Karyopherins in the Remodeling of Extracellular Matrix: Implications in Tendon Injury
Connor Diaz1, Finosh G. Thankam2, Devendra K. Agrawal2*
1University of Missouri School of Medicine, Springfield Clinical Campus, Springfield, MO 65807, USA
2Department of Translational Research, Western University of Health Sciences, Pomona, California 91766, USA
*Corresponding Author: Devendra K. Agrawal, Department of Translational Research, Western University of Health Sciences, Pomona, California 91766, USA.
Received: 30 August 2023; Accepted: 06 September 2023; Published: 14 September 2023
Article Information
Citation:
Connor Diaz, Finosh G. Thankam, Devendra K. Agrawal. Karyopherins in the Remodeling of Extracellular Matrix: Implications in Tendon Injury. Journal of Orthopedics and Sports Medicine. 5 (2023): 357-374.
View / Download Pdf Share at FacebookAbstract
Rotator Cuff Tendinopathies (RCT) are debilitating conditions characterized by alterations in the extracellular matrix (ECM) of the shoulder tendon, resulting in pain, discomfort, and functional limitations. Specific mediators, including HIF-1 α, TGF-β, MMP-9 and others have been implicated in the morphological changes observed in the tendon ECM. These mediators rely on karyopherins, a family of nuclear proteins involved in nucleo-cytoplasmic transport; however, the role of karyopherins in RCT remains understudied despite their potential role in nuclear transport mechanisms. Also, the understanding regarding the precise contributions of karyopherins in RCT holds great promise for deciphering the underlying pathophysiological mechanisms of the disease and potentially fostering the development of targeted therapeutic strategies. This article critically discusses the implications, possibilities, and perspectives of karyopherins in the pathophysiology of RCT.
Keywords
<p>Exportins; Importins; Karyopherins; Rotator Cuff Tendinopathies; Shoulder tendon</p>
Article Details
1. Introduction
Rotator Cuff Tendinopathies (RCT) are associated with pain, discomfort, and functional impairment owing to changes or destruction of the extracellular matrix (ECM) of the shoulder tendon [1]. Also, specific mediators such as HIF-1α [2], TGF-β1 [3], and MMP-9 [4] are linked to the observed changes in the morphology of tendon ECM. Transcription of these mediators largely relies on the activity and function of a family of nuclear proteins, the karyopherins. Karyopherins, such as importins, are crucial for the nucleo-cytoplasmic bidirectional transport of specific transcriptional mediators facilitating the movement of cargo into and out of the nucleus [5,6]. Importantly, the regulation of such bidirectional nucleo-cytoplasmic transport is crucial for the tissue homeostasis; however, the information on karyopherins especially the importin sub-types in relation to RCT is limited. On this background, this review comprehends the implications and perspectives of karyopherin sub-types in the pathophysiology of RCT.
2. Karyopherin Biology
Karyopherins represent a family of transporter proteins aiding to shuttle cargo between nucleus and cytoplasm comprising two major families, importin and exportin. Importin functions on transporting specific biomolecules between the cytoplasm and the nucleus mediated by specific nuclear localization signal (NLS) [7]. Structurally, importin exists as heterodimer composing importin-α and importin-β subunits. Importin-α was discovered as a 60kD cytosolic protein in Xenopus eggs that exhibited 44% sequence identity to the nuclear pore complex in yeast [8]. Eventually, the cDNA construct of importin-a was further cloned and studied in E coli system unveiling the conduits of nucleocytoplasmic transport [9]. Importin-β was identified by Michael Rexach and Dirk Görlich as 90kD protein subunit that dissociated from importin-a using affinity chromatography on nickel (II)-nitrilotriacetic acid-Sepharose [10]. These findings confirmed that the 60kD importin-a and 90kD importin-β subunits function synergistically in targeting substrate import at the nuclear envelopes of mammalian cells [11].
Exportin is responsible for the nucleocytoplasmic transport of cellular cargo from the nucleus to the cytoplasm [12]. The major cellular function of exportin is to transport mature mRNA and 5’ and 3’ end processed tRNA from the nucleus to the cytoplasm [13]. Multiple sub-types of exportin exist and the X-ray structures of exportin-5 and exportin-1 have been determined [14]. In the cargo-free state, Exportin-5 is stabilized by a ring-shaped closed conformation through interactions between C-terminal anchors and N-terminal heat repeats. Upon the interaction of exportin with RanGTP, the C-terminal anchors are removed resulting in the conformational change in the N-terminal heat repeats, and thus creating an open space cargo binding [14]. Exportin-1 transports molecules with highly variable nuclear export signals (NES) [15]. X-ray crystallography of exportin-1 bound to cargo containing different NES has revealed that exportin-1 adopts a wide range of diverse conformations which vary from loop-like to all-helix depending on the invariant NES-binding groove [16].
Karyopherins are classified into karyopherin-α (KPNA) and karyopherin-β families (KPNB) and each member can recognize its own unique set of cargo and RNA [17]. KPNA orthologues are further subdivided into subfamilies α1, α2 and α3 based on differences found within their amino acid sequence. Each subfamily further contains a unique set of genes and proteins [18]. The KPNA subfamilies have been broken into their respective genes in Table 1 along with their current identified interactions. Similarly, the KPNB family contains 18 orthologues which are divided into three groups based on whether they function as exportins, importins, or for bidirectional transport [19]. This family has subfamilies and genes with respective interactions are given in Table 2.
|
α1 |
α2 |
α3 |
|||
|
Gene |
Interactions |
Gene |
Interactions |
Gene |
Interactions |
|
KPNA2 |
CDKN1B HSPA4 ITK STAT1 SV40gp6 SV40gp6 IXNIP APOBECI ARLAA ARLSA BRCA1 CASP2 CDA CHD3 CHEK2 CREBBP CSNK1A1 EP300 EPB41 H1-0 HAP1 |
KPNA3 |
CDKN1B COIL FAM50B KPNB1 NFKB1 RCC1 IGM2 ACACA ACE2 ACIN1 ACTRIA AKIRIN2 ANKRD11 APEX1 APOL6 ARNT ARRB1 ARRB2 ATG16L1 ATM ATOSB B2M BAHD1 BAP1 BARD1 |
KPNA1 |
ANP32A ELAVL1 H2BC21 H3-4 LEF1 LMO4 PAX5 RAG1 RUNX1T1 SRP19 STAT1 STAT3 TAF9 UBR5 ADNP ADNP2 AICDA ALKBH5 ANP32A ANP32B ANP32E APEX1 ARHGEF7 ARID2 ARNT |
|
KPNA7 |
H1-2 IKBKG LARP7 MEPCE SLEN11 TWIST1 USP1 |
KPNA4 |
IGM2 RECOL TGM2 ACLY ACTC1 ACTRIA ACTR1B ADNP AFF2 AHSA1 AICDA AKIRIN2 APP ARMC8 ARRB2 B2M BAP1 BCOR C9orf40 CALCOCO2 CANX CBX5 CCAR1 CCNT1 CCT2 |
KPNA5 |
ANP32A BRMS1 COKN1B ADNP AIPL1 ANP32A ANP32B ANP32E ARNT ATM ATOSB BRMS1 CHD4 CHD8 CUL3 DCPS E2F4 EPAS1 FBL FBXO7 FOXE1 GATAD2B H3C1 HIF1A HNRNPC |
|
KPNA6 |
ANP32A BRCA1 CSE1L KPNB1 RELB STAT1 STAT TAF9 ACTC1 ALKBH5 ANKRD11 ANP32A ANP32B ANP32E ARK2N ARMC8 ARNI BAP1 BCOR BIRC3 BZW1 CACUL1 CARD19 CBX1 CBX3 |
||||
Table1: KPNA subfamilies with their associated genes and interactions.
KPNA consists of 3 subfamilies – α1, α2, α3. Subfamily α1 contains genes KPNA1 and KPNA7. Subfamily α2 contains KPNA3 and KPNA4. Subfamily α3 contains KPNA2, KPN5, and KPNA6. The interactions currently identified in the National Library of Medicine National Center for Biotechnology Information for each gene have been included in the table. Information regarding KPNA1 can be found at https://pubchem.ncbi.nlm.nih.gov/gene/KPNA1/human. Information regarding KPNA2 can be found at https://www.ncbi.nlm.nih.gov/gene/3838. Information regarding KPNA3 can be found at https://www.ncbi.nlm.nih.gov/gene/3839. Information regarding KPNA4 can be found at https://www.ncbi.nlm.nih.gov/gene/3840. Information regarding KPNA5 can be found at https://www.ncbi.nlm.nih.gov/gene/3841. Information regarding KPNA6 can be found at https://www.ncbi.nlm.nih.gov/gene/23633. Information regarding KPNA7 can be found at https://www.ncbi.nlm.nih.gov/gene/10527.
Table 2: KPNB subfamilies with their associated genes and interactions.
KPNB consists of 3 subfamilies based off their direction of nucleocytoplasmic transport – importin, exportin, or bidirectional transport. Like the KPNA family, each subfamily contains a unique set of genes. The interactions currently identified in the National Library of Medicine National Center for Biotechnology Information for each gene have been included in the table. Information regarding XPO1 can be found at https://pubchem.ncbi.nlm.nih.gov/gene/XPO1/human. Information regarding XPO2 can be found at https://www.ncbi.nlm.nih.gov/gene/171621. Information regarding XPOT can be found at https://pubchem.ncbi.nlm.nih.gov/gene/XPOT/human. Information regarding XPO-5 can be found at https://www.ncbi.nlm.nih.gov/gene/57510. Information regarding KPNB1 can be found at https://pubchem.ncbi.nlm.nih.gov/gene/KPNB1/human. Information regarding TNPO1 can be found at https://pubchem.ncbi.nlm.nih.gov/gene/TNPO1/human. Information regarding TNPO2 can be found at https://www.ncbi.nlm.nih.gov/gene/30000. Information regarding TNPO3 can be found at https://www.ncbi.nlm.nih.gov/gene/23534. Information regarding IPO-5 can be found at https://www.ncbi.nlm.nih.gov/gene/3841. Information regarding IPO-7 can be found at https://www.ncbi.nlm.nih.gov/gene/10527. Information regarding IPO-8 can be found at https://www.ncbi.nlm.nih.gov/gene/10526. Information regarding IPO-9 can be found at https://www.ncbi.nlm.nih.gov/gene/55705. Information regarding IPO-11 can be found at https://www.ncbi.nlm.nih.gov/gene/51194. Information regarding IPO-4 can be found at https://www.ncbi.nlm.nih.gov/gene/79711. Information regarding IPO-13 can be found at https://www.ncbi.nlm.nih.gov/gene/9670.
3. Structure of Importin
Importin-α has distinct domains; the N-terminal domain is the Impβ1-binding (αIBB) domain and C-terminal domain or the central domain. The central domain has two major binding grooves created by ten tandem flexible armadillo motifs of three α helices encoded by ~40 amino acid residues [20,21]. The consecutive stacking of these motifs creates a spherical shaped molecule that has the appearance of a groove [20,22]. Additionally, within the C-terminal domain, there are two grooves: a major and minor groove. These grooves function as receptors and the binding site for the NLS of cargo and are lined with tryptophan and asparagine residues which in turn are surrounded by acidic amino acids [20]. These grooves recognize and interact with the basic (positively charged) NLS through hydrophobic and electrostatic interactions [23]. Between the two binding grooves is a linker-binding region that interacts with the NLS backbone [23]. Importantly, the changes in residues within these three regions affect the nuclear localization and binding of the NLS to importin-α [24,25].
Similarly, importin-α is composed of 18-20 amino acid residues of the HEAT repeat motif composing of huntingtin, elongation factor 3, the 65kD alpha regulatory subunit of phosphatase 2A (PP2A) and the yeast PI3-Kinase TOR 1 [26]. These HEAT motifs contain two antiparallel alpha helices linked together by a turn in which the secondary structure of the polypeptide chain reverses its direction. These helices stack together to form mature importin-α [27]. A major function of importin-α is to interact with nucleoporins by weak, transient bonds between the surface hydrophobic pockets and the FG repeats found within the nucleoporin [28].
4. Nuclear Localization Signals
While all cargo transported into the nucleus by importin are mediated by NLS, these signals are not necessarily coded by the same sequence. Thus, importin-α recognizes two specific classes of NLS: classical (cNLS) and non-classical. cNLS bears both monopartite and bipartite signals. Monopartite NLS consist of one stretch of basic amino acids while bipartite NLS contain two stretches of basic amino acids [29-31]. Within bipartite NLS, the two basic stretches are separated by an amino acid “linker” region that allows the NLS to maintain its functionality and to withstand point mutations and insertions [22,30,32,33]. Both basic domains are necessary for the integrity of bipartite NLS to retain their functionality in nuclear targeting.
Monopartite NLS are traditionally represented by the SV40 large T antigen NLS (126-PKKKRRV-132) while bipartite NLS are represented by the nucleoplasmin NLS (155-KRPAATKKAGQAKKKK-170) [23]. Structural [22,32,33] and thermodynamic [34] studies have identified the key structural/chemistry requirements for cNLS transport. Monopartite cNLS requires a most critical Lys-155 that is stabilized by Gly-161, Thr-166, and Asp-203 [20]. The major binding groove of importin-α is located near the N-terminus and binds monopartite NLS and the larger stretch of basic residues composing the bipartite NLS [23]. The minor groove of importin-α, located closer to the C-terminal, binds the basic residues composing the bipartite NLS [23]. cNLS binds to the grooves of importin-α in an extended conformation in which their main chains bind antiparallel to the importin-α chain [23].
Furthermore, there are other types of non-classical NLS that differ from the binding chemistry of classical NLS. While nuclear import of cNLS-cargo requires the presence of both importin-α and importin-b, non-classical NLS typically are recognized solely by the importin-α family without the importin-α [35]. For example, parathyroid hormone-related protein (PTHrP) has been imported to the nucleus through a direct interaction with importin-b [36]; however, in some cases, importin-α plays a role in non-classical NLS cargo import. Also, the non-classical NLS of the influenza virus nucleoprotein binds specifically and exclusively to the minor groove of importin-α with low affinity [37].
5. Cytoplasm to Nucleus
Eukaryotic cells regulate the bidirectional transport of cargo between the cytoplasm and the nucleus through nuclear pore complexes (NPCs) which regulates the transport of cargo both into and out of the nucleus [38]. NPCs are composed of approximately 30 distinct proteins called nucleoporins (Nups) forming the central constituents of the complex. The fundamental structural components of NPCs include the inner pore ring, which is bound to both the inner and outer nuclear envelope; the nuclear and cytoplasmic rings, which are fused to the inner pore ring; the nuclear basket; and cytoplasmic filaments [39]. Within the inner pore ring is a central channel which allows for the passive diffusion of small molecules, while preventing the diffusion of large macromolecules [35]. For large molecules to enter the nucleus, the C-terminal region of importin-α recognizes cargo containing an NLS and the N-terminus of importin-α bind to the central domain of importin-β [40]. Subsequently, importin-α binds to both the tagged cargo and importin-β, forming a heterodimer. The recognition and binding between the two importin molecules are attributed to importin beta binding (IBB) domain [40]. Once the heterodimer is intact, importin-β interacts with FG-repeats of nucleoporins (Phe-Gly sequence repeats) which facilitates the diffusion of the heterodimer-cargo complex through the NPC to the nucleus [41]. The FG-repeats within the nucleoporins are enriched with phenylalanine and glycine residues and separated by hydrophilic linkers. Also, the FG repeats provide direct and specific interactions with the transport receptors, which provide the structural basis for the translocation of the cargo across the nuclear envelope [41].
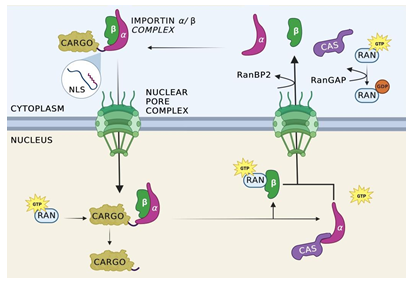
Figure 1: Import cycle for importin-α/β.
Nuclear import is initiated with the binding of the alpha and beta subunits of importin to cellular cargo tagged with an NLS. Once the complex is formed, importin-β interacts with the FG-repeats of a nucleoporin, allowing for the complex to pass into the nucleus. Once inside the nucleus, Ran-GTP binds allosterically to the importin complex, causing the complex to dissociate and releasing the cargo from importin into the nuclear space. Importin-α is recycled back to the cytosol through the mediation of cellular apoptosis susceptibility protein (CAS). Importin- β is transported separately without the aid of an additional protein. Once the complex has been exported from the nucleus and into the cytoplasm, Ran Binding Proteins facilitate the separation of Ran-GTP from importin-β and importin-β, resetting the cycle.
Once the heterodimer-cargo complex has diffused into the nucleus, dissociation of the nuclear import complex occurs to release the cargo within the nucleus. Ran-GTP binds allosterically to both importin-α and importin-β present in the complex, causing a conformational change which facilitates the release of the cargo from importin [21]. Importin-α recycles back to the cytosol through the mediation of cellular apoptosis susceptibility protein (CAS) [42]. However, the Ran-GTP complex remains bound to importin-α, preventing importin-α from binding to additional targets. Once the complex has been exported from the nucleus and into the cytoplasm, Ran Binding Protein (RanBP2) facilitates the separation of Ran-GTP from importin allowing importin-β to regain its function in the cytoplasm. Additionally, this separation provides access to Ran GTPase activating proteins (RanGAP) which facilitate the hydrolysis of the guanine nucleotides in Ran-GTP to produce Ran-GDP [21]. Importin-β is transported out of the nucleus independently and in the cytoplasm, Ran Binding Proteins separates Ran-GTP from importin-β, freeing the molecule for subsequent transport cycles. The import cycle is schematically shown in Figure 1.
6. Nucleus to Cytoplasm
Cargo transport from the nucleus to the cytoplasm generally reverses the nuclear import process. Within the nucleus, distinct macromolecules need to be transported from the nucleus to the cytoplasm. This is essential for processes such as DNA replication and RNA synthesis. These macromolecules are transported from the nucleus to the cytoplasm after being tagged with a Nuclear Export Sequence (NES) which is recognized by the nucleo-cytoplasmic transporter, exportin. Studies have shown that Crm1 (exportin 1) is a necessary element for nuclear export process. Crm1 acts as a NES receptor that facilitates the diffusion of macromolecules across the nuclear envelope [43]. In addition, Crm1 forms a leptomycin B-sensitive complex, which provides a co-operative binding site for Ran-GTP and the NES [44]. Additional studies conducted on budding or fission yeast with mutated Crm1 genes revealed nuclear export was inhibited. This data further supports the requirement of Crm1 in the adequate function of the nuclear export process. Importantly, Crm1 binds to several different nucleoporins including Nup214 [45] confirming their major role in the adequate functioning of the nuclear export system.
Ran-GTP binds to exportin through its N-terminal domain which is inevitable for the exportin to bind the cargo [15,46-48]. Importantly, the interaction between Ran-GTP and exportin occur only in the presence of cargo leading to form a trimeric export [49]. Moreover, RanBP1 and RanBP2 play a crucial role in the release of Ran-GTP from the nuclear export complex. RanBP1 and/or RanBP2 bind to Ran-GTP in the export complex on the cytoplasmic side of the NPC resulting in a conformational change in Ran-GTP which leads to the dissociation of Ran-GTP from Crm1 releasing the cargo from Crm1, and concurrently, the dissociation of Crm1 from the NPC [50]. Furthermore, RanBP2 and RanBP1 stimulate RanGAP, which triggers the cleavage of Ran-GTP to Ran-GDP and renders the reaction irreversible as Crm1 has limited affinity to Ran-GDP [51-53]. This process completes the export of cargo from the nucleus to the cytoplasm. Once completed, Ran and Crm1 recycle back to the nucleus through distinctive mechanisms [54]. The transport factor, NTF2, facilitates the import of cytosolic Ran-GDP back into the nucleus, restoring the concentrations of Ran 52, 57–59. NTF2 interacts with the FG repeats allowing the translocation of Ran back into the nucleus where Ran Guanine Nucleotide Exchange Factor (RanGEF/RCC1) facilitates the dissociation of NTF2 from Ran-GDP by converting Ran-GDP to Ran-GTP. RanGEF is strictly found in the nucleus while RanGAP is localized within the cytoplasm creating a compartmentalized asymmetry of Ran-GTP across the nuclear envelope for the directionality of nucleo-cytoplasmic transport. This export process is shown in Figure 2.
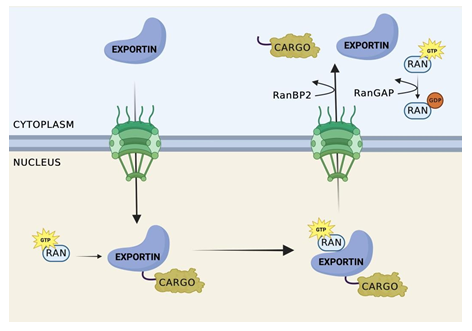
Figure 2: Nuclear Export Cycle.
Exportin recognizes cargo containing a NES. Once recognized, cooperative binding of RanGTP and cargo occurs, forming a trimeric export complex. Exportin then facilitates the transfer of the trimeric complex through NPC to the cytoplasmic side of the nucleus. The complex remains attached to the cytoplasmic side of the NPC until RanBP1 and/or RanBP2 bind to Ran-GTP on the complex on the cytoplasmic side of the NPC. The binding of Ran Binding Proteins to Ran-GTP leads to a conformational change in the trimeric complex, resulting in the dissociation of Ran-GTP from Crm1. This dissociation releases the cargo from Crm1, and concurrently, the dissociation of Crm1 from the NPC.
7. Perspectives of karyopherins in RCT
Karyopherins may play a critical role in the signaling pathways of rotator cuff tendinopathies (RCTs) by regulating the nucleocytoplasmic transport of key mediators, including HIF-1α [2], TGF-β [3], and MMP-9 [4], which have been associated with changes in tendon extracellular matrix (ECM) remodeling. Specifically, karyopherins facilitate the nuclear import of Smad proteins in the TGF-β/Smad3 pathway, thereby regulating gene transcription and downstream signaling [3]. Additionally, karyopherins are involved in the nuclear transport of HIF-1α, an essential protein for hypoxic conditions and tissue repair [55,56]. Importin proteins also contribute to the transportation of NF-κβ, a transcription factor implicated in various inflammatory conditions and tissue remodeling in various diseases, influencing the expression of pro-inflammatory genes and MMPs [57]. Interestingly, the function of specific importins and exportins could be modulated by immunomodulators, such as vitamin D [58-60]. By understanding the interplay between karyopherins and these signaling pathways, valuable insights into the underlying mechanisms of RCT pathology can be gained, leading to the development of therapeutic strategies aimed at enhancing healing and reducing fibrosis.
8. TGF- β/Smad3 Pathway and Karyopherins
Following RCT, the tendon fibrosis hinders tendon healing via the activation of transforming growth factor-β (TGF-β) signaling as an integral role in this process. Importantly, the TGF-β/Smad3 pathway promotes the proliferation and collagen deposition by tendon fibroblasts while inhibiting the fibroblast apoptosis [61]. Interestingly, TGF-β upregulation in the tendon fibroblasts [62,63] increases the phosphorylation of Smad2, Smad3, and the conversion of fibroblasts into myofibroblasts [64,65]. Initially, myofibroblasts contribute to tendon healing by contracting, remodeling the ECM, forming fibrous scars, and improving injury stability [66,67]. However, their sustained presence in later phases leads to tendon fibrosis and impaired function [68]. Therefore, targeting TGF-β activation is essential to reduce tendon fibrosis and promote healing from RCT [69] where the Smad family of proteins serve as key regulators in the downstream propagation of TGF-β [70]. This family includes nine proteins, namely Smad1 to Smad9, divided into three categories: R-Smads (receptor-regulatory Smads), Co-Smads (Co-mediating Smads), and I-Smads (inhibitory Smads). R-Smads are activated by TGF-β receptors [71-73], Co-Smad4 forms complexes with activated R-Smads and plays a regulatory role in TGF-β signal transduction [74]. I-Smads reside in resting cell nuclei and enter the cytoplasm upon TGF-β stimulation to interfere with R-Smad recruitment and phosphorylation and Co-Smad complex formation [75]. Therefore, I-Smads act as negative regulators to the TGF-β signal transduction pathway.
Smad proteins consist of two conserved domains connected by a proline-rich non-conserved intermediate linking region. The N-terminal Mad homolog domain 1 (MH1) acts as a transcription factor, while the C-terminal Mad homolog domain 2 (MH2) facilitates protein-protein interactions. When TGF-β receptors are activated, R-Smad proteins undergo phosphorylation at their C-terminals [76] facilitating the translocation of R-Smads and Co-Smad complex to the nucleus to regulate gene transcription [76-79]. Karyopherins play a crucial role in the nuclear transport of Smads. For instance, the N-terminal conserved MH1 domain of Smad 3 interacts with importin-β1 for nuclear import [3]. This interaction with karyopherins, therefore, serves as the final regulatory mediator of the TGF-β signaling cascade. The Smad complex enters the nucleus and bind to TGF-β-responsive sequences (TBRS) (between -174 and -84 bp) of downstream transcription start site of COL1α1 gene [80].
Studies have demonstrated the significance of the TGF-β/Smad3 pathway in tendon healing. In a SiRNA rat model of rotator cuff injury, inhibition of Smad3 led to improved bone-tendon junction structure, collagen fiber continuity, and organization compared to controls [81]. Similarly, Smad 3 (-/-) mice exhibited better range of motion and reduced scar formation following flexor digitorum longus tendon repair [82]. These findings suggest that modulation of the TGF-β /Smad3 pathway enhance healing after RCT. Unfortunately, the information regarding the involvement of TGF-β and/or Smad translocation by karyopherins in RCT are currently unavailable. However, it is logical that the dysregulation of nuclear transport of Smad proteins occurs following RCT, contributing to increased TGF-β activity and subsequent fibrosis that hampers healing. The possible mechanisms of TGF-β-karyophrenes axes age depicted in Figure 3. Further research is warranted along this direction and to design potential techniques to modulate the Smad- karyopherin interaction for therapeutic purposes.
9. HIF-1α and Karyopherins
Millar et al. [83] and Mosca et. al. [2] reported that hypoxia regulated early tendinopathy with an increased level of HIF-1α in apoptotic supraspinatus cells, suggesting a potential role of HIF-1α RCT pathology [2,83,84]. Under normoxic conditions, HIF-1α undergoes proline hydroxylation leading to its proteolysis via polyubiquitination whereas hypoxia inhibits the hydroxylation stabilizing the HIF-1 complex [85]. HIF-1α upregulates collagen genes through activation of NOTCH1, NOTCH1 ligand (JAGGED1), and hairy and enhancer of split-1 (HES1) [86]. Upon binding of HIF-1, the presenilin proteases (PSEN1/2) cleave the intracellular domain of NOTCH to form the Notch Intracellular Domain (NICD) [87] for import into the nucleus by importin-α3, -4, and -7 [56]. In the nucleus, NICD interacts with immunoglobulin k J region (RBP-Jk) and Mastermind-like (MAML) proteins which in turn trigger the transcription of HES and HEY genes (mammalian counterparts of drosophila HES1) [88,89]. Interestingly, overexpression of NICD in transgenic mice inhibited osteoblast differentiation and reduced production of type I collagen, resulting in severe osteosclerosis [90]. Additionally, the HIF-1α/NOTCH pathway suppressed the expression of MMP1 and MMP13 and increased expression of TIMP1. MMP1 and MMP13 are the key regulators in ECM turnover and are crucial for ECM degradation [91,92]. In addition, HIF-1α promotes expression of pro-angiogenic factors, including vascular endothelial growth factor (VEGF), which stimulates the growth of new blood vessels into injured tissue contributing to tissue repair by improving the oxygen and nutrient [93]. However, the invasion of endothelial cells during angiogenesis has been associated with potential weakening of tendon stability [94]. Therefore, the contribution of angiogenesis to the healing process in RCT remains a topic of debate.
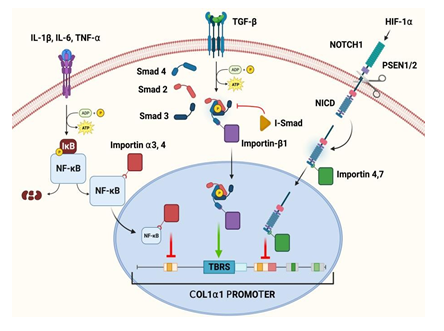
Figure 3: Pathways of key mediators in the regulation of type-1 collagen and the role of specific karyopherins in these pathways.
These studies highlight the important role which HIF-1α plays in the healing process of RCT. HIF-1α is exported to the cytoplasm by Exportin 1 (CRM1) using a cNES regulated by MAPK-dependent phosphorylation of two serine residues [55]. Hence, karyopherins are crucial for HIF-1α signaling in RCT. Moreover, the disruptions in the import, export, or both may lead to aberrant gene expression patterns and compromised cellular responses aggravating the RCT pathology/healing responses. The possible mechanisms of HIF-1α-karyopherin axes are depicted in Figure 3. However, the information regarding the association of HIF-1α signaling and karyopherin biology are obscure warranting further investigations. Consequently, understanding the interaction between HIF-1α and karyopherins would open novel avenues to enhance RCT healing processes.
10. NF-ΚΒ and Importin
Following RCT, NF-κβ activation occurs in response to the convergence of pro-inflammatory chemokines including TNF-α, IL-1β, IL-6, and IL-18 released by inflammatory cells [86]. The activation of NF-κβ results in the transcription of pro-inflammatory genes which leads to the production of additional cytokines, chemokines, and adhesion molecules that leads to the recruitment and activation of immune cells to the site of injury [86]. These recruited immune cells release additional inflammatory mediators that perpetuate the inflammatory cascade. In addition, NF-κβ regulated the expression of MMPs. Evidently, NF-κβ activation increases the expression of MMPs, such as MMP-1, MMP-3, and MMP-13, in human tendon induced with TNF-α or IL-1Β [94-97]. Additionally, a specific binding site for NF-κβ has been identified in the promoter gene of MMP-9, which is responsive to TNF-α [98]. Additionally, NF-κβ maintain the balance between tendon fibroblast proliferation and apoptosis and inhibit caspase 8 activation, thereby blocking apoptosis in different cell types in the tendon [99]. Moreover, NF-κβ suppresses apoptosis through activation of the Bcl-2 family member A1/Bfl-1 which reduces the release of proapoptotic cytochrome C from mitochondria [100]. Interestingly, this study aligned with our previously recorded findings of higher expression of Bcl-2 in cultured hypoxic tenocytes [101]. In the context of RCT, NF-κβ activation promotes tendon fibroblast proliferation during the early proliferative phase of tendon healing. However, when NF-κβ activity is downregulated, fibroblast proliferation ceases, and the cells undergo apoptosis. Sustained activation of NF-κβ lead to excessive fibroblast proliferation, which contribute to fibrosis and impaired tendon function [102].
NF-κβ transcription factors are composed of 5 subunits - p65 (RelA), ReIB, c-Rel, p50, and P52. All 5 subunits contain an N-terminal Rel homology domain that bears the NLS, dimerization, and DNA binding domains [103]. In the resting state, these subunits are sequestered in the cytoplasm by an inhibitor protein called Iκβ, which masks the nuclear localization signal (NLS). Upon stimulation with signals like IL-1, IL-6, TNF-α, and TREM-1 during RCT and other pathologies [97,104-106], a multi-subunit Iκβ kinase (IKK-B) complex phosphorylates Iκβa, targeting it for degradation via the ubiquitin pathway exposing the NLS on NF-κβ to translocate to the nucleus. Importin-α3 and importin-α4 have been identified as the karyopherins responsible for mediating the nuclear import of NF-κβ in TNF-α-challenged cells [103]. In the nucleus, NF-κβ inhibit the COL1α1 and COL1α2 genes in fibroblasts [107]. The reduction in collagen type I (COL1) expression is the basis of RCT pathology [80]. Overall, NF-κβ plays a dual role in RCT (Figure 3), mediating inflammation and influencing tissue repair and remodeling processes. NF-κβ activation leads to the production of pro-inflammatory mediators and MMPs and modulates tendon fibroblast behavior [108-113]. Understanding the complex interplay of inflammatory chemokines, NF-κβ, and importin in RCT could provide novel insights into therapeutic interventions to accelerate healing and mitigate fibrosis.
11. Summary and Future Perspectives
The role of karyopherins in the context of ECM remodeling in RCT is yet to be unveiled and underscores several intriguing avenues for future research. Further investigations are warranted to analyze the specific alterations in karyopherin expression and function in the context of RCT [113-116]. This may entail comparing the levels of importin and exportin subtypes in healthy tendons and RCT-affected tendons. A comprehensive understanding of karyopherin dysregulation would provide insights into the pathogenesis of RCT and potentially guide the development of targeted therapeutic interventions. Secondly, given the documented association between ECM remodeling and RCT, the scientific data regarding the involvement of karyopherins in the ECM homeostasis is missing. Karyopherins play a pivotal role in regulating the expression and activity of key mediators such as HIF-1α [2], TGF-β [3], and MMP-9 [4], which are recognized for their influence on ECM morphology. Hence, the detailed understanding regarding the role of karyopherins in ECM remodeling in RCT is beneficial in modulating the activity or expression of specific karyopherins to restore nucleo-cytoplasmic transport and ameliorate the pathological changes associated with RCT. This may encompass the development of small molecules or peptides designed to selectively target and regulate the function of karyopherins in the context of RCT. Finally, an in-depth understanding of the role of karyopherins in RCT may present opportunities for the development of diagnostic biomarkers. Through screening of specific karyopherin subtypes in patient specimens would pave the way to predict the individuals at risk of developing RCT, monitor disease progression, and determine post-RCT prognosis. Consequently, further research is warranted to validate the utility of karyopherins as diagnostic markers and to establish their correlation with clinical parameters and outcomes. In conclusion, continued research on karyopherins, their subtypes, and implications in the pathophysiology of RCT holds significant promise for future investigations. These perspectives would contribute to an enhanced understanding of the molecular mechanisms underlying RCT and facilitate the development of targeted therapeutic interventions and diagnostic approaches for this debilitating condition.
Declarations:
Funding: The research work of FGT was supported by the startup research funds from Western University of Health Sciences. DKA is supported by research grants R01 HL144125 and R01 HL147662 from the National Institutes of Health, USA.
Conflict of Interest:
All the authors have read the manuscript and declare no conflict of interest. No writing assistance was utilized in the production of this manuscript.
Author Contributions:
CD, FT, and DKA – conceptualization, manuscript preparation, edits, proof reading, preparation of figures, formatting, and finalization of the article.
References
- Fang W, Sekhon S, Teramoto D, et al. Pathological alterations in the expression status of rotator cuff tendon matrix components in hyperlipidemia. Mol Cell Biochem 478 (2022): 1887-1898.
- Mosca MJ, Carr AJ, Snelling SJB, et al. Differential expression of alarmins—S100A9, IL-33, HMGB1 and HIF-1α in supraspinatus tendinopathy before and after treatment. BMJ Open Sport Exerc Med 3 (2017): e000225.
- Kurisaki A, Kose S, Yoneda Y, et al. Transforming Growth Factor-β Induces Nuclear Import of Smad3 in an Importin-β1 and Ran-dependent Manner. Mol Biol Cell 12 (2001): 1079-1091.
- Seki M, Uzuki M, Ohmoto H, et al. Matrix metalloproteinase 9 (MMP-9) in patients with rheumatoid arthritis. Jpn J Rheumatol 7 (1997): 197-209.
- Cagatay T, Chook YM. Karyopherins in Cancer. Curr Opin Cell Biol 52 (2018): 30-42.
- Wing CE, Fung HYJ, Chook YM. Karyopherin-mediated nucleocytoplasmic transport. Nat Rev Mol Cell Biol 23 (2022): 307-328.
- Wu J, Corbett AH, Berland KM. The Intracellular Mobility of Nuclear Import Receptors and NLS Cargoes. Biophys J 96 (2009): 3840-3849.
- Holzer G, Antonin W. Nuclear Pore Complex Assembly Using Xenopus Egg Extract. Methods Mol Biol Clifton NJ 2502 (2022): 51-66.
- Görlich D, Prehn S, Laskey RA, et al. Isolation of a protein that is essential for the first step of nuclear protein import. Cell 79 (1994): 767-778.
- Görlich D, Kostka S, Kraft R, et al. Two different subunits of importing cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol CB 5 (1995): 383-392.
- Enenkel C, Blobel G, Rexach M. Identification of a Yeast Karyopherin Heterodimer That Targets Import Substrate to Mammalian Nuclear Pore Complexes. J Biol Chem 270 (1995): 16499-16502.
- Fu X, Liang C, Li F, et al. The Rules and Functions of Nucleocytoplasmic Shuttling Proteins. Int J Mol Sci 19 (2018): 1445.
- Arts G-J, Kuersten S, Romby P, et al. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J 17 (1998): 7430-7441.
- Yamazawa R, Jiko C, Choi S, et al. Structural Basis for Selective Binding of Export Cargoes by Exportin-5. Structure 26 (2018): 1393-1398.e2.
- Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science 278 (1997): 141-144.
- Fung HYJ, Fu S-C, Chook YM. Nuclear export receptor CRM1 recognizes diverse conformations in nuclear export signals. eLife 6 (2017).
- Cautain B, Hill R, de Pedro N, Link W. Components and regulation of nuclear transport processes. Febs J 282 (2015): 445-462.
- Pumroy RA, Cingolani G. Diversification of importin-α isoforms in cellular trafficking and disease states. Biochem J 466 (2015): 13-28.
- Pasha T, Zatorska A, Sharipov D, et al. Karyopherin abnormalities in neurodegenerative proteinopathies. Brain 144 (2021): 2915-2932.
- Conti E, Uy M, Leighton L, et al. Crystallographic Analysis of the Recognition of a Nuclear Localization Signal by the Nuclear Import Factor Karyopherin α. Cell 94 (1998): 193-204.
- Oka M, Yoneda Y. Importin α: functions as a nuclear transport factor and beyond. Proc Jpn Acad Ser B Phys Biol Sci 94 (2018): 259-274.
- Fontes MRM, Teh T, Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α1. J Mol Biol 297 (2000): 1183-1194.
- Lange A, Mills RE, Lange CJ, et al. Classical Nuclear Localization Signals: Definition, Function, and Interaction with Importin α. J Biol Chem 282 (2007): 5101-5105.
- Gross S, Moore C. Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I. Proc Natl Acad Sci U S A 98 (2001): 6080-6085.
- Leung SW, Harreman MT, Hodel MR, et al. Dissection of the karyopherin alpha nuclear localization signal (NLS)-binding groove: functional requirements for NLS binding. J Biol Chem 278 (2003): 41947-41953.
- Andrade MA, Bork P. HEAT repeats in the Huntington’s disease protein. Nat Genet 11 (1995): 115-116.
- Yoshimura SH, Hirano T. HEAT repeats – versatile arrays of amphiphilic helices working in crowded environments? J Cell Sci 129 (2016): 3963-3970.
- Bayliss R, Littlewood T, Stewart M. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell 102 (2000): 99-108.
- Kalderon D, Richardson WD, Markham AF, et al. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311 (1984): 33-38.
- Robbins J, Dilworth SM, Laskey RA, et al. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64 (1991): 615-623.
- Dingwall C, Laskey RA. Nuclear targeting sequences--a consensus? Trends Biochem Sci 16 (1991): 478-481.
- onti E, Kuriyan J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Struct Lond Engl 1993 8 (2000): 329-338.
- Fontes MRM, Teh T, Toth G, et al. Role of flanking sequences and phosphorylation in the recognition of the simian-virus-40 large T-antigen nuclear localization sequences by importin-alpha. Biochem J 375 (2003): 339-349.
- Hodel AE, Harreman MT, Pulliam KF, et al. Nuclear Localization Signal Receptor Affinity Correlates with in Vivo Localization in Saccharomyces cerevisiae. J Biol Chem 281 (2006): 23545-23556.
- Mattaj IW, Englmeier L. Nucleocytoplasmic Transport: The Soluble Phase. Annu Rev Biochem 67 (1998): 265-306.
- Lam MH, Briggs LJ, Hu W, et al. Importin beta recognizes parathyroid hormone-related protein with high affinity and mediates its nuclear import in the absence of importin alpha. J Biol Chem 274 (1999): 7391-7398.
- Nakada R, Hirano H, Matsuura Y. Structure of importin-α bound to a non-classical nuclear localization signal of the influenza A virus nucleoprotein. Sci Rep 5 (2015).
- Kabachinski G, Schwartz TU. The nuclear pore complex – structure and function at a glance. J Cell Sci 128 (2015): 423-429.
- Beck M, Hurt E. The nuclear pore complex: understanding its function through structural insight. Nat Rev Mol Cell Biol 18 (2017): 73-89.
- Moroianu J, Blobel G, Radu A. The binding site of karyopherin alpha for karyopherin beta overlaps with a nuclear localization sequence. Proc Natl Acad Sci U S A 93 (1996): 6572-6576.
- Terry LJ, Wente SR. Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J Cell Biol 178 (2007): 1121-1132.
- Macara IG. Transport into and out of the Nucleus. Microbiol Mol Biol Rev 65 (2001): 570-594.
- Nguyen KT, Holloway MP, Altura RA. The CRM1 nuclear export protein in normal development and disease. Int J Biochem Mol Biol 3 (2012): 137-151.
- Fornerod M, Ohno M, Yoshida M, et al. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90 (1997b): 1051-1060.
- Port SA, Monecke T, Dickmanns A, et al. Structural and Functional Characterization of CRM1-Nup214 Interactions Reveals Multiple FG-Binding Sites Involved in Nuclear Export. Cell Rep 13 (2015): 690-702.
- Ribbeck K, Lipowsky G, Kent HM, et al. NTF2 mediates nuclear import of Ran. EMBO J 17 (1998): 6587-6598.
- Englmeier L, Olivo JC, Mattaj IW. Receptor-mediated substrate translocation through the nuclear pore complex without nucleotide triphosphate hydrolysis. Curr Biol CB 9 (1999): 30-41.
- Black BE, Holaska JM, Lévesque L, et al. NXT1 is necessary for the terminal step of Crm1-mediated nuclear export. J Cell Biol 152 (2001): 141-155.
- Ullman KS, Powers MA, Forbes DJ. Nuclear export receptors: from importin to exportin. Cell 90 (1997): 967-970.
- Li Y, Zhou J, Min S, et al. Distinct RanBP1 nuclear export and cargo dissociation mechanisms between fungi and animals. eLife 8 (2019): e41331.
- Xu L, Massagué J. Nucleocytoplasmic shuttling of signal transducers. Nat Rev Mol Cell Biol 5 (2004): 209-219.
- Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol 8 (2007): 195-208.
- Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol 9 (2008): 464-477.
- Yoneda Y, Hieda M, Nagoshi E, et al. Nucleocytoplasmic protein transport and recycling of Ran. Cell Struct Funct 24 (1999): 425-433.
- Mylonis I, Chachami G, Paraskeva E, et al. Atypical CRM1-dependent nuclear export signal mediates regulation of hypoxia-inducible factor-1alpha by MAPK. J Biol Chem 283 (2008): 27620-27627.
- Huenniger K, Krämer A, Soom M, et al. Notch1 signaling is mediated by importins alpha 3, 4, and 7. Cell Mol Life Sci 67 (2010): 3187-3196.
- Aggarwal A, Agrawal DK. Importins and Exportins Regulating Allergic Immune Responses. Mediators Inflamm 2014 (2014): 476357.
- Agrawal T, Gupta GK, Agrawal DK. Calcitriol Decreases Expression of Importin α3 and Attenuates RelA Translocation in Human Bronchial Smooth Muscle Cells. J Clin Immunol 32 (2012): 1093-1103.
- Agrawal T, Gupta GK, Agrawal DK. Vitamin D supplementation reduces airway hyperresponsiveness and allergic airway inflammation in a murine model. Clin Exp Allergy J Br Soc Allergy Clin Immunol 43 (2013): 672-683.
- Chen S, Swier VJ, Boosani CS, et al. Vitamin D Deficiency Accelerates Coronary Artery Disease Progression in Swine. Arterioscler Thromb Vasc Biol 36 (2016): 1651-1659.
- Zhou H, Jiang S, Li P, et al. Improved tendon healing by a combination of Tanshinone IIA and miR-29b inhibitor treatment through preventing tendon adhesion and enhancing tendon strength. Int J Med Sci 17 (2020): 1083-1094.
- Tomasek JJ, Gabbiani G, Hinz B, et al. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Biol 3 (2002): 349-363.
- Brunner G, Blakytny R. Extracellular regulation of TGF-β activity in wound repair: growth factor latency as a sensor mechanism for injury. Thromb Haemost 92 (2004): 253-261.
- Wipff P-J, Rifkin DB, Meister J-J, et al. Myofibroblast contraction activates latent TGF-β1 from the extracellular matrix. J Cell Biol 179 (2007): 1311-1323.
- Voleti PB, Buckley MR, Soslowsky LJ. Tendon Healing: Repair and Regeneration. Annu Rev Biomed Eng 14 (2012): 47-71.
- Wynn TA, Vannella KM. Macrophages in Tissue Repair, Regeneration, and Fibrosis. Immunity 44 (2016): 450-462.
- Pakshir P, Hinz B. The big five in fibrosis: Macrophages, myofibroblasts, matrix, mechanics, and miscommunication. Matrix Biol 68–69 (2018): 81-93.
- Garg K, Corona BT, Walters TJ. Therapeutic strategies for preventing skeletal muscle fibrosis after injury. Front Pharmacol 6 (2015).
- Li Y, Liu X, Liu X, et al. Transforming growth factor-β signalling pathway in tendon healing. Growth Factors 40 (2022): 98-107.
- Feng X-H, Derynck R. Specificity and Versatility In Tgf-Β Signaling Through Smads. Annu Rev Cell Dev Biol 21 (2005): 659-693.
- Xu X, Dai K, Tang T. The role of Smads and related transcription factors in the signal transduction of bone morphogenetic protein inducing bone formation. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi Zhongguo Xiufu Chongjian Waike Zazhi Chin J Reparative Reconstr Surg 17 (2003): 359-362.
- Nakagawa T, Li JH, Garcia G, et al. TGF-beta induces proangiogenic and antiangiogenic factors via parallel but distinct Smad pathways. Kidney Int 66 (2004): 605-613.
- Li J, Tang X, Chen X. Comparative effects of TGF-β2/Smad2 and TGF-β2/Smad3 signaling pathways on proliferation, migration, and extracellular matrix production in a human lens cell line. Exp Eye Res 92 (2011): 173-179.
- Wang G, Li C, Wang Y, et al. Cooperative Assembly of Co-Smad4 MH1 with R-Smad1/3 MH1 on DNA: A Molecular Dynamics Simulation Study. PLoS ONE 8 (2013a).
- Kim MS, Jin W. TrkB-Induced Inhibition of R-SMAD/SMAD4 Activation is Essential for TGF-β-Mediated Tumor Suppressor Activity. Cancers 12 (2020): 1048.
- Massagué J, Chen Y-G. Controlling TGF-β signaling. Genes Dev 14 (2000): 627-644.
- Dijke P ten, Miyazono K, Heldin C-H. Signaling inputs converge on nuclear effectors in TGF-β signaling. Trends Biochem Sci 25(2000): 64-70.
- Attisano L, Wrana JL. Smads as transcriptional co-modulators. Curr Opin Cell Biol 12 (2000): 235-243.
- Massagué J, Wotton D. Transcriptional control by the TGF-β/Smad signaling system. EMBO J 19 (2000): 1745-1754.
- Thankam FG, Dilisio MF, Gross RM, et al. Collagen I: a kingpin for rotator cuff tendon pathology. Am J Transl Res (2018b): 3291-3309.
- Wang Y, Zhou Z, Liu Y, et al. Inhibition of Smad3 promotes the healing of rotator cuff injury in a rat model. J Orthop Res Off Publ Orthop Res Soc 39 (2021): 204-218.
- Katzel EB, Wolenski M, Loiselle AE, et al. The impact of Smad3 loss of function on scarring and adhesion formation during tendon healing. J Orthop Res Off Publ Orthop Res Soc 29 (2011): 684-693.
- Millar NL, Reilly JH, Kerr SC, et al. Hypoxia: a critical regulator of early human tendinopathy. Ann Rheum Dis 71 (2012): 302-310.
- Thankam FG, Chandra I, Diaz C, et al. Matrix regeneration proteins in the hypoxia-triggered exosomes of shoulder tenocytes and adipose-derived mesenchymal stem cells. Mol Cell Biochem 465 (2020): 75-87.
- Huang LE, Gu J, Schau M, et al. Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci U S A 95 (1998): 7987-7992.
- Liu T, Zhang L, Joo D, et al. NF-κB signaling in inflammation. Signal Transduct Target Ther 2 (2017): 17023.
- Wang H, Tian Y, Wang J, et al. Inflammatory Cytokines Induce NOTCH Signaling in Nucleus Pulposus Cells: Implications in Intevertebral Disc Degeneration. J Biol Chem 288 (2013b): 16761-16774.
- Retief E, Parker MI, Retief AE. Regional chromosome mapping of human collagen genes alpha 2(I) and alpha 1(I) (COLIA2 and COLIA1). Hum Genet 69 (1985): 304-308.
- Ghosh AK. Factors involved in the regulation of type I collagen gene expression: implication in fibrosis. Exp Biol Med Maywood NJ 227 (2002): 301-314.
- Engin F, Yao Z, Yang T, et al. Dimorphic effects of Notch signaling in bone homeostasis. Nat Med 14 (2008): 299-305.
- Burrage PS, Mix KS, Brinckerhoff CE. Matrix Metalloproteinases: Role in Arthritis. Front Biosci-Landmark 11 (2006): 529-543.
- Lee Y-A, Choi HM, Lee S-H, et al. Hypoxia differentially affects IL-1β-stimulated MMP-1 and MMP-13 expression of fibroblast-like synoviocytes in an HIF-1α-dependent manner. Rheumatology 51 (2012): 443-450.
- Petersen W, Unterhauser F, Pufe T, et al. The angiogenic peptide vascular endothelial growth factor (VEGF) is expressed during the remodeling of free tendon grafts in sheep. Arch Orthop Trauma Surg 123 (2003): 168-174.
- Vincenti MP, Brinckerhoff CE. Transcriptional regulation of collagenase (MMP-1, MMP-13) genes in arthritis: integration of complex signaling pathways for the recruitment of gene-specific transcription factors. Arthritis Res 4 (2002): 157-164.
- Liacini A, Sylvester J, Qing Li W, et al. Induction of matrix metalloproteinase-13 gene expression by TNF-α is mediated by MAP kinases, AP-1, and NF-κB transcription factors in articular chondrocytes. Exp Cell Res 288 (2003): 208-217.
- Thankam FG, Dilisio MF, Agrawal DK. Immunobiological factors aggravating the fatty infiltration on tendons and muscles in rotator cuff lesions. Mol Cell Biochem 417 (2016a): 17-33.
- Thankam FG, Dilisio MF, Dietz NE, et al. TREM-1, HMGB1 and RAGE in the Shoulder Tendon: Dual Mechanisms for Inflammation Based on the Coincidence of Glenohumeral Arthritis. PLOS ONE 11 (2016b): e0165492.
- Yan C, Wang H, Boyd DD. KiSS-1 Represses 92-kDa Type IV Collagenase Expression by Down-regulating NF-κB Binding to the Promoter as a Consequence of IκBα-induced Block of p65/p50 Nuclear Translocation*. J Biol Chem 276 (2001): 1164-1172.
- Wang CY, Mayo MW, Korneluk RG, et al. NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science 281 (1998): 1680-1683.
- Wang C-Y, Guttridge DC, Mayo MW, et al. NF-κB Induces Expression of the Bcl-2 Homologue A1/Bfl-1 To Preferentially Suppress Chemotherapy-Induced Apoptosis. Mol Cell Biol 19 (1999): 5923-5929.
- Thankam FG, Chandra IS, Kovilam AN, et al. Amplification of Mitochondrial Activity in the Healing Response Following Rotator Cuff Tendon Injury. Sci Rep 8 (2018a): 17027.
- Best KT, Nichols AEC, Knapp E, et al. NF-κB activation persists into the remodeling phase of tendon healing and promotes myofibroblast survival. Sci Signal 13 (2020): eabb7209.
- Fagerlund R, Kinnunen L, Köhler M, et al. NF-κB Is Transported into the Nucleus by Importin α3 and Importin α4 *. J Biol Chem 280 (2005): 15942-15951.
- Jimenez SA, Varga J, Olsen A, et al. Functional analysis of human alpha 1(I) procollagen gene promoter. Differential activity in collagen-producing and -nonproducing cells and response to transforming growth factor beta 1. J Biol Chem 269 (1994): 12684-12691.
- Beauchef G, Bigot N, Kypriotou M, et al. The p65 Subunit of NF-κB Inhibits COL1A1 Gene Transcription in Human Dermal and Scleroderma Fibroblasts through Its Recruitment on Promoter by Protein Interaction with Transcriptional Activators (c-Krox, Sp1, and Sp3). J Biol Chem 287 (2012): 3462-3478.
- Thankam FG, Roesch ZK, Dilisio MF, et al. Association of Inflammatory Responses and ECM Disorganization with HMGB1 Upregulation and NLRP3 Inflammasome Activation in the Injured Rotator Cuff Tendon. Sci Rep 8 (2018c): 8918.
- Rippe RA, Schrum LW, Stefanovic B, et al. NF-kappaB inhibits expression of the alpha1(I) collagen gene. DNA Cell Biol 18 (1999): 751-761.
- Fornerod M, Deursen J van, Baal S van, et al. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J 16 (1997a): 807-816.
- Fukuda M, Asano S, Nakamura T, et al. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390 (1997): 308-311.
- Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15 (1999): 607-660.
- Lott K, Cingolani G. The Importin β Binding Domain as a Master Regulator of Nucleocytoplasmic Transport. Biochim Biophys Acta 1813 (2011): 1578-1592.
- Ossareh-Nazari B, Gwizdek C, Dargemont C. Protein Export from the Nucleus. Traffic 2 (2001): 684-689.
- Quimby BB, Lamitina T, L’Hernault SW, et al. The mechanism of ran import into the nucleus by nuclear transport factor 2. J Biol Chem 275 (2000): 28575-28582.
- Stade K, Ford CS, Guthrie C, et al. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90 (1997): 1041-1050.
- Watanabe M, Fukuda M, Yoshida M, et al. Involvement of CRM1, a nuclear export receptor, in mRNA export in mammalian cells and fission yeast. Genes Cells Devoted Mol Cell Mech 4 (1999): 291-297.
- Weis K, Mattaj IW, Lamond AI. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science 268 (1995): 1049-1053.


 Impact Factor: * 5.3
Impact Factor: * 5.3 Acceptance Rate: 73.64%
Acceptance Rate: 73.64%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 