Complete Remission of the Severe Advanced Stage Cancer by miRNA-Mediated Transcriptional Control of Bcl-xL with Huaier Therapy Compared to the Conventional Chemotherapy with Platinum (II) Complex
Manami Tanaka1*, Tomoo Tanaka1, Fei Teng2, Hong Lin2, Ning Li3, Zhu Luo3, Sotaro Sadahito4, Toshiyuki Suzuki5, Ding Wei6, Zhengxin Lu7
1Bradeion Institute of Medical Sciences, Kanagawa, Japan
2BGI-Shenzhen, Shenzhen, China
3BGI-Japan, Kobe, Japan
4Department of Surgery, Kameda-Morinosato Hospital, Kanagawa, Japan
5Department of Surgery, Oiso Hospital, Tokai University School of Medicine, Kanagawa, Japan
6Japan Kampo NewMedicine, Tokyo, Japan
7QiDong Gaitianli Medicines, Jiangsu Province, China
*Corresponding author: Manami Tanaka, Bradeion Institute of Medical Sciences, Co. Ltd., Itado 433-1,
Isehara, Kanagawa 259-1145, Japan
Received: 27 March 2021; Accepted: 03 April 2021; Published: 12 April 2021
Article Information
Citation:
Manami Tanaka, Tomoo Tanaka, Fei Teng, Hong Lin, Ning Li, Zhu Luo, Sotaro Sadahito, Toshiyuki Suzuki, Ding Wei, Zhengxin Lu. Complete Remission of the Severe Advanced Stage Cancer by miRNA-Mediated Transcriptional Control of Bcl-xL with Huaier Therapy Compared to the Conventional Chemotherapy with Platinum (II) Complex. Archives of Clinical and Biomedical Research 5 (2021): 230-261.
View / Download Pdf Share at FacebookAbstract
Background: The severe advanced stage cancer has scarce choice of treatment except pain control. Conventional chemotherapy is commonly not applicable for these patients from severe adverse events and toxicity.
Objective: We have initiated genome-scope project for precise comprehension of anti-cancer effects of Huaier, with a comparison to the results by the treatment with platinum (II) complex (FOLFOX6, FOLFIRINOX, and Cisplatin: CDDP).
Methods: The RNA samples from the obtained peripheral blood from the volunteer patients were analyzed by total RNA and non-coding small RNA sequencing on BGISEQ-500 Platform. KEGG pathway classification was hired for the analysis of the obtained information of transcripts in the present study (https://www.genome.jp/kegg/).
Results: Here we present the successful remission and recovery from the severe advanced stage of 1) oesophageal squamous cell carcinoma, 2) pancreatic adenocarcinoma, and 3) colorectal adenocarcinoma. The complete remission was obtained in the patients treated with Huaier only, and Huaier and FOLFIRINOX combined therapy, although poor prognosis was predicted in these patients if only treated with platinum (II) complex chemotherapy, such as Cisplatin: CDDP, FOLFOX, or FOLFIRINOX. The efficacy of the treatment could be easily and typically confirmed by the improvement of the image analysis with a significant decrease of tumor markers such as CEA, CA19-9, and DUPAN-2 by the time course of Huaier administration.
The KEGG pathway classification indicated the key for the recovery was the inhibition of Bcl-xL, which altered and rescued impaired function of multiple signal transduction pathways enhancing apoptosis, especially through the intrinsic mitochondria-mediated pathway. These changes were caused by the miRNA-mediated transcriptional control as reported. In contrast, platinum (II)
Keywords
<p>Huaier (<em>Trametes robiniophila murr</em>); Platinum (II) complex (FOLFOX6, FOLFIRINOX, and Cisplatin: CDDP); KEGG signaling pathway characterization; miRNA-medicated transcription control; Bcl-xL (B-cell lymphoma-extra large)</p>
Article Details
1. Introduction
The severe advanced stage cancer has scarce choice of treatment, except pain control and supplement of nutrition, which results in grim prognosis [1, 2]. Data on molecular level of each patient and controlling agents are strongly lacking to determine the combination of chemotherapeutic agents for their efficacy and safety. Huaier (Trametes robiniophila murr) has been introduced its significant anti-cancer efficacy, and more importantly, without any adverse events and/or toxicity [3-8]. We have proved the molecular basis for Huaier effects based on the rescue of disrupted Hippo signalling pathway in Drosophila mutants, especially through the rescue of transcriptional dysregulation [5, 9]. From 2018, we proceeded the investigation as a clinical research to analyze every genomic and genetic event happening to the Huaier-administered cancer patients on the time course of treatment [7, 8]. Total RNA sequencing and also small non-coding RNA sequencing revealed d quantitative and qualitative genomic and genetic alterations and modifications in Huaier-treated cancer patients; 1) 92,427 SNP per sample (ranging from 48,994 to 146,102), whereas 22,688 in total among normal healthy individuals [10],27,801 splicing events per sample (those changes did not show any significant correlation to the specific cancer, which corresponding to 3) the quantitative changes in translation at maximum 85% (23,210/27,447 per person) of the total transcripts. These changes were estimated as a result of alteration in the transcriptional factors (66 kinds of transcription families coding 7,664 transcriptional factors, mean 1,115 per person) and their modified control on massive rescue of signal transduction functions. These results indicated that Huaier can restore biological function by miRNA-mediated transcription control [7, 8].
Interestingly, Huaier effects as “minimum-essential” to the required level for the purpose (cure of the diseases) in a dose-dependent manner, as a spontaneous reaction according to individual genomic potential. The general observation and evaluation of recovered physiological condition resembled to the status as “rewinding the life time”, not to say “rejuvenescence” first purposed by the ancient expedition on the search for natural products for immortality directed by the ancient Chinese Emperor.
In contrast, the common conventional therapy using molecular-targeting pharmaceuticals often causes long-lasting severe adverse events than therapeutic efficacy, even though elongated prognosis by months [11-13]. It is obvious that surgical dissection is the best treatment to begin with, however, elimination of tumor mass remains some possibility of occult metastasis (40% in stage II colorectal cancer, and 60% in stage III). Even though surgical dissection was not applicable at the time of diagnosis, it is designated to have better prognosis to remove tumor mass (including metastatic lesion) with reasonable period of chemotherapy and/or radiotherapy (especially for locally advanced or metastatic colorectal cancer) [14, 15].
In the present study, we analysed molecular characteristics in the severe advanced stage cancer patient treated with Huaier, and completely recovered from 1) oesophageal squamous cell carcinoma, 2) colorectal adenocarcinoma, and successfully recovered to the remission stage to be applicable for surgical dissection of the main lesion of 3) pancreatic adenocarcinoma. The molecular characterization of these cases was compared with the patients treated with conventional chemotherapy containing platinum complex [11-13], such as FOLFOX [11] or FOLFIRINOX [13]. The same strategy using the information obtained by total RNA- and small non-coding RNA sequencing were used for a search to identify the key molecule and/systems to determine the fate of the patients [7, 8]. As a result, two points should be emphasized in the present study.
First, the key molecule to determine the prognosis of those severe advanced stage patient is Bcl-xL (B-cell lymphoma-extra large), encoded by the Bcl2-like 1 gene, is a transmembrane molecule in the mitochondria [16-19]. Huaier rescued the impairment of apoptotic pathway through the intrinsic mitochondria-mediated pathway. The other factors such as NF B, TGF , BRCA2, and p53 genes were minor regulators, unlike to the findings in the other cancers related to excretory organs such as prostate cancer [7, 8].
The second point is the molecular basis of the severe adverse events by platinum (II) complex containing anti-cancer drugs [11-13]. These complexes inhibit DNA replication, transcription, consequently, cellular proliferation, with strong the severe adverse events such as nausea, vomiting, anorexia, haemolysis, toxicity to many organs, neurotoxicity, tinnitus. The molecular basis for these symptoms and disorders including onset time were still remained unknown. The sensory nerve disturbance was well known symptoms realized very soon after the administration, however, the molecular basis to cause this disorder has not been reported yet.
In addition, severe inhibition of signal transfer in central nervous systems has not been clearly defined yet, either. The present study provides the detailed information for these symptoms with the molecular changes. The changes and modifications in the central nervous system were found to influence to the long-term depression system, too. Thus, the present study provides successful remission/recovery of the severe advanced stage cancer by Huaier therapy, and the effective molecular monitoring revealed the onset of the severe adverse events by platinum (II) complex anti-cancer drugs.
2. Materials and Methods
2.1 Project design and patients’ profile
Huaier compounds were provided by the manufacturer for this purpose with a strict control on transfer to Japan, good condition for maintenance, and provision to the patient volunteers [7, 8]. The volunteer patients and the normal control person in the present study were described basically 20g per day Huaier administration, with an exception of 60 g per day to the patient of pancreatic cancer with combined therapy with FOLFIRINOX [13].
From the past experiences in our clinical research started 2018, the results clealy indicated that the usual dose of 20g was not enough for pancreatic cancer with or after excessive administration of conventional chemotherapy. In Chinese Hospitals, they use 60g for breast cancer and pancreatic cancer, according to the malignancy of the diseases. We decided to follow this Chinese original quantity to cope with the patient of pancreatic adenocarcinoma, diagnosed by MRI. The duration of Huaier administration was at least 3 months, and continued according to the choice of each patient, until the complete recovery was confirmed. The profiles and medical characteristics of the patients were summarized in Table 1.
Table 1: Clinical features of cancer patients and normal controls. Huaier administration was 20g per day, except Patient No. 6 (60g per day).
The present study was specifically focused on the severe advanced cancer patients, which was strictly conducted according to the guidelines of the Declaration of Helsinki and the principles of good clinical practice. Written informed consent was obtained from the patients. This clinical research was applied according to the Consolidated Standards of Clinical Research Trials guidelines and was applied to the Japanese Medical Association on 9th February 2018, and approved on 5th March, 2018 (ID: JMA-IIA00335).
The project has been strictly conducted with a monthly review by the ethics committee consisted by the experts on Medicine, Nursing, Laws, Pharmaceutics and Business Community (first committee held on 9th February, 2018) [7, 8]. The patient status was assessed according to his or her medical history, complete physical examination by a physician, and complete blood tests including tumour markers with image analysis. All patients underwent MRI examination regularly, once in 2-3 months interval. They are still having Huaier compound as their own choice, but the quantity varies among individuals (3-20 g per day). Periodical assessments of QOL and clinical tests are also being monitored at present.
2.2 RNA extraction, miRNA library construction, and mRNA library construction
The obtained peripheral blood samples introduced in the present study were designated to be analyzed by RNA extraction, miRNA library construction, and mRNA Library Construction in BGI Shenzhen, followed by the construction of whole transcriptome library for sequencing on BGISEQ-500 Platform, followed by whole transcriptome mapping [7, 8, 20, 21]. The detailed methodology was described preciously, and the protocols including bioinformatics workflow were provided and demonstrated at BGI website: http://www.bg itechsolutions.com/).
Briefly, we first removed the reads mapped to rRNAs and get raw data, then we filtered the low-quality reads (More than 20% of the bases qualities are lower than 10), reads with adaptors and reads with unknown bases (N bases more than 5%) to get the clean reads. After reads filtering, we mapped those clean reads only to reference genome. After genome mapping, to reconstruct transcripts, and with genome annotation information, novel transcripts were identified, and the coding ability of those new transcripts were predicted. Then we map those clean reads onto reference genome (hg38_UCSC _20180118), followed with novel gene prediction, SNP and INDEL calling and gene splicing detection.
We identify single nucleotide polymorphisms (SNP) and insertions and deletions (INDL) variants for each sample. This process supported importing multiple samples for comparison and show the distribution of reads in the exon, intron, UTR, intergenic areas based on the annotation results. Finally, we identify DEGs (differentially expressed genes) between samples and do clustering analysis and functional annotations.
2.3. Bioinformatics workflow
Bioinformatics Work was processed in BGI, Shenzhen, China, as escribed previously [7, 8]. The detailed protocols were provided and demonstrated at BGI website: http://www.bgitechsolutions.com/). In the transcriptome analysis, all samples had library construction and sequenced on the MGISEQ-2000 and BGISEQ-500 platform. The obtained novel genes have been deposited to The NCBI GEO (GSE157086). With quantitative analysis of DEGs, we performed Gene Ontology (GO) classification and functional enrichment just as the same as reported [7, 8].
2.4 Small RNA analysis
We have also performed total non-coding small RNA sequencing on the same BGISEQ-500 Platform, and the length of the small RNAs were found to be between 18 to 30 nucleotides. To make unique small RNA mapped to only one annotation, we follow priority rule: miRNA>piRNA>Rfam>other sRNA. After sRNA annotation, those unknown tags will be used to predict novel sRNA based on their architectural features.
Thus, novel miRNA, piRNA, and siRNA were predicted [7, 8]. The obtained novel small nuclear non-coding RNA have been uploaded to the former deposited information; The NCBI GEO (GSE157086).
3. Results
3.1 Characterization of volunteer patients and normal controls
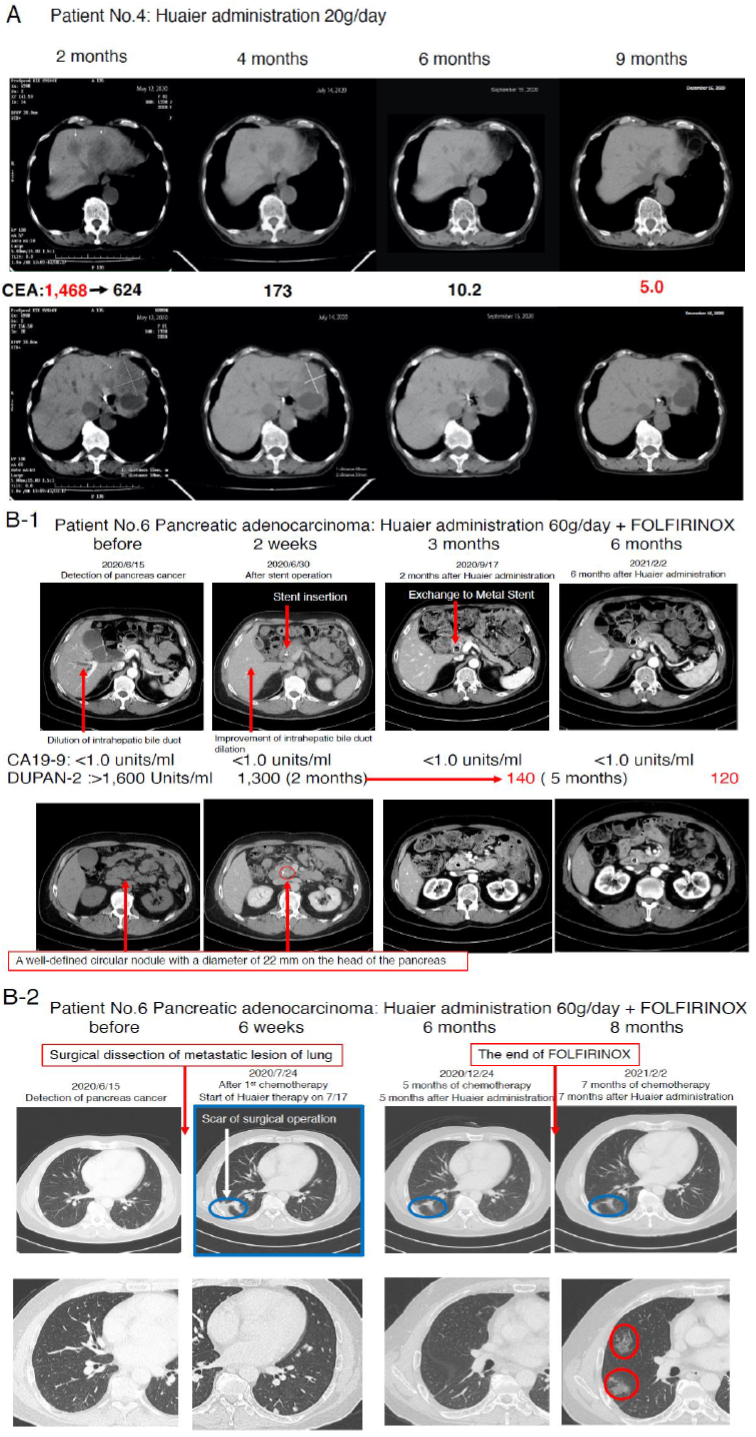
We compared the process and the efficacy of Huaier on the severe advanced stage, mainly diagnosed as TNM Stage IV patients according to the time course of administration. The medical characteristics of the 8 volunteer patients were summarized in Table 1. Figure 1 demonstrated the CT image to show the process of recovery in the patient No. 4 (Huaier 20g per day) and No. 6 (Huaier 60g per day). MRI analysis was not used in those patients, since no need in the patient No.4, and N.6 inserted metal stent tube for biliary drainage.
As shown in Figure 1 Panel A, the tumor marker CEA was good indicator to indicate the recovery process of colorectal cancer. In contrast, patient No. 5 (after 6 months of FOLFOX6 [12]), showed 447 Units/ml before Huaier administration, and 3,010 at 30 days after Huaier treatment (CA125 215 units/ml, CA19-9
3,290 Units/ ml, alpha-FP 3.3 ng/ml). Patient No.6 did not show significant increase in CA19-9 titers from the diagnosed time throughout the observation period (less than 1.0 units/ml), however, excessive amount of DUPAN-2 [22] was detected (more than 1,600 Units/ml, over measurement limits) before any treatment (Figure 1 Panel B).
After 2 months of Huaier and FOLFIRINOX treatment [13], it began to decrease to 1,300, followed by 140 units/ml (within normal limits) after 5 months, 120 Units/ml at 7 months. The metastatic lesions in both lungs were shown in Figure 1 Panel B2. The partial dissection of metastatic lesion was repaired well by the time course (blue circle), but suddenly multiple interstitial lesions (red circle) were detected just at the same time of the stoppage of FOLFIRINOX [13]. These changes were very similar to those often observed in the patients with anticoagulants for the treatment of arrhythmia. We considered the new lung lesions were the results from micro bleeding as a side effect of FOLFILRINOX [13].
Figure 1C1 and Figure 1C2 movies show angiography of the patient No.6 from every direction, confirming no angiogenesis inside/around the cancer lesion (caput pancreatis), which located at the adjacent area of the metal stent. Huaier treatment with 60g/day began at the same time of FOLFIRINOX [13] administration (for 6 months). We paid special attention to maintain total protein content over 7.0 g/dl, and albumin 4.0 g/dl, with liver function data such as AST (COT), ALT (GPT), and LDH isozyme staying within normal limits.
Since many papers have reported significant Huaier effects on liver dysfunction, the blood test titers and CT images of the liver were well maintained normal status (except the metal stent for biliary drainage). The CA19-9 titer in the patient No.7 was 3716.0 at the time of Huaier administration, after FORFIRINOX and Gemcitabine [14]. The patient No.6 successfully applied for surgical dissection of the main lesion. As for oesophageal squamous cell carcinoma, Patient No.1 and 2 refused to have any chemotherapy for fear of strong toxicity, and only took Huaier 20g per day. Physiological status of both patients was maintained in very good condition, and one year later, University attached Hospitals examined thoroughly by blood test, MRI, and endoscopic examination and declared complete recovery. We could not follow the fate of the patient No.3 after 30 days of Huaier administration, since he switched to the chemotherapy by Cisplatin: CDDP and 5-FU [11] combination.
3.2 Summary of RNA editing events; SNP and Differently spliced gene variants indicated no significant relationship to the fate of the patients
The quantitative analysis of > 7.0 GB RNA sequencing of each sample were performed as previously [6, 7]. The results provided estimated transcribed genes of >27,000; mean number of 27, 447 per sample, and the average mapping ratio with reference genome is 89.64 % (varies from 71.49% to 94.51%) as previously reported. The obtained additional novel genes and small nuclear non-coding RNA obtained in the present study have been up-loaded to the genes already deposited to the NCBI GEO (GSE157086) [6, 7].
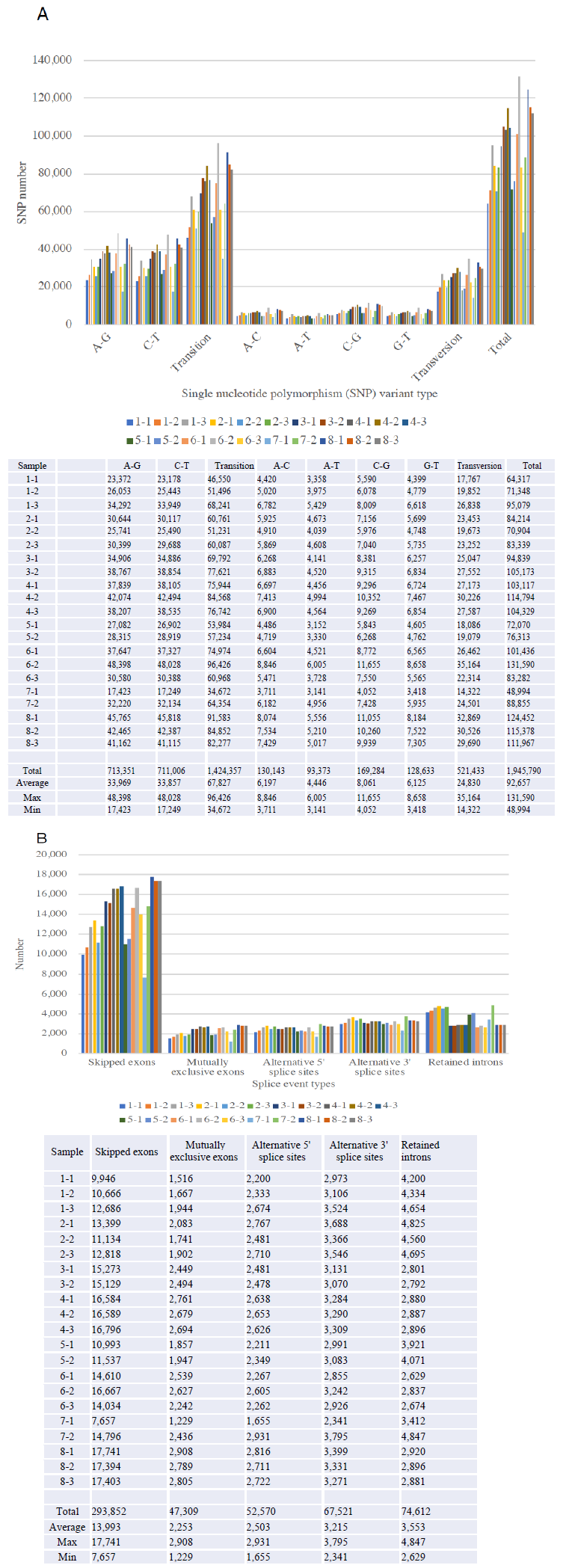
Figure 2 demonstrated the SNP variants (Panel A) and splicing events (Panel B) compared between the samples obtained before and after 1month, and 3 months, and the follow-up monitoring after 1year of Huaier administration (see Table 1 for sampling time). We identified total 1,945,790 SNP variant types in 21 samples used in the present study, and 92,657 Single Nucleotide Polymorphism (SNP) variants per sample in mean number (ranging from 48,994 to 131,590), whereas 22,688 in total among normal healthy individuals [10]. As for the SNP variations, A-G > C-T transitions were the most common mutations (713,351 and 711,006 respectively, 73.2%), followed by A-C > A-T (130,143 and 93,373, respectively, 11.5%), C-G > G-T (169,284 and 128,633 respectively, 15.3%) transversions, which is consistent with the previous reports on oesophageal squamous carcinoma cells [20]. These ratios found no significant differences among samples, but the similar gross increase of SNP variants, as reported previously were also detected in those patients in the present study. The incidence ratio was three times higher in transition, which is much higher than that identified in oesophageal squamous carcinoma cells, and any other cancer cells analyzed before. We also detect the level of differentially splicing genes (DSG) compared before and after Huaier administration. The summary of splicing variation comparison is shown in Figure 2. DSGs are regulated by alternative splicing (AS), which allows the production of a variety of different isoforms from one gene only. Changes in relative abundance of isoforms, regardless to the expression change, indicates a splicing-related mechanism. We detected five types of AS events, including skipped exon (SE), alternative 5’ splicing site (A5SS), alternative 3’ splicing site (A3SS), mutually exclusive exons (MXE), and retained intron (RI). Approximately total 535,864 splicing events in 21 samples were observed, in which were 293,852 SE (54.8%); 47,309 MXE (8.8%); 52,570 A5SS (9.8%); 67,521 A3SS (12.6%); and 74,612 RI (13.9%), respectively. Mean numbers of each event per sample (= per person) were 13,993 (ranging from 7,657 to 17,741) SE; 2,253 (ranging from 1,229 to 2,908) MXE; 2,503 (ranging from 1,655 to 2,931) A5SS; 3,215 (ranging from 2,341 to 3,795) A3SS; and 3,553 (ranging from 2,629 to 4,847) RI, respectively. These ratios found no significant differences among samples, and the distribution and the numbers in each alteration was almost the same as the result reported previously [7, 8]. Distribution of both SNP and INDL (>2,000,000 per sample) location was identified 40 to 45 % in exons, and up to 60% including the adjacent area. Thus, in contrary to the previous reports, these RNA editing events did not randomly scatter among the whole genome, nor any correlation to cancer origin or stages [7, 8].
3.3 miRNA-mediated alterations in transcription control: Analysis of differentially expressed small RNAs (DSGs) and their correlation to the differentially expressed genes (DEGs)
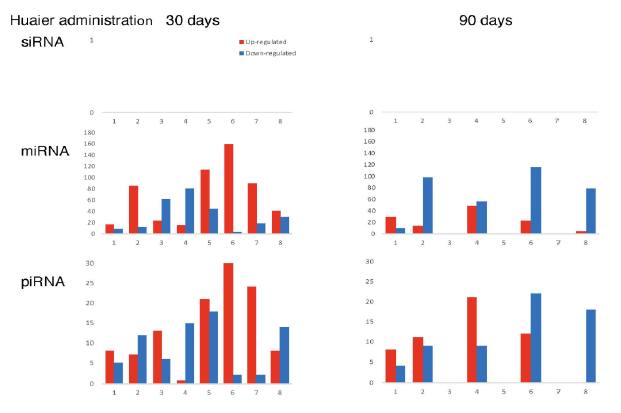
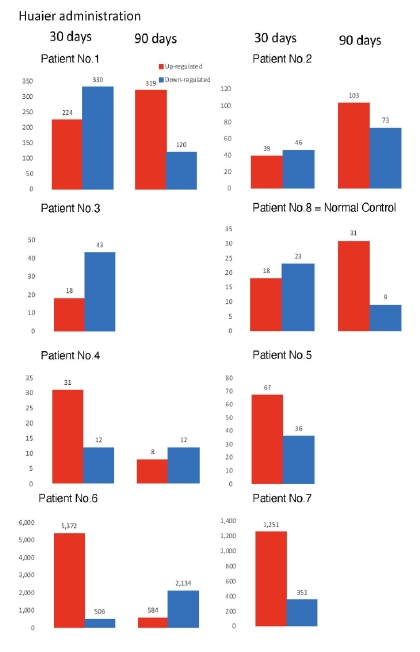
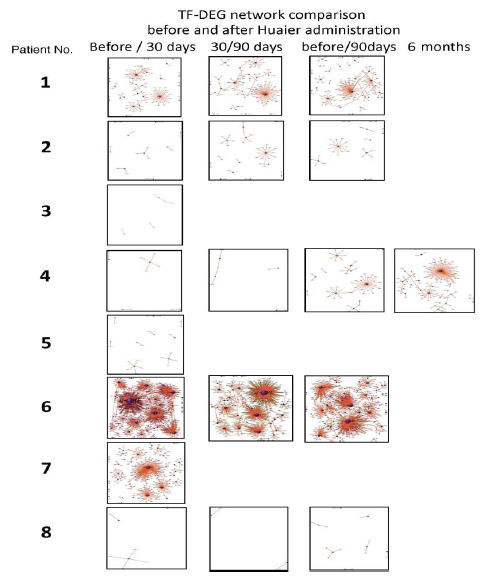
The numbers of detected small non-coding RNA for each sample were summarized in Figure 3. The structure of miRNAs and the sequences of the novel small RNAs were deposited to the NCBI GEO (GSE157086), and shown and stored in precursor.tar.gz. Comparison of up- and down-regulated transcriptomes by numbers was shown in Figure 4. Each bar represents the comparison before and after 30 days, 30 days and 90 days after Huaier administration. Up-regulation was indicated in red bars, and down-regulation in blue bars. The relationship of those quantitative and qualitative DEG alterations to encoding transcription factors (TF) was clearly demonstrated by the TF-DEG network changes(Figure 5). These functional linkage map of TF-DEG network were most convenient form to comprehend the level of genetic changes and the course of recovery. The results indicated the changes in gene expression by Huaier administration were resulted in, and also caused by altered transcription control. Small RNA alterations shown in Figure 3 correlated with the changes in DEGs (Figure 4) and resulting Transcriptional Factor (TF)-DEG network (Figure 5). A comparison between Figs. 3 and 4 well demonstrated miRNA-mediating gene silencing effects. At the same time, this silencing effect of miRNA was well demonstrated in the patients No. 1-5, treated only by Huaier. A long history of FOLFOX6 treatment (patient No. 5 and 7) showed the different pattern, which drastically increased up-regulation of the genes in number. For example, the significant changes in up- and down-regulated DEGs were in inverse proportion to those observed in quantitative mi- and pi-RNA changes, such as significant increase of up-regulated miRNAs in the patient No. 4 resulted in the significant decrease of both up- and down-regulated DEGs. The changes between miRNA and piRNAs typically observed in the numbers (see scale bars in Figure 3). Interestingly, no alterations or modifications in siRNAs were found in these patients. Thus, miRNA-mediated were directly associated with the drastic up-regulation of transcription factor families, which resulted in the modifications in transcripts [7, 8]. We could not detect any alterations in siRNA in the patients and normal control. In the present study, no significant siRNA function was identified so far. Piwi-interacting RNA (piRNA) is the largest class of small non-coding RNA molecules (containing about 24 nucleotides in the present study) expressed in animal cells [23]. PiRNAs were reported distinct from microRNA (miRNA) in size (26-31) nucleotides as opposed to 21–24 nt, lack of sequence conservation, increased complexity. However, as shown in Figure 3, the dynamic statistics of piRNA revealed similar movement as miRNAs, although the total numbers of the known and the novel piRNAs were much smaller compared with miRNAs.
Figure 5: Genome-scope view of TF-DEG network in each patient by the time course of Huaier administration. The red and green dots represent the up-regulated and down-regulated DEGs, respectively. Purple ball represents transcription factor, the greater the node the more DEGs the transcription factor regulate.
3.4 Genome-scope view revealed the key molecule Bcl-xL [16-18] to determine the prognosis
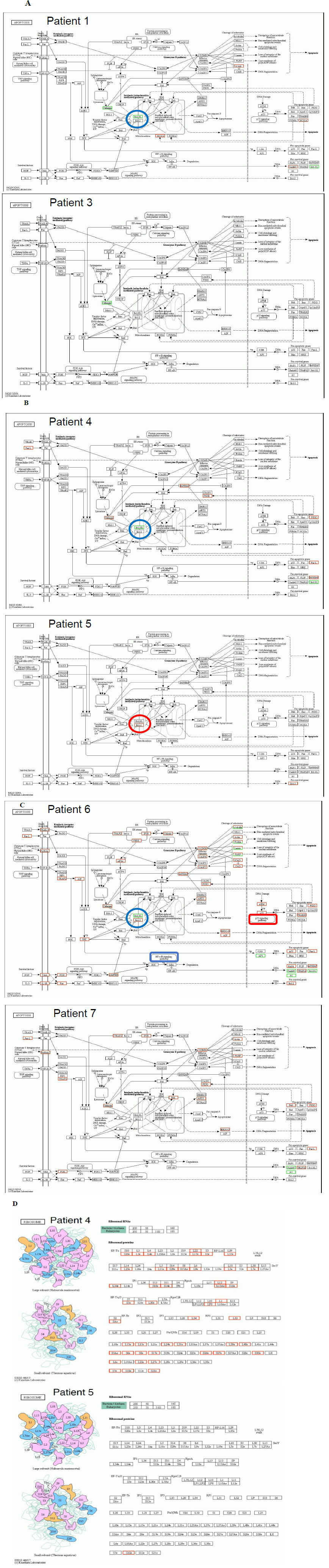
Figure 6 demonstrated detailed factors and molecules with up/down regulation according to the KEGG pathway map (https://www.genome.jp/kegg/), after 30 days of Huaier administration [21]. Here we need typical pattern recognition by genome-scope view, and compare the molecule dynamics involved in pathway functions. As shown in Figure 6, down regulation of Bcl-xL (B-cell lymphoma-extra large), a transmembrane molecule in the mitochondria [16-18], is the key molecule to determine the prognosis in oesophageal squamous cell carcinoma, colorectal carcinoma, and pancreatic adenocarcinoma. The spontaneous recovery in the patients were based on the apoptotic function to prevent cancer cell survival via intrinsic mitochondria-mediated pathway [17]. For example, ribosomal molecular activity was significantly increased in patient No.4, completely recovered from liver metastasis, even though the total number of up-regulated genes were very limited in the low level compared with Patient 5. These Bcl-xL functions were more enhanced in pancreatic adenocarcinoma, and consequent p53 and NF B pathway control were also enhanced in Patient No.6. This result also supported the fact that the efficacy of Bcl-xL on apoptotic function was highly superior to that of Bcl-2, by 5 to 10 times [18].
Figure 6: Genome-scope pattern recognition of Huaier effects on each biophisiological systems by KEGG biological pathways. The comparison of the Bcl-xL down-regulation as a key to determine the prognosis of Stage IV cancer, Panel A; oesophageal squamous cell carcinoma, Panel B; colorectal adenocarcinoma, and Panel C; pancreatic adenocarcinoma. Since a broad range of enriched pathway with various up- and down-regulation of transcripts involved, the major alterations in Bcl-xL and apoptotic signaling pathway was demonstrated in the Figure. In Panel D, the consequent quantitative changes in ribosome were shown as the practical solution to the complete recovery in the patient 4. In Panel C, down-regulation of NF B signaling pathway and up-regulation of p53 signaling pathway were highlighted to show the significant differences between the patients with successfully remission (No.6) and poor prognosis (No. 7).
3.5 Molecular basis for the severe adverse events by Platinum (II) complex involved in conventional chemotherapy; Cisplatin: CDDP, FOLFOX6, and FOLFORINOX
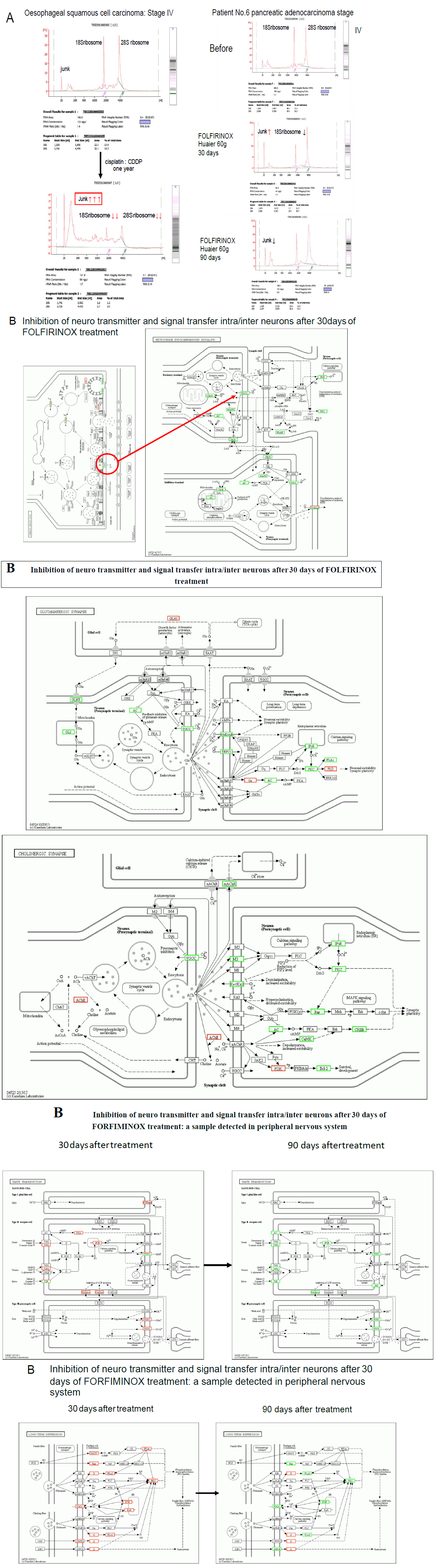
Figure 7 demonstrates the molecular alterations by continuous administration of platinum (II) complex [11-13] to the cancer patients. Panel A is the typical effect of Cisplatin: CDDP [12] on RNA synthesis in the TNM Stage IV oesophageal squamous cell carcinoma with multiple metastasis in both lung. The clinical course and the efficacy of Huaier on this patient was reported in the previous report (Patient No. 5 in ref. 7). This patient had complete remission of the disease after 1.5 year’s treatment with Huaier. At the time of remission, RNA synthesis was normal and the alteration and modification for normal homeostasis was successfully recovered. Then 1 year of chemotherapy (Cisplatin: CDDP, Bevacizumab, etc., administration by two weeks’ interval) was started to cope with complete elimination of metastatic lesion judged by CT images in both lung, and the resulted destruction of RNA synthesis were observed (left column in Panel A), which could not applied for further investigation as a source material. The similar destruction was observed in the course of treatment in the Patient No.6 with Huaier and FOLFIRINOX [13], at 30 days after administration. However, the destruction effect supposed by platinum (II) complex recovered at 90 days after the combined therapy (Figure 7, Panel A, right column). This patient showed significant peripheral neuropathy within couple of weeks after FOLFIRINOX therapy, with strong nausea and dysphagia. These symptoms became less severe except peripheral neuropathy, but after 90 days, we detected massive inhibition in transcription related to the signal transfer in central and peripheral nervous systems as shown in serial Panel Bs in Figure 7. Not only in nervous systems, the influence of platinum (II) complex reached to phycological systems, to initiate long-term depression in the patients.
There have not been defined criteria about the molecular basis of the severe adverse events on central and peripheral nervous systems yet [13]. The strong inhibition of transcription and resulted down-regulation of the molecules in every signaling pathway were clearly noted. The present study provides valuable information on systematic understanding of the onset time and molecular basis for the severe adverse events by FOLFIRINOX treatment. We thus decided to withdraw FOLFIRINOX therapy from the patient No.6, and the improved physical status enabled the patient to have successful surgical dissection of the original cancer lesion (caput pancreatis).
Figure 7: Genome-scope pattern recognition of the severe adverse events of platinum (II) complex to nervous system. Panel A: typical destructed RNA patterns in the samples of oesophageal squamous cell carcinoma patient with both lung metastasis (multiple) after one year administration of Cisplatin: CDDP [12] to the oesophageal squamous cell carcinoma patient with both lung metastasis (multiple), with a comparison to extracted RNAs from pancreatic adenocarcinoma patients. Huaier rescues degradation of RNAs after one month of administration. Series of Panel Bs: Molecular basis of at random damage of signal transfer of inter/intra central and peripheral nervous systems and in the patient No.6 with FOLFIRINOX [13].
4. Discussion
4.1 The present study demonstrates
- The patients in the severe advanced stage of cancer can be recovered by the treatment inducing strong inhibition of Bcl-xL [16-19]. The rescued apoptotic functions seemed to be responsible for the specific cell death in occult metastasis in the peripheral blood to prevent the recurrence and relapse of cancer.
- Huaier administration contributes to alter and modify in genomic and genetic status for normal apoptotic signal transfer via intrinsic mitochondria-mediated pathway, which resulted in successful recovery/remission without any side effects and toxicity.
- The surgical dissection of tumor mass, or complete removal of cancer apparently supports the recovery.
- The molecular basis for the severe adverse events by platinum (II) complex therapy was destruction of RNA synthesis resulted in massive down-regulation of transcripts in every signaling pathway, especially in signal transfer in central and peripheral nervous systems, and 30 days to 90 days.
In the present study, it is surprising to see the minimum-essential genomic flexibility and capability caused enough alteration and modification for recovery. Even though the genomic potential was not so large in the Patient No.4, it was enough to recover all the dysfunctions caused by colorectal cancer (of course, surgical dissection was performed 3 years ago). At the beginning, the average number of RNA editing events such as SNP and INDL showed that Huaier administration induced approximately 4 times higher movability of the genome, ranging 2 times to 7 times higher than those detected as identified among normal population [10]. The statistics of splicing events resulted in expanded the plasticity and flexibility, and consequently enhanced the genomic capability to manage or cope with the following processed in transcription and translation. These results brought over 24,344 novel transcripts, and also caused the drastic rearrangements in number of up/down-regulated transcripts. These changes in transcripts were successfully demonstrated as Transcription Factor (TF)-Differentially Expressed Genes (DEG) lineage map (see Figure 5).
The duration period required for the recovery was dependent on the physiological status in each individual, and also the dose of Huaier. This observation was identical to the clinical observation reported previously. Interestingly, the patients No.1 and 2 with multiple metastasis was successfully recovered judged by CT images and endoscopic examination, without any significant increase of both up- and down-regulated transcripts. The clinical examinations showed a good coincidence to the results obtained here, with a significant improvement/elimination of metastatic lesion, and that without any more progression of the disease.
In the patient No.4, haemoglobin counts were improved from 6.0 to 9.8 g/dl. miRNAs also play crucial roles in the regulation of complex enzymatic cascades including the haemostatic blood coagulation system. Large scale studies of functional miRNA targeting have recently uncovered rationale therapeutic targets in the haemostatic system [24, 25]. Many novel miRNAs significantly contributed to cancer recovery, and further investigation of these molecules in vitro and in vivo are expected [24-27]. miRNAs have been reported to typically silence genes by repression of translation, and with broader specificity than siRNA (no changes at all in the present study), to functions in RNA silencing and post-transcriptional regulation of gene expression. The novel molecules were deposited to NCBI GEO: GSE157086.
The consequent restoration of transcription control seemed to be the major molecular mechanisms induced by Huaier administration. However, it is not true to the patients with platinum (II) complex therapy. They did not demonstrate typical mi-RNA-mediated transcription control, merely indicates the massive up-regulation of the transcripts which were inhibited randomly before. The severe adverse events of platinum (II) complex, starting at least 30 days after administration, should be taken into consideration to the effective therapy to cancer patients, and hopefully, with adequate dose of Huaier administration. We are suggesting 20g of Huaier per day before one month of surgical treatment (colorectal cancer), up to 3 months for endocrine tumors (pituitary, thyroid, and adrenal gland). Pre-treatment with Huaier has been reported as significantly effective especially to breast cancer.
5. Conclusion
The present study provided the importance of normal or enhanced function of Bcl-xL is required for the recovery from the most severe stage of cancer. The effect was originated from a drastic alteration and modification of RNA events, followed by mi-RNA mediated transcriptional control toward a large-scale alteration in DEGs. Although the individual genomic potential to undergo those changes and modulations is the key for cancer recovery, minimum-essentials were enough to recover. Since the surgical dissection of major tumor mass was obviously recommended as a first choice of cancer treatment if applicable, the elimination of occult metastasis after surgical dissection should be emphasized, too. These effects are spontaneous, and dose- and duration-dependent. It is the major reason why Huaier does not cause any side effects and/or toxicity.
Acknowledgements
The authors wish to thank cancer patient volunteers and many healthy volunteers kindly collaborated with the present study. We also wish to thank Prof. Dr. Tongbiao Zhao, Professor of Stem Cell and Immunology, Institute of Zoology, Chinese Academy of Sciences, China, for critical review and the comments on the project scheme; the reviewing committee for the medical ethics and safety monitoring of the project. The present study was grant-in-aid from QiDong Gaitianli Medicines Co., Ltd. and Japan Kampo NewMedicine, Co., Ltd.
Author Contributions
T.T., M.T., designed the study from the clinical observation of the cancer patients with Huaier treatment (as a complementary therapy), and managed the sampling and clinical assessment of the patient volunteers, statistically analyzed the data, and drafted the manuscript. F.T., H.L., managed total RNA and small nuclear RNA sequencing and conducted systematic analysis of the data. S.S., and T. S., contributed clinical diagnosis and treatment of the patients, together with the assessment of QOL and the effects of Huaier administration, Z.L., D.W., contributed to the provision of Huaier granules and clinical evaluation of the data, especially focused on immunological evaluation.
Conflict of Interest
The authors have no competing interest to declare.
Author information
Readers are welcome to comment on the paper. Correspondence should be addressed to T.T. (ttanaka@bradeion.com) and M.T. (manami-tanaka@bradeion.com), and those researchers contributed equally to this work. Requests for Huaier extract and commercially-available granules should be addressed to D.W. (teii@newkampo.co.jp) and M.T. (manami-tanaka@bradeion.com).
References
- World Cancer Report. International Agency for Research on Cancer, World Health Organization (2014).
- Bray F, ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin 68 (2018): 394-424.
- Song X, Li Y, Zhang H, et al. The anticancer effect of Huaier (Review). Oncol. Rep 34 (2015): 12-21.
- Wang X, Wang N, Cheung F, et al. Chinese medicines for prevention and treatment of human hepatocellular carcinoma: current progress on pharmacological actions and mechanisms. J Integr Med 13 (2015): 142-164.
- Chen Q, Shu C, Laurence AD, et al. Effect of Huaier granule on recurrence after curative resection of HCC: A multicentre, randomised clinical trial. Gut 67 (2018): 2006-2016.
- Tanaka T, Suzuki T, Nakamura J, et al. Huaier Regulates Cell Fate by the Rescue of Disrupted Transcription Control in the Hippo Signaling Pathway. Arch Clin Biomed Res 1 (2017): 179-199.
- Tanaka M, Tanaka T, Teng F, et al. Huaier Induces Cancer Recovery by Rescuing Impaired Function of Transcription Control Based On the Individual Genomic Potential. Arch Clin Biomed Res 4 (2020): 817-855.
- Tanaka M, Tanaka T, Teng F, et al. Anti-cancer effects of Huaier on prostate cancer; miRNA-mediated transcription control induced both inhibition of active progression and prevention of relapse. J. Alternative Compl Integr Med 7 (2021): 146-155.
- Mo JS, Park JW, Guan KL. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep 25 (2014): 642-656.
- Peng Z, Cheng Y, Tan BC, et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol 30 (2012): 253-260.
- Andre T, Boni C, Mpinedji-Boudiaf, et al. Oxaplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. New Eng J Med 350 (2004): 2343-2351.
- George B. Kauffman. Michele Peyrone (1813–1883) Discoverer of Cisplatin. Platinum Metals Review 54 (2010): 250-256.
- Conroy T, Drsseigne F, Ychou M, et al. FOLFIRINOX cersus Gemcitabine for metastatic pancreatic cancer. New Eng J Med 364 (2011): 1817-1825.
- Mecedo FI, Ryon E, maithel SK, et al. Survival outcomes associated with clinical and pathological response following neoadjuycant FOLFIRINOX or Gemcitabine/Nac-paclitaxel chemotherapy in resected pancreatic cancer. Annals Sur 270 (2019): 400-413.
- Barnes CA, Chavez MI, Tsai S, et al. Survival of patients with borderline resectable pancreatic cancer who received neoadjucant therapy and surgery. Surgery 166 (2019): 277-285.
- Korsmeyer SJ. Regulators of Cell Death. Trends in Genetics 11 (1995): 101-105.
- Finucane DM, Bossy-Wetzel E, Waterhouse NJ, et al. Bax-induced Caspase Activation and Apoptosis via Cytochromec Release from Mitochondria Is Inhibitable by Bcl-xL. J Biol Chem 274 (1999): 2225-2233.
- Fiebig AA, Zhu W, Hollebach C, et al. Bcl-xL is qualitatively different from and ten times more effective than Bcl-2 when expressed in a breast cancer cell line. BMC Cancer 6 (2006): 213.
- Wyld L, Bollantuono I, Tohkonia T, et al. Senescence and Cancer: A Review of Clinical Implications of Senescence and Senotherapies. Cancers 12 (2020): e2134.
- Song Y, Li L, Ou Y, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature 509 (2014): 91-95.
- Kanehisa M, Araki M, Goto S, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res 36 (2008): 480-484.
- Metzgar RS, Rodriguez N, Finn OJ, et al. Detection of a pancreatic cancer-associated antigen (DU-PAN-2 antigen) in serum and ascites of patients with adenocarcinoma. Proc Natl Acad Sci USA 81 (1984): 5242-5246.
- Seto AG, Kingston RE, Lau NC. The Coming of Age for Piwi Proteins. Mol Cell 26 (2007): 603-609.
- Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell 136 (2009): 215-233.
- Nourse J, Braun J, Lackner K, et al. Large-scale identification of functional microRNA targeting reveals cooperative regulation of the hemostatic system. J Thrombosis and Haemostasis 16 (2018): 2233-2245.
- Griffiths-Jones S. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34 (2006): D140-D144.
- Teruel-Montoya R, Rosendaal FR, Martínez C. MicroRNAs in hemostasis. J. Thrombosis and Haemostasis 13 (2015): 170-181


 Impact Factor: * 5.8
Impact Factor: * 5.8 Acceptance Rate: 71.20%
Acceptance Rate: 71.20%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 