Towards homeostasis: Correlations in the Gut Microbiome of Young Overweight Females following a Yeast Nutritional Supplement
Peter Joller1*, Sophie Cabaset2
1Dr. Joller BioMedical Consulting, 8057 Zurich, Switzerland
2Bio-Strath® AG, Zurich, 8008 Switzerland
*Corresponding author: Peter Joller, BioMedical Consulting, 8057 Zurich, Switzerland
Received: 23 July 2021; Accepted: 02 August 2021; Published: 17 September 2021
Article Information
Citation:
Peter Joller, Sophie Cabaset. Towards homeostasis: Correlations in the Gut Microbiome of Young Overweight Females following a Yeast Nutritional Supplement. Archives of Clinical and Biomedical Research 5 (2021): 714-723.
View / Download Pdf Share at FacebookAbstract
Background: Previous scientific studies showed that the yeast-based dietary supplement affected the gut and the bowel movement significantly. To find reasons concerning this matter we devised this gut microbiome study.
Methods: At the beginning and after three weeks of supplementation the stool samples of the 26 female probands were analyzed by shotgun metagenomics sequencing. The results were assessed by multiple statistical methods. The initial percentage of a certain bacterium was compared with the difference at the end of the study using correlation statistics.
Results: Sixty-eight gut bacteria were present in all probands in different percentages. The overall difference from day 1 to day 21 in all participants was more than ±20%. The in-depth analysis of these differences revealed a correlation between the initial value and increase or decline at the end in the sense that high values for a certain bacterium recessed and low values expanded in the direction of an average.
Conclusions: Within three weeks the yeast-based nutritional supplement influenced the bacteria of the gut microbiome in a tendency towards mean values. This could be due to immunological actions and/or quorum sensing.
Keywords
<p>BMI; Fecal collections; OMNIgene®âˆ™GUT; Shotgun Metagenomic Sequencing; Yeast supplement; Homeostasis</p>
Article Details
1. Introduction
Life without a microbiota is not conceivable and eukaryotic life became only possible through the help of bacteria. All life on earth should be framed in the context of microbial evolution [1]. The gut microbiome is not only responsible for gaining additional energy from food and protecting us from pathogens but also for instructing the immune system, quorum sensing, gene transfer, as well as producing amino acids, neurotransmitters, hormones, vitamins and mucus. Nonetheless, many diseases and syndromes are induced by members of the microbiota. There is still no consensus on how a healthy gut microbiome looks like [2]. Apart from pathogenic and beneficial microbes, also the quantity and composition of the microbiome affect our well-being [3]. Moreover, the diversity and complexity of the microbial populations vary greatly between individuals [4]. Further, functional redundancy in the microbiota with different species using the same niches is necessary for a balanced, homeostatic survival. There must be a cooperative interaction with other strains in the gut [5]. Dysbiosis, toxic effects, inflammation, and infectious diseases are contingent on the number of bacteria and the composition of the microbiome. Health depends on a subtle balance between the host response and its colonizers [1]. In reality, the residents of the large bowel cannot be divided into good and evil. When present in too high numbers, facultative pathogens like Escherichia coli can cause sepsis, Bacteroides cause abscesses, Enterococci cause endocarditis, Clostridioides perfringens cause gas gangrene. Inversely, indigenous bacteria are potential pathogens: Bacteroides, Enterobacteriaceae, Enterococci, and Clostridioides histolyticum [6]. Besides that, specific pathogens such as Serpulina, Fusobacteria, Enterobacteriaceae, or Gardnerella may lead to serious diseases. Even alleged beneficial microorganisms like Akkermansia muciniphila in too high quantities can be harmful [7,8]. Quorum sensing in the gut microbiota involving autoinducers is a key issue [9].
2. Materials and Methods
2.1 Study design
2.1.1 Participants: 26 healthy females, 25-35 years of age exhibiting a BMI (Body Mass Index) between 30 and 35 were selected in a specialized medical institution (Adimed - Zentrum für Adipositas- und Stoffwechselmedizin Winterthur, Switzerland).
The ethics committee approved the study and the participants signed an informed consent. The inclusion criteria were gender (female); age (between 25 and 35 years); BMI (from 30 to 35); linguistic competence in German and handy users. Excluded were persons with nutrition-therapy dependent diseases and other serious ailments requiring therapy; participants who were on a diet during the last half year or took medication for weight loss prior to the study. Also, enrollment in any weight loss scheme and continual consumption of pre- or probiotics were arguments for exclusion. To keep the statistical basis as homogenous as possible only women of a certain age and BMI were recruited.
Ethics approval BASEC ID 2016-02162 (15.08.2017); ClinicalTrials.gov NCT03223987, BS2016.1
2.1.2 Study conduct: Three-week single-arm pre/post pilot intervention. Entrants underwent three visits at Adimed. During the run-in phase, the content of the trial was explained to the study participants. The informed consent form was signed before undergoing a physical examination. Then the participants were given the tubes for fecal collection. In the subsequent visit the females brought back their first stool collection. They were given the nutritional supplement for the subsequent three weeks. At the end of the study, they brought back their second fecal sample and the remaining quantity of their dietary supplement. The personal diet over the whole study time was monitored with a smartphone-based nutrition protocol (oviva Schweiz, www.oviva.com/ch). Participants were instructed not to depart from the normal nutrition during the entire study. No deviations from the rules were observed.
2.1.3 Stool sample collection: Fecal samples were saved at home with the help of an OMNIgene®•GUT tube (DNA Genotek Inc. 3000 - 500 Palladium Drive, Ottawa, ON, Canada) [10].
2.1.4 Dietary supplement: Strath® liquid consists of a plasmolyzed herbal yeast preparation containing over 60 vital substances (among others vitamins, minerals, enzymes, amino acids, and oligo-elements). Strath® is sold worldwide for 60 years (Bio-Strath® AG, Zurich, Switzerland, https://bio-strath.com/). Constituents: 83% Plasmolyzed Herbal Yeast (Saccharomyces cerevisiae MEYEN), 9% Malt extract, 5% Orange syrup, 3% Honey. Supplement dosage in the study: 3 x 5ml daily (morning, midday, and evening 15 minutes before the meals) for three weeks. Compliance was confirmed by measuring the quantity of dietary supplement left after three weeks of intake. No discrepancies from the rules were found.
2.2 Shotgun metagenome sequencing
The extracted fecal samples were analyzed by shotgun metagenomic sequencing (CoreBiome, St. Paul, MN, USA). Extensive method specification was summarized in a prior publication [11].
2.2.1 Statistics: Raw Taxonomy Tables, Filtered Taxonomy Tables, Strain Coverage Tables, Beta Diversity including Species Bray-Curtis Distance Matrix, and Alpha Diversity were calculated by CoreBiome. Downstream analysis was done with Analyse-it® for Excel 5.40 build 7149.20136 (Leeds, United Kingdom). Correlations were computed with bimodal Pearson’s r and the Benjamini-Hochberg Procedure for False Discovery Rate (FDR). Significance level α=5%; confidence interval 95% two-sided; Fisher’s Z Method. No missing data were present.
3. Results
Since the determination of the gut microbiome composition with shotgun metagenomic sequencing cannot reveal the function of the individual bacteria, it is difficult to assign a clear role to the microbes found. This at the moment is only possible by cultivation [12]. Not only the genome but also the quantity of a particular bacterium determines whether it is a friend or foe. Therefore, the principal focus in this publication is a possible interpretation of the negative and positive bacterial population changes in the individual participant we found in our pilot study.
We calculated the correlation of the initial percentages in the entirety of 68 individual bacterial species with the difference after 3 weeks supplementation in all probands. Twenty-five of 68 species showed a statistically significant correlation between these percentages.
3.1. Correlation results
Table 1: A) Species showing a significant correlation between the initial percentage of the individual bacterium and its percentual change after 3 weeks in all participants; B) Species revealing no statistical significance in the correlation analysis; C) Some bacterial species were not analyzable because of too many samples with percentages close to zero or with only minor changes during the trial.
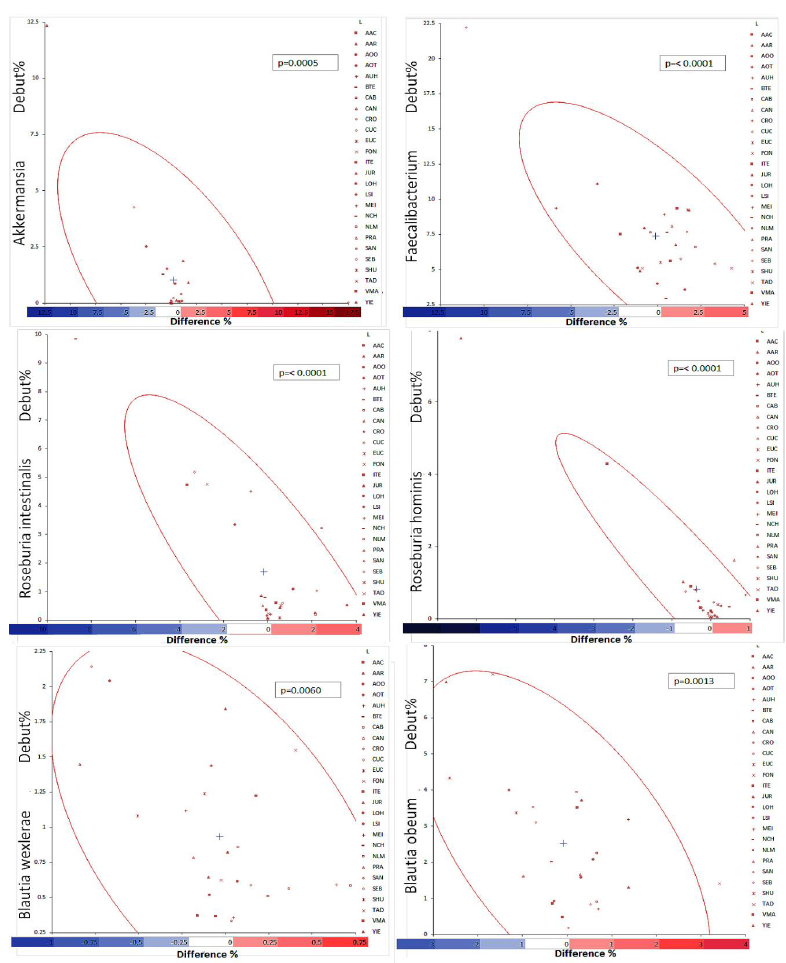
For some bacteria of the gut microbiome, there is something like a consensus about the positive influence on the carrier. This applies for example to Akkermansia muciniphila [13-15], Faecalibacterium prausnitzii [16-18], Roseburia intestinalis [19-21], Roseburia hominis [22-24], Blautia wexlerae [25-27] and Blautia obeum [28-30]. For these species, the correlation diagrams are reproduced in Figure 1.
3.2. Correlation diagrams
Figure 1: Correlation diagrams of 6 potentially beneficial bacterial species in all participants. They show the initial percentage of the bacterium (y-axis) in percent of all bacteria found in a proband correlated with the difference (x-axis) in percent in the same proband after three weeks of treatment. The density ellipse equals 95% CI. The three-letter code refers to the participants.
4. Discussion
In earlier publications of our study [11, 31] we reported an improvement by the supplement in the fields of composition of the bacteria, in the Firmicutes to Bacteroidetes ratio (F/B), in the amount of butyrate producing bacteria, in significantly lowering blood pressure and betterment of the metabolic syndrome.
Since we could then just describe the outcome, we wanted to find reasons for these effects. Therefore, we reanalyzed statistically our results of the shotgun metagenomic sequencing. When computing the correlations of the initial percentage of a specific bacterium and the percentual change for this bacterium at the end of the study in the whole group we found for 25 bacterial species a highly significant correlation in the sense that initially high amounts diminished and low values increased.
This suggests a trend to homeostasis which would be a highly important feature in understanding and treating dysbiosis of the gut microbiome. To date, we did not find a similar observation linked to a dietary intervention in the literature.
Alleged beneficial bacteria like Akkermansia muciniphila in an increased amount can be detrimental [8, 9]. Also, Faecalibacterium prausnitzii can be associated with unwanted symptoms [32, 33], same for Roseburia intestinalis, Roseburia hominis [34, 35], Blautia wexlerae, and Blautia obeum [36, 37]. These observations were done in persons treated for a medical condition, while our participants were healthy. Hence there might be a skew in our study.
The quantitative range of a certain bacterium and the overall composition in healthy people is not known [15]. As stated before, the diversity and complexity of the microbial populations vary greatly between individuals [4], and for a bacterium to be effective, there must also be collaboration with other strains in the gut [5]. Quorum sensing [7] is therefore a crucial performance in the gut microbiota. As bacterial density cannot be sensed directly [16] there must be another approach.
One method of numerically adjusting the population sizes of the bacterial species is quorum sensing. Quorum sensing is a cell-to-cell communication process that enables bacteria to collectively modify behavior in response to changes in the cell density and species composition of the surrounding microbial community. Quorum sensing is a chemical communication (by auto inducers) among bacteria and can partially shape the composition of the microbiota [7, 38-41].
Auto inducers are secreted by the bacteria themselves [17]. Bivar Xavier states that quorum sensing can influence the balance of the major bacterial groups in the gut and promote recovery of gut homeostasis [40]. Also, the innate and the adaptive immune systems are involved in adjusting the numbers of certain bacteria [41-43]. How the food supplement based on herbal yeast is connected to quorum sensing and/or alterations in the immune system remains currently unsolved.
Our study sample involved only 26 persons, which could be non-representative. Nevertheless, the proportion of the significant correlations in our results is too high to be at random.
5. Conclusion
In a study in healthy obese young women [11, 31] taking 3x5ml of a plasmolyzed herbal yeast preparation per day for three weeks, we found a modification of more than a ± 0.5 log2 fold change in the gut microbiome, an increase in butyrate producing species, a normalization of the F/B ratio and a statistically significant lowering of the blood pressure.
It became evident in the subsequent statistical workup that this change in the quantity of the bacteria showed a statistically significant correlation in 25 of the 68 gut bacteria found with shotgun metagenome sequencing.
The transformation is based on altering initially high bacterial quantities to medium values and low quantities to average values during the study. Quorum sensing and/or immunological mechanisms might play a role in this adaptation.
Declarations
Author Contributions
PJ: Did the study design, statistics and draft writing. SC: Involved in the participant work-up, enhancing and correcting the draft. Both authors read and approved the final manuscript.
Funding
The study was supported by Bio-Strath® AG, Zurich, Switzerland, www.bio-strath.com
Conflict of Interest
Peter Joller received financial support from Bio-Strath AG. Sophie Cabaset is an employee of Bio-Strath AG
Review Boards
Trial Registration: BASEC ID 2016-02162 (15.08.2017); ClinicalTrials.gov NCT03223987, BS2016.1
Informed Consent
Informed Consent was obtained in writing from all participants.
Data Availability Statement
All data concerning shotgun metagenomic sequencing are with CoreBiome (CoreBiome, St. Paul, MN, USA). All data underlying the downstream statistics can be demanded from the corresponding author.
References
- Godoy-Vitorino F. Human microbial ecology and the rising new medicine. Ann. Transl. Med 7 (2019): 342-342.
- Eisenstein M. The hunt for a healthy microbiome. Nature 577 (2020).
- Mills S, Stanton C, Lane JA, Smith GJ, Ross RP. Precision nutrition and the microbiome, part I: Current state of the science. Nutrients 11 (2019): 1-45.
- Huttenhower C et al. Structure, function and diversity of the healthy human microbiome. Nature 486 (2012): 207-214.
- Klemashevich C, Wu C, Howsmon D, Alaniz RC, Lee K, et al. Rational identification of diet-derived postbiotics for improving intestinal microbiota function. Curr. Opin. Biotechnol 26 (2014): 85-90.
- Swidsinski A, Loening-Baucke V. Functional structure of intestinal microbiota in health and disease. Hum. Microbiota How Microb. Communities Affect Heal. Dis (2013): 211-254.
- Geerlings S, Kostopoulos I, de Vos W, Belzer C. Akkermansia muciniphila in the Human Gastrointestinal Tract: When, Where, and How? Microorganisms 6 (2018): 75.
- Keshavarzian A, et al. Colonic bacterial composition in Parkinson’s disease. Mov. Disord 30 (2015): 1351-1360.
- Kai Papenfort B, Bassler. Quorum-Sensing Signal-Response Systems in Gram-Negative Bacteria. Nat Rev Microbiol 14 (2016): 576-588.
- Ilett EE. et al. Gut microbiome comparability of fresh-frozen versus stabilized-frozen samples from hospitalized patients using 16S rRNA gene and shotgun metagenomic sequencing. Sci. Rep 9 (2019): 1-11.
- Joller P, Cabaset S, Maurer S. Influence of a Food Supplement on the Gut Microbiome in Healthy Overweight Women. Funct. Foods Heal. Dis 10 (2020): 428.
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, et al. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med 6 (2009).
- Cani PD, de Vos WM. Next-generation beneficial microbes: The case of Akkermansia muciniphila. Front. Microbiol 8 (2017): 1-8.
- Ottman N, Geerlings SY, Aalvink S, de Vos WM, Belzer C. Action and function of Akkermansia muciniphila in microbiome ecology, health and disease. Best Pract. Res. Clin. Gastroenterol 31 (2017): 637-642.
- Depommier C, et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat. Med 25 (2019): 1096-1103.
- Heinken A, Khan MT, Paglia G, Rodionov DA, Harmsen HJM, et al. Functional metabolic map of Faecalibacterium prausnitzii, a beneficial human gut microbe. J. Bacteriol 196 (2014): 3289-3302.
- Blanco G, Sánchez B, Fdez-Riverola F, Margolles A, Lourenço A. In silico approach for unveiling the glycoside hydrolase activities in faecalibacterium prausnitzii through a systematic and integrative large-scale analysis. Front. Microbiol 10 (2019): 1-12.
- Miquel S, et al. Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol 16 (2013): 255-261.
- Mirande C, et al. Dietary fibre degradation and fermentation by two xylanolytic bacteria Bacteroides xylanisolvens XB1AT and Roseburia intestinalis XB6B4 from the human intestine. J. Appl. Microbiol 109 (2010): 451-460.
- La Rosa SL, et al. The human gut Firmicute Roseburia intestinalis is a primary degrader of dietary β-mannans. Nat. Commun 10 (2019): 1-14.
- Devillard E, McIntosh FM, Duncan SH, Wallace RJ. Metabolism of linoleic acid by human gut bacteria: Different routes for biosynthesis of conjugated linoleic acid. J. Bacteriol 189 (2007): 2566-2570.
- Zhang J, et al. Beneficial effect of butyrate-producing Lachnospiraceae on stress-induced visceral hypersensitivity in rats. J. Gastroenterol. Hepatol 34 (2019): 1368-1376.
- Moens F, et al. A four-strain probiotic exerts positive immunomodulatory effects by enhancing colonic butyrate production in vitro. J. Pharm 555 (2019): 1-10.
- Machiels K, et al. A decrease of the butyrate-producing species roseburia hominis and faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63 (2014): 1275-1283.
- Hirakawa A, et al. The Nested Study on the Intestinal Microbiota in GENKI Study with Special Reference to the Effect of Brown Rice Eating. J. Obes. Chronic Dis 03 (2019): 1-13.
- Benítez-Páez A, Gómez del Pugar EM, López-Almela I, Moya-Pérez A, Codoñer-Franch P, et al. Depletion of Blautia Species in the Microbiota of Obese Children Relates to Intestinal Inflammation and Metabolic Phenotype Worsening. mSystems 5 (2020): 1-13.
- Kasai C, et al. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analysed by terminal restriction fragment length polymorphism and next-generation sequencing (2015): 1-10.
- Huang L, et al. Analysis of microbiota in elderly patients with Acute Cerebral Infarction. PeerJ 2019 (2019).
- Chénard T, Malick M, Dubé J, Massé E. The influence of blood on the human gut microbiome. BMC Microbiol 20 (2020): 1-10.
- Mullish BH, et al. Microbial bile salt hydrolases mediate the efficacy of faecal microbiota transplant in the treatment of recurrent Clostridioides difficile infection. Gut 68 (2019): 1791-1800.
- Joller-Jemelka HI, Joller P, Cabaset S. Auswirkungen auf das Mikrobiom. Schweizer Zeitschrift für Ernährungsmedizin 1 (2021): 15-18.
- Codoñer FM, et al. Gut microbial composition in patients with psoriasis. Sci. Rep 8 (2018): 1-7.
- Song H, Yoo Y, Hwang J, Na YC, Kim HS. Faecalibacterium prausnitzii subspecies-level dysbiosis in the human gut microbiome underlying atopic dermatitis. J. Allergy Clin. Immunol (2016).
- Ruff WE, Vieira SM, Kriegel MA. The Role of the Gut Microbiota in the Pathogenesis of Antiphospholipid Syndrome. Curr. Rheumatol. Rep 17 (2014): 1-10.
- Robinson H, Barrett H, Gomez-Arango L, McIntyre HD, Callaway L, et al. Ketonuria is associated with changes to the abundance of roseburia in the gut microbiota of overweight and obese women at 16 weeks gestation: A cross-sectional observational study. Nutrients 11 (2019): 1-10.
- Bard JM, et al. Relationship between intestinal microbiota and clinical characteristics of patients with early stage breast cancer. FASEB J (2015).
- Luo XM, et al. Gut microbiota in human systemic lupus erythematosus and a mouse model of lupus. Appl. Environ. Microbiol 84 (2018): 1-10.
- Falony G, et al. Population-level analysis of gut microbiome variation. Science 352 (2016): 560-564.
- Drees B, Reiger M, Jung K, Bischofs IB. A modular view of the diversity of cell-density-encoding schemes in bacterial quorum-sensing systems. Biophys. J 107 (2014): 266-277.
- Thompson JA, Oliveira RA, Djukovic A, Ubeda C, Xavier KB. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep 10 (2015): 1861-1871.
- Pietschke C, et al. Host modification of a bacterial quorum-sensing signal induces a phenotypic switch in bacterial symbionts. Proc. Natl. Acad. Sci. USA 114 (2017): E8488-E8497.
- Davies DG, et al. The Involvement of Cell-to-Cell Signals in the Development of a Bacterial Biofilm Published by: American Association for the Advancement of Science Stable URL: http://www.jstor.org/stable/2895685 REFERENCES Linked references are available on JSTOR for t. Science 280 (1998): 295-298.
- Truax AD, et al. The Inhibitory Innate Immune Sensor NLRP12 Maintains a Threshold against Obesity by Regulating Gut Microbiota Homeostasis. Cell Host Microbe 24 (2018): 364-378.


 Impact Factor: * 5.8
Impact Factor: * 5.8 Acceptance Rate: 71.20%
Acceptance Rate: 71.20%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 