-Menten kinetics at a rate Kobs where Kobs = Kinact * [I] / (KI + [I]) where [I] is the unbound concentration of inhibitor, Kinact is the maximal rate of inactivation and KI is the concentration at 50% Kinact. The model assumes unbound liver concentration = unbound plasma concentration.
The rate of change in the relative abundance of active CYP3A4, 3A4active, is therefore:
d[3A4active]/dt = Kdeg-Kdeg*3A4active-Kobs
The rate of change in the relative abundance of active 3A5, 3A5active, is therefore:
d[3A5active]/dt = Kdeg-Kdeg*3A5active-Kobs
All ordinary differential equations were solved simultaneously in Berkeley Madonna 8.3.18 across a suitable time period with time interval set to 0.02h. The PULSE function in Berkeley Madonna was used to simulate repeat dose schedules (input into absorption compartment of an amount A, n times, at a dose interval, Tau).
2.5.3 Initial Validation
Data, derived by the initial model, was compared with published predictions generated by Simcyp, using clinical data for imatinib, obtained from the publication of Filppula et al [11]. The initial model compared favourably with the Simcyp model, which accurately predicted plasma concentrations of imatinib compared with actual clinical data for imatinib (400mg QD dosing) [11].
The model was further validated using in vitro and in vivo PK data from non-human primates (NHP), as NHP CYP3A MBI was also observed in vitro and drug accumulation was observed after 28 day repeat dosing in NHP. The model was based on known pharmacokinetic parameters (Cl and Vss), obtained from single dose i.v. data and measured KI and kinact for CYP3A, obtained from in vitro NHP studies. Plasma concentrations, after 28 day repeat dosing in NHP, were adequately predicted with the developed model.
2.5.4 Human Model
The starting dose for this FIH clinical trial was proposed as 50mg once a day followed by a dose escalation phase with proposed once daily levels of 100, 200, 400, 600 and 800mg.
These dose levels were simulated using the MBI model with the parameters shown below. The fraction of clearance that is hepatic (Fraction_hepatic) was set to 0.67 to match the best fit to the NHP data and assumes 67% of drug is cleared through hepatic metabolism. Similarly, all hepatic clearance is assumed to be metabolic.
The following inputs were used:
- Kinact Human CYP3A4 = 0.067min-1 (measured)
- KI Human CYP3A4 = 3.3µM (measured)
- Kinact Human CYP3A5 = 0.038min-1 (measured)
- KI Human CYP3A5 = 18.6µM (measured)
- Kdeg CYP3A4 = 0.0193 h-1 [12]
- Fraction metabolised by CYP3A4 = 0.82 and by CYP3A5 = 0.18 (estimated from CYP phenotyping with CLint for each enzyme scaled to µL/min/mg microsomal protein)
- Fraction of hepatic clearance that is metabolic (Fraction_hepmetab) = 1.0 (matched to the NHP model, in line with <1% parent compound detected in rat bile)
- Fraction_hepatic = 0.67 (matched to the NHP model)
- Initial total clearance = 5.2mL/min/kg (allometry)
- Volume of distribution = 4.7L/kg (allometry)
- Liver blood flow = 20.7mL/min/kg
- Bodyweight = 60 kg with dose in mg (total)
- Fa and Fg = 1.0 (same as NHP)
- Rate of absorption = 1.1h-1 (same as NHP)
- Fraction unbound in plasma = 0.034 (measured, assume blood:plasma =1.0)
- Dose interval = 24h
3. Results
3.1 Predicted Cmax and AUC on Day 1 and Day 14 using the MBI model
The predicted human Cmax and AUC are shown in Table 1. Based on AR, the ratio of AUC Day14/AUC Day 1, predicted drug accumulation varies from 1.7 to 2.9 for the 50 mg to 400 mg doses respectively.
Table 1: Predicted Cmax and AUC on Day 1 and Day 14, and the Accumulation ratio (AR)
|
Day 1 |
Day 14 |
||||
|
Dose (mg) |
Cmax (ng/mL) |
AUC (ng/mL.h) |
Cmax (ng/mL) |
AUC (ng/mL.h) |
AR |
|
50 |
123 |
1772 |
188 |
2934 |
1.7 |
|
100 |
247 |
3607 |
436 |
7244 |
2 |
|
200 |
495 |
7448 |
1042 |
18443 |
2.5 |
|
400 |
997 |
15721 |
2434 |
45174 |
2.9 |
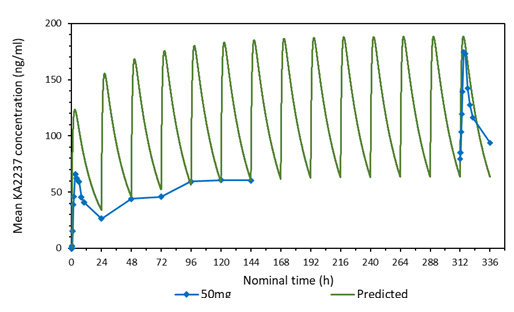
3.2 Comparison of the predicted and mean plasma-concentration time profile after daily administration of 50 mg KA2237
A comparison of the predicted and mean plasma-concentration time profile of KA2237after daily oral doses of 50 mg KA2237 is shown in Figure 1. The dynamic MBI model predicted that steady state would be reached by day 7 and drug accumulation after 14 days daily administration of 50 mg KA2237 would be around 1.7-fold. The model demonstrated good prediction of full profile plasma concentrations on days1 and 14 and Cmin values on days 2-7. However, actual accumulation on day 14 was 2.8-fold. Based on this modelling, prediction of further drug exposure was done using this approach to assist safe dose escalation.
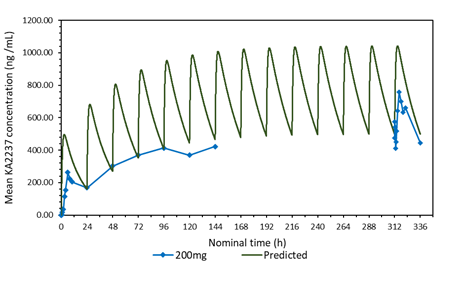
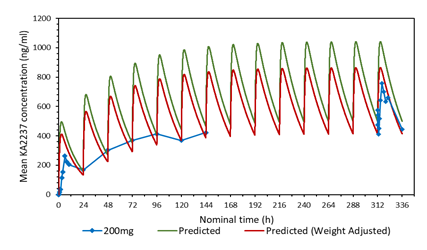
3.3 Comparison of the predicted and mean plasma-concentration time profile after daily administration of 200 mg KA2237. Effect of adjusting the model for body weight of subjects.
A plot of the predicted and actual mean plasma concentrations of KA2237 after daily oral doses of 200 mg is shown in Figure 2a. The model gave a good prediction of Cmin on days 2-5, but tended to over predict the plasma concentrations of the full profiles on days1 and 14 and the Cmin on days 6-7. One possible explanation for this over prediction of plasma concentrations was the body weights of subjects in this cohort, which ranged from 59.6 - 82.1 kg, with a mean of 72.28 kg. The model assumed a body weight of 60 kg for all subjects for added safety, as this would give higher plasma concentrations. The predicted plasma concentrations were adjusted for the mean body weight of 72.28 kg for subjects in this cohort. A plot with body weight adjusted to 72.28 Kg, compared to the original model using 60 kg and the actual mean plasma concentrations found in the subjects after administration of 200 mg KA2237, is shown in Figure 2b. The model, using the adjusted body weight, gave a better prediction of plasma concentrations especially for Cmin on days 6-7 and the full profile on day 14.
3.4 Plasma concentration - time profiles in subjects after all administered doses of KA2237
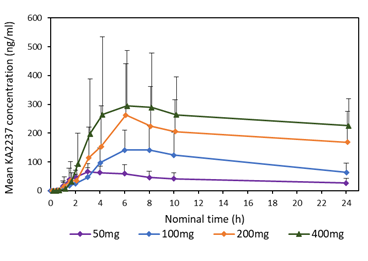
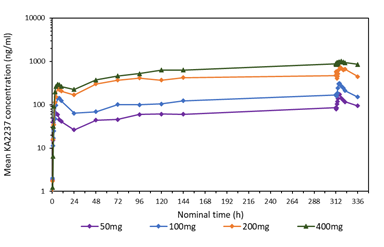
A comparison of the mean plasma concentration-time profiles after single dose of KA2237 on day 1, is shown in Figure 3 for all administered doses. On Day 1, there was a relatively rapid increase in plasma concentrations of KA2237 to peak values followed by a generally slow decline with KA2237 still quantifiable in plasma at 24h post-dose. Mean plasma concentration-time profiles after each dose of KA2237 over 14-day dosing are shown in Figure 4. On Day 14, all subjects had quantifiable levels of KA2237 in plasma at pre-dose. Plasma concentrations increased with increasing dose in an approximately linear manner up to the 200 mg dose, but the increase appeared less than linear between the 200 mg to 400 mg doses. Steady state appeared to be reached by Day 7, based on Cmin pre-dose concentrations. Plasma concentrations were higher on Day 14 compared with Day 1, for all doses, suggesting drug accumulation after multiple dosing.
3.5 PK parameters of KA2237
The main PK parameters for KA2237 calculated from plasma concentration-time data using NCA are summarised in Table 2.
Table 2: PK parameters of KA2237 calculated from plasma concentration-time data using NCA
|
Parameter/ |
Tmax (h) |
|
Cmax (ng/mL) |
Half-life (h) |
AUCtau (ng.h/mL) |
R (AUCtau) |
|||
|
Dose |
|
|
|||||||
|
|
Day 1 |
Day 14 |
Day 1 |
Day 14 |
Day 1 |
Day 14 |
Day 1 |
Day 14 |
D14/D1 |
|
50 mg |
|||||||||
|
n |
6 |
4 |
6 |
4 |
6 |
3 |
6 |
4 |
4 |
|
Mean* |
3.5 |
3.6 |
83.5 |
188 |
22.3 |
31 |
933.58 |
2827.91 |
2.8 |
|
SD |
24.81 |
90.56 |
12.52 |
3.02 |
405.41 |
1372.05 |
0.51 |
||
|
CV % |
45.4 |
18.2 |
29.7 |
48.2 |
56.2 |
9.8 |
43.4 |
48.5 |
18.2 |
|
100 mg |
|||||||||
|
n |
3 |
3 |
3 |
3 |
2 |
3 |
3 |
3 |
3 |
|
Mean* |
6 |
4 |
147.67 |
320 |
19.5 |
26.3 |
2169 |
4994.82 |
2.7 |
|
SD |
79.97 |
90.5 |
10.83 |
18.63 |
1282.53 |
1786.28 |
1.55 |
||
|
CV% |
19 |
24.7 |
54.2 |
28.3 |
55.4 |
71 |
59.1 |
35.8 |
57.2 |
|
200 mg |
|||||||||
|
n |
6 |
5 |
6 |
5 |
3 |
3 |
5 |
5 |
4 |
|
Mean* |
6 |
6.1 |
301 |
810.6 |
13.2 |
19.6 |
4397.97 |
14252.75 |
3.3 |
|
SD |
156.1 |
576.1 |
5.64 |
11.44 |
1978.33 |
12684.36 |
1.44 |
||
|
CV% |
95.4 |
47 |
51.9 |
71.1 |
42.8 |
58.5 |
45 |
89 |
43.6 |
|
400 mg |
|||||||||
|
n |
5 |
3 |
5 |
3 |
2 |
1 |
5 |
3 |
3 |
|
Mean* |
6 |
6.2 |
412.2 |
1069.7 |
22.9 |
31.1 |
5510.21 |
22086 |
3.9 |
|
SD |
187.73 |
791.09 |
13.12 |
NC |
2711.47 |
16922.01 |
1.51 |
||
|
CV% |
96.3 |
42.2 |
45.5 |
74 |
57.3 |
NC |
49.2 |
76.6 |
38.8 |
*median for Tmax; n=number evaluable in cohort; NC=not calculated
PK parameters were variable, however this may be due, in part, to the large variation in body weight between the subjects, 59.6 -129.7 kg, and that dosing was not based on body weight. Tmax was also highly variable, but this may reflect on the type of food intake prior to dosing, solid or liquid food supplement, and the exact timing of food intake. A food effect study will be done to fully evaluate this. Absorption of KA2237 was relatively rapid after all administered doses, with median Tmax ranging from 3.5 to 6.2 h. Mean Cmax increased with increasing dose and was higher on Day 14 compared with Day 1 for all doses, suggesting some drug accumulation. The mean apparent elimination half-life was around 20-30 h and was similar after all administered doses and did not appear to change after multiple dosing. This half-life is compatible with once daily dosing regimens. Mean AUCtau increased with increasing dose and was higher on Day 14 compared with Day 1, again suggesting some drug accumulation. Drug accumulation, measured as the ratio of AUCtau Day 14 / AUCtau Day 1, was modest and was 3.9-fold after 400 mg KA2237, the highest dose administered. Drug accumulation after 200 mg KA2237, the MTD and RP2 dose was 3.3-fold. Both Cmax and AUCtau increased approximately in proportion to the increase in dose from 50mg to 200mg, but appeared to be less than proportional from 200-400mg, both after single and repeat dosing.
3.6 Comparison of KA2237 accumulation ratio (AR) predicted by modelling, based on half-life and dosing interval, and AUC ratio.
Along with the AUC ratio, drug accumulation can also be calculated using the elimination half-life of the drug and the dosing interval, and assumes no inhibition:
AR=1/(1-e-k*tau) where k is the elimination rate constant, and tau is the dosing interval.
A comparison of the KA2237 accumulation ratio (AR) predicted by modelling, based on elimination half-life and dosing interval, and the AUCtau Day 14/AUCtau Day 1 ratio is shown in Table 3.
Table 3: Comparison of KA2237 accumulation ratio (AR) predicted by modelling, based on half-life and dosing interval, and AUC ratio
|
Dose (mg) |
Accumulation Ratio (AR) |
||
|
Model Predicted |
Half-life, Dosing interval |
AUCtau Day 14 / AUCtau Day 1 |
|
|
50 |
1.7 |
1.9 |
2.8 |
|
100 |
2 |
1.7 |
2.7 |
|
200 |
2.5 |
1.7 |
3.3 |
|
400 |
2.9 |
1.9 |
3.9 |
Around half the accumulation of KA2237, when administered once a day, could be explained solely on elimination half-life and dosing interval, this would suggest that CYP3A MBI may be less important than suggested from in vitro studies. However, at the highest dose of KA2237 administered, a further 2-fold of accumulation is likely to be due to CYP3A MBI. Model predicted AR was lower than that actually observed based on the AUC ratio.
4. Discussion
Results on safety and tolerability of KA2237 in this clinical study have been previously published by Nastoupil et al [12,13]. KA2237 was shown to be safe with an overall objective response rate of 37% in this refractory patient population. The alteration of drug-metabolizing enzyme activities can occur by several mechanisms including reversible inhibition, mechanism-based inactivation, and induction. For mechanism-based inhibitors, static mechanistic methodologies describing the prediction of in vivo drug-drug interactions (DDIs), particularly for CYP3A have been published [14, 15, 16, 8]. These studies, amongst others, formed the basis of the publication of Vieira et al [17] which has been included in the most recent FDA guidance on in vitro drug interaction studies [18].
More complex dynamic mechanistic models have been developed to predict the consequence of CYP3A4 inhibition. Zhang et al [19] used physiologically based PK (PBPK) modelling to study diltiazem a substrate of CYP3A4 which also displays MBI of CYP3A4. This model included parameters for the activity of CYP3A4 in gut wall and liver and accurately predicted plasma concentrations of diltiazem in human, after single and multiple dosing, and the effect on the AUC of midazolam, a sensitive CYP3A4 substrate. Filppula et al [11] constructed a mechanistic dynamic PBPK model to predict the effect of CYP3A4 MBI on imatinib pharmacokinetics. The model was validated by interaction simulations using ketoconazole, rifampicin, and ritonavir and was shown to be in good agreement with published studies. Thus, the in vitro data for CYP3A4 generated reliable predictions of its role in imatinib pharmacokinetics. Results showed that autoinhibition of CYP3A4 led to an increased role of CYP2C8 in imatinib metabolism.
In the present study a mechanistic dynamic model was developed to predict the potential effects of human CYP3A MBI on the repeat dose PK of KA2237 in patients. Our initial model compared favourably with the published model of Filppula [11], which was constructed within the Simcyp Population-Based Simulator (V11.00; Simcyp Limited, UK). We were fortunate that we could further validate our model using in vitro and in vivo PK data from NHP which also showed CYP3A MBI. Plasma concentrations, after 28 day repeat dosing in NHP, were adequately predicted with the further refined model. The successful prediction of the consequences of MBI in NHP, using appropriate in vitro and in vivo parameters as input to the model, gave greater confidence in the development and predictability of the human model.
The starting dose of 50 mg in this study was derived using the methodology described in the FDA guidance document [9]. This dose was simulated in the human MBI model and plasma concentration-time data generated. The dynamic MBI model predicted that after daily administration of 50 mg KA2237, steady state would be reached by day 7 and drug accumulation after 14 days administration would be around 1.7-fold. A comparison of the model-generated plasma concentrations with the actual concentrations, obtained after the 50 mg dose, demonstrated that there was excellent prediction of full profile plasma concentrations on days1 and 14 and of Cmin values on days 2-7. However, the MBI model slightly underpredicted accumulation, which, based on AUC ratio, was 2.8-fold. Based on this successful modelling, prediction of further doses was done using this approach to assist safe dose escalation.
A comparison of the predicted and actual mean plasma-concentration time profile after daily administration of 200 mg KA2237 showed that the model tended to over predict plasma concentrations. One possible explanation for this over prediction was that the model, in line with FDA recommendations, assumed a body weight of 60 kg for all subjects. The body weights of subjects in the 200 mg cohort ranged from 59.6 - 82.1 kg, with a mean of 72.28 kg. The model was adjusted to this mean body weight and gave a better prediction of plasma concentrations. The FDA recommendation for the use of a 60 kg body weight for predictions is a safe approach as it will, in most cases, lead to estimated plasma concentrations that are over predictions rather than underpredictions, which could have an impact on safety.
PK parameters of KA2237 were variable, this could be due, in part, to the small number of patients in each dosing cohort, the different pathophysiology of patients and the large variation in body weight between the subjects (59.6 -129.7 kg) as dosing was not based on body weight. The possible effect of the intake of solid food compared with liquid food supplements on drug absorption was evident in the variability of Tmax. However, absorption of KA2237 was relatively rapid after all administered doses, with median Tmax ranging from 3.5 to 6.2 hrs. Both mean Cmax and AUCtau increased approximately in proportion to the increase in dose from 50mg to 200mg, but appeared to be less proportional from 200-400mg.The mean apparent elimination half-life of 20-30 h did not appear to be dose dependent or dependent on the number of days of dosing. This half-life is compatible with once daily dosing regimens. Drug accumulation after 200 mg KA2237, the MTD and RP2 dose, was modest and around 2.5,1.7 and 3.3-fold, based on MBI model predictions; elimination half-life and dosing interval; and AUC ratios, respectively. Accumulation assessed by half-life and dosing interval assumes no inhibition, this suggests that the small additional accumulation, observed with the MBI model and AUC ratios, was likely to be due to reduced KA2237 clearance due to MBI.
Pre-clinical studies demonstrated that biliary and renal clearance of KA2237 is low, therefore all clearance was considered metabolic. The model assumed that all hepatic metabolism was through CYP3A, however, in the initial model developed using NHP data, the best-fit was obtained when the fraction of clearance that is hepatic was set to 0.67 and this value was also used in the human model. Accumulation was highly sensitive to this parameter, as with increasing dose of KA2237 clearance would be reduced due to time-dependent inhibition of CYP3A. With the availability of actual clinical data, the model was re-run using the data obtained from the 200 mg dose and the fraction of clearance that is hepatic set to 1.0. This assumed that all clearance was hepatic through metabolism by CYP3A. Using this value of 1.0, gave an accumulation value of 7-fold compared with 2.5-fold using the original value for the fraction of clearance that is hepatic of 0.67. These results indicated that not all of KA2237 metabolism is through CYP3A and that alternative pathways may be involved.
With the wide commercial availability of recombinant human CYPs and human liver microsomes, cytochrome P450 enzymes can be readily studied in early drug development. The assessment of potential drug inhibition of human CYPs, using these in vitro methodologies, is now standard practice. However, other enzymes may also be involved in drug metabolism, but, due to the lack of available models, they have not been as extensively studied and can sometimes be overlooked. Turnover of KA2237 was observed in human hepatocytes and this appeared to be higher than in microsomes. As hepatocytes contain both CYPs and cytosol enzymes, these cytosol enzymes could act as an alternative pathway to the auto-inhibition of CYP3A by KA2237. KA2237 has nitrogen containing heterocycles and many drugs with this chemotype have been shown to be substrates for aldehyde oxidase [20]. More work needs to be done to fully characterise the major enzymes involved in KA2237 metabolism, but, based on the modelling results and actual clinical results, it would appear that MBI of CYP3A is less important than originally assessed.
5. Conclusion
KA2237 is a selective dual inhibitor of PI3K β/δ with a PK profile that allows once a day oral dosing. KA2237 exhibits CYP3A MBI, but at the RP2 daily dose of 200 mg the accumulation, based on AUC, is modest at 3.3-fold and approximately half of this may be explained by the accumulation due to half-life and dosing interval (1.7-fold). An appropriate mechanistic dynamic model was developed to assess the extent of accumulation prior to the start of FIH studies. This, along with the close monitoring of drug levels and clinical safety data during the on-going study, led to the safe conduct of the study and the achievement of required plasma concentrations for activity. The findings of this study support the further evaluation of KA2237.
References
- Vanhaesebroeck B, Stephens L, Hawkins P. PI3K signalling: the path to discovery and understanding. Nature reviews 13 (2012): 195-203.
- Gopal AK, Kahl BS, de Vos S et al. PI3Kδ inhibition by idelalisib in patients with relapsed indolent lymphoma. N Engl J Med 370 (2014): 1008-18.
- Dreyling M, Campo E, Hermine O et al. Newly diagnosed and relapsed mantle cell lymphoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 28 (2017): iv62-iv71.
- Flinn IW, van der Jagt R, Kahl B et al. First-line treatment of patients with indolent non-hodgkin lymphoma or mantle-cell lymphoma with bendamustine plus rituximab versus R-CHOP or R-CVP: results of the BRIGHT 5-year follow-up study. J Clin Oncol 37 (2019): 984-991.
- Lu Y, Yi Y, Liu P et al. Common human cancer genes discovered by integrated gene-expression analysis. PLoS 2 (2007).
- Wee S, Wiederschain D, Maira SM et al. PTEN-deficient cancers depend on PIK3CB. Proc Natl Acad Sci U S A 105 (2008): 13057-62.
- Ni J, Liu Q, Xie S et al. Functional characterization of an isoform-selective inhibitor of PI3K-p110β as a potential anticancer agent. Cancer Discov 2 (2012): 425-433.
- Galetin A, Burt H, Gibbons L, Houston J B. Prediction of time-dependent CYP3A4 drug-drug interactions: impact of enzyme degradation, parallel elimination pathways, and intestinal inhibition Drug Metab Dispos 34 (2006): 166-175.
- Food and Drug Administration, Guidance for Industry: ICH S9 Nonclinical Evaluation for Anticancer Pharmaceuticals (2010).
- Rowland Yeo K, Walsky RL, Jamei M, Rostami-Hodjegan A, Tucker GT. Prediction of time-dependent CYP3A4 drug-drug interactions by physiologically based pharmacokinetic modelling: impact of inactivation parameters and enzyme turnover. Eur J Pharm Sci 43 (2011): 160-173.
- Filppula AM, Neuvonen M, Laitila J, Neuvonen PJ, Backman JT. Autoinhibition of CYP3A4 leads to important role of CYP2C8 in imatinib metabolism: variability in CYP2C8 activity may alter plasma concentrations and responses. Drug Metab Dispos 41 (2013): 50-59.
- Nastoupil LJ, Neelapu SS, Davis RE et al. Results of a First in Human, Dose Ascending, Phase I Study Examining the Safety and Tolerability of KA2237, an Oral PI3K p110β/δ Inhibitor, in Patients with Relapsed/Refractory (R/R) B-cell Lymphoma. Blood 134 (2019): 4099.
- Nastoupil LJ, Neelapu SS, Davis RE et al. Preclinical and phase I studies of KA2237, a selective and potent inhibitor of PI3K β/δ in refractory B cell lymphoma. Leukemia & Lymphoma 62 (2021):3452-3462.
- Mayhew BS, Jones DR, Hall SD. An in vitro model for predicting in vivo inhibition of cytochrome P450 3A4 by metabolic intermediate complex formation. Drug Metab Dispos 28 (2000): 1031-1037.
- Yamano K, Yamamoto K, Katashima M et al. Prediction of midazolam-CYP3A inhibitors interaction in the human liver from in vivo/in vitro absorption, distribution, and metabolism data. Drug Metab Dispos 29 (2001): 443-452.
- Ito K, Ogihara K, Kanamitsu S, Itoh T. Prediction of the in vivo interaction between midazolam and macrolides based on in vitro studies using human liver microsomes. Drug Metab Dispos 31 (2003): 945-954.
- Vieira LT, Kirby B, Ragueneau-Majlessi I et al. Evaluation of various static in vitro-in vivo extrapolation models for risk assessment of the CYP3A inhibition potential of an investigational drug. Clin Pharmacol Ther 95 (2014): 189-198.
- Food and Drug Administration, Guidance for Industry: In Vitro Drug Interaction Studies Cytochrome P450 Enzyme- and Transporter-Mediated Drug Interactions January (2020).
- Zhang X, Quinney SK, Gorski JC, Jones DR, Hall SD. Semiphysiologically based pharmacokinetic models for the inhibition of midazolam clearance by diltiazem and its major metabolite. Drug Metab Dispos 37 (2009): 1587-1597.
- Kitamaru S, Sugihara K, Ohta S. Drug-metabolizing ability of molybdenum hydroxylases. Drug Metab Pharmacokinet 21 (2006): 83-98.


 Impact Factor: * 3.3
Impact Factor: * 3.3 Acceptance Rate: 74.39%
Acceptance Rate: 74.39%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 