miR-425-5p, a SOX2 target, regulates the expression of FOXJ3 and RAB31 and promotes the survival of GSCs
Arlet María Acanda de la Rocha1,2,3,12, Marisol González-Huarriz1,2,3, Elizabeth Guruceaga1,6, Nicole Mihelson4, Sonia Tejada-Solís1,2,7, Ricardo Díez-Valle1,2,7, Naiara Martínez-Vélez1,2,3, Juan Fueyo8, Candelaria Gomez-Manzano8,9, Marta M. Alonso1,2,3, John Laterra4,5,10,11 and Hernando López-Bertoni4,5*
1The Health Research Institute of Navarra (IDISNA), Spain
2Program in Solid Tumors and Biomarkers, Foundation for the Applied Medical Research, Spain
3Department of Pediatrics, University Hospital of Navarra, Pamplona, Spain
4Hugo W Moser Research Institute at Kennedy Krieger, Baltimore, MD, USA
5Department of Neurology, The Johns Hopkins School of Medicine, Baltimore, MD, USA
6Bioinformatics Unit, Center for Applied Medical Research, Pamplona, Spain
7Department of Neurosurgery, University Hospital of Navarra, Pamplona, Spain
8Department of Neuro-Oncology, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA
9Department of Genetics, The University of Texas MD Anderson Cancer Center, Houston, Texas, USA
10Department of Neuroscience, The Johns Hopkins School of Medicine, Baltimore, MD, USA
11Department of Oncology, The Johns Hopkins School of Medicine, Baltimore, MD, USA
12Department of Environmental Health Sciences. Robert Stempel College of Public Health & Social Work. Florida International University, USA
*Corresponding author: Hernando López-Bertoni, Hugo W Moser Research Institute at Kennedy Krieger, Baltimore, USA
Received: 05 June 2020; Accepted: 15 June 2020; Published: 18 June 2020
Article Information
Citation:
Arlet María Acanda de la Rocha, Marisol González-Huarriz, Elizabeth Guruceaga, Nicole Mihelson, Sonia Tejada-Solís, Ricardo Díez-Valle, Naiara Martínez-Vélez, Juan Fueyo, Candelaria Gomez-Manzano, Marta M. Alonso, John Laterra and Hernando López-Bertoni. miR-425-5p, a SOX2 target, regulates the expression of FOXJ3 and RAB31 and promotes the survival of GSCs. Archives of Clinical and Biomedical Research 4 (2020): 221-238.
View / Download Pdf Share at FacebookAbstract
Glioblastoma (GBM) is the most common malignant primary brain tumor in adults and prognosis is poor despite maximum therapeutic efforts. GBM is composed of heterogeneous cell populations, among which the glioma stem-like cells (GSCs) play an important role in tumor cell self-renewal and the ability to initiate and drive tumor growth and recurrence. The transcription factor SOX2 is enriched in GSCs where it controls the stem cell phenotype, invasion and maintenance of tumorigenicity. Therefore, understanding the molecular mechanisms governed by SOX2 in GSCs is crucial to developing targeted therapies against this resistant cell population. In this study, we identified and validated a miRNA profile regulated by SOX2 in GSCs. Among these miRNAs, miR-425-5p emerged as a significant robust candidate for further study. The expression of miR- 425-5p was significantly enriched in clinical GBM specimens compared with a human brain reference sample and showed a positive correlation with SOX2 expression. Using a combination of in silico analyses and molecular approaches, we show that SOX2 binds to the promoter of miR-425-5p. Loss of function studies show that repressing miR-425-5p expression in multiple GSCs inhibited neurosphere renewal and induced cell death. More importantly, miR-425-5p inhibition extended survival in an orthotopic GBM mouse model. Finally, combining several bioinformatics platforms with biological endpoints in multiple GSC lines, we identified FOXJ3 and RAB31 as high confidence miR-425-5p target genes. Our findings show that miR-425-5p is a GBM stem cell survival factor and that miR-425-5p inhibition function is a potential strategy for treating GBM.
Keywords
<p>Glioblastoma; miR-425-5p; SOX2; Glioma stem cells; Apoptosis</p>
Article Details
Introduction
Glioblastoma (GBM) is the most common malignant and aggressive primary brain tumor in adults, and despite the advances in multiple treatments, the median survival for patients is only 12-20 months [1,2]. GBM is characterized by a strong vascular proliferation, resistance to apoptosis, and diffuse infiltration of the brain making complete surgical resection difficult [3]. At the cellular level GBM is composed of heterogeneous cell populations and among them is a unique subset of tumor cells with stem cell properties, known as glioma stem-like cells (GSCs) [4]. GSCs display the ability to self-renew and differentiate into distinct lineages and are hypothesized to initiate and drive tumor growth and recurrence. As a result, GSCs plays a critical role in GBM progression and recurrence after therapy [5].
The transcription factor SOX2 plays a critical role in stem cell maintenance and is one of the factors necessary to reprogram somatic cells towards pluripotency [6,7]. Genetic and epigenetic mechanisms, including gene amplification and promoter hypomethylation respectively, leads to the up-regulation of SOX2 gene expression in multiple cancer types, including GBM [8]. SOX2 dysregulation affects plasticity and differentiation of GSCs [9]. Moreover, SOX2 is enriched in human GSCs where it controls stemness, migration, invasion and maintenance of tumorigenicity [8,10].
We recently reported that SOX2 controls a network of coding and non-coding RNAs in GSCs and identified a subset of long non-coding RNAs (lncRNAs) as critical down-stream mediators of SOX2 function [11]. In this study, we expand upon these findings by further investigating the miRNA profile regulated by SOX2 in GSCs. We uncover that miR-425-5p is a novel transcriptional target of SOX2 required for GSC survival. Down-regulation of miR-425-5p was found to interfere with GSC neurosphere formation, cell survival and cell-cycle progression, leading to cell death via apoptosis. In vivo studies in a GBM orthotopic model shows that inhibition of miR-425-5p results in a significant increase in the overall median survival time of mice. This study identified miR-425-5p as a novel regulator of GSC survival, providing future novel diagnostic and therapeutic strategies for GBM.
Materials and Methods
Cell Culture
The adult neurosphere lines GSC-11 and GSC-23, a kind gift of Dr. Lang at UT MD Anderson Cancer Center, were established from acute cell dissociation of human GBM surgical specimens and maintained in our lab. The neurosphere lines GBM1A and GBM1B were originally derived and characterized by Vescovi and co-workers [12]. All neurosphere cell lines were cultured and maintained in serum-free medium containing Dulbecco's modified Eagle's medium/nutrient mixture F12 (1:1, vol/vol) (Thermo Fisher Scientific Inc, Waltham, MA) supplemented with 1% of Penicillin/Streptomycin (Lonza, Verviers, Belgium), B27 10x (Thermo Fisher Scientific Inc, Waltham, MA) and 20 ng/ml of both EGF and FGF (Sigma-Aldrich, St Louis, MO) according to the procedures described by Galli [12].
The human embryonic kidney 293FT (HEK293FT) cell line was purchased from ATCC and were maintained in Dulbecco's modified Eagle/F12 medium (1:1, vol/vol) and supplemented with 10% FBS (Fetal Bovine Serum, Thermo Fisher Scientific Inc, Waltham, MA). All the cell lines were grown at 37°C in a humidified incubator with 5% CO2. All cell models were authenticated by STR sequencing.
Patient Samples and Ethic Statement
GBM tissues were obtained from surgical procedures performed at the Department of Neurosurgery of the University Hospital of Navarra. Tissue samples were resected during surgery and immediately frozen in liquid nitrogen for subsequent investigation. We collected 30 human GBM specimens. Written informed consent was obtained from all subjects, specifying that sample collection was for experiment purposes, and approved by the Ethics Committee of the University Hospital of Navarra (Ref. 123/2014).
MicroRNA Array
To study the set of miRNAs regulated by SOX2, we subjected samples to a miRNA array analysis. Total RNA from scrambled and SOX2-knock down GSC-11 cells was reverse transcribed into cDNA using Taqman miRNA Reverse Transcription Kit (Applied Biosystems) and loaded onto a Human miRNA array panel containing 384 wells according to manufacturer’s protocol (Applied Biosystems). Quantitative miRNAs expression data were normalized with U6b housekeeping gene and quantified using ABI 7700 sequence detection system (Applied Biosystems, Foster City, CA). The microarray data from this study have been submitted to Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE115086.
For analysis of miRNA targets induced by SOX2, total RNA including small RNA was extracted from GBM1A and GBM1B expressing exogenous SOX2 (GBM1A-SOX2) using miRNeasy kit (Qiagen). The cDNA was synthesized using miScript II RT kit (Qiagen) and used to perform microarray analysis using a brain cancer miRNA PCR Array (SABiosciences, Frederick, MD, USA) according to manufacturer’s instructions. Array data were analyzed using miScript miRNA PCR array data analysis tools (Qiagen, SABiosciences).
Lentivirus Generation and Cell Transduction
For the production of lentiviral particles, we used the 2nd-generation lentiviral system according to Addgene instructions. Lentiviral expression vectors LentimiRa-Off-hsa-miR-425-5p vector to inhibit the expression of miR- 425-5p or pLenti-III-mir-GFP as a Scramble control vector were purchased from ABM (Applied Biological Materials Inc, Richmond, BC, Canada); psPAX2 packaging plasmid and pMD2.G envelope plasmid were obtained from Addgene (Addgene, Cambridge, MA). Co-transfection of the lentiviral packaging/envelope plasmids and transfer vector was performed in HEK239FT cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). The lentiviral particles in supernatant were collected at 48–72 h and used to transduce cells.
GSC-23, GSC-11, GBM1A and GBM1B cells were transduced with lentiviral particles. Briefly, a total of 1.5 × 104 cells were seeded in a 6-well cell culture plate and infected with lentiviral medium containing viral particles and polybrene (1 µg/mL), supplemented with appropriate medium. The following morning cells were spun down and resuspended in fresh neurosphere medium. GFP expression was assessed by fluorescence microscope fluorescence.
qRT–PCR and miRNA Expression
Total RNA was isolated from harvested cells with Trizol reagent according to the manufacturer`s instructions (Life Technologies, California, USA). RNA samples were quantified using Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific) and stored at -80ºC.
For miRNA analysis, total RNA including small RNA was extracted using miRNeasy Kit from Qiagen and 1μg of total RNA was used as a template to generate cDNA using miScript II RT Kit (Qiagen). To measure levels of pre-miRNAs, total RNA was extracted using miRNeasy Kit and cDNA was synthesized from total RNA (1 μg) using gene-specific primers and High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, ThermoFisher). The gene-specific primers included a mixture of 10 μm each of the antisense primers to the selected miRNA and U6 RNA. Following an 80°C denaturation step and 60°C annealing, the cDNA was reverse transcribed for 45 min at 60°C. cDNA was diluted 1:20 before detection using Power SYBR Green PCR Kit from Applied Biosystems (ThermoFisher). Primer sequences are listed below:
|
Gene |
Forward primer (Fw) |
Reverse primer (Rv) |
|
U6 |
CTCGCTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT |
|
miR-7 |
TGGAAGACTAGTGATTTTGTTGT |
AGACTGTGATTTGTTGTCGATT |
|
miR-16 |
GCAGCACGTAAATATTGGCGT |
CAGCAGCACAGTTAATACTGGAGA |
|
miR18a |
TAAGGTGCATCTAGTGCAGATAG |
GAAGGAGCACTTAGGGCAGT |
|
miR-20a |
GCACTAAAGTGCTTATAGTGCAG |
GTACTTTAAGTGCTCATAATGCA |
|
miR-128a |
TGGATTCGGGGCCGTAG |
AAAGAGACCGGTTCACTGTGAG |
|
miR-128b |
GGAAGGGGGGCCGATA |
AAAGAGACCGGTTCACTGTGAG |
|
miR-425-5p |
ATGACACGATCACTCCCGTTG |
GGGCGGACACGACATTC |
Table 1: pre-miRNA Primer Sequences
Neurosphere Formation and Forced Differentiation Assay
To assess neurosphere size, cells were dissociated into single cells and cultured in ultra-low attachment flasks (1.5 × 104 cells/ml). After 10 days, neurospheres were embedded in 1% agarose and stained with 0.1% Wright stain solution for 1–2 h at 37°C. Cells were washed four times with phosphate-buffered saline (PBS) and incubated at 4°C overnight (in PBS) before quantification. Spheres larger than 100 μm were quantified using computer-assisted image analysis (MCID™ Analysis Software).
Forced differentiation was performed according to the method of Galli and co-workers [12] with some modifications. Briefly, the neurosphere cells were plated onto Matrigel in FGF-containing neurosphere medium (no EGF) for 2 days and subsequently grown in 1% FBS without EGF/FGF for 5 days.
Cell Cycle Analysis
GSC-23 cells were collected 7 days after transduction with miR-425-5p or Scramble lentivirus, washed with PBS 1X, fixed by adding ice-cold 70% ethanol and store at 4°C for at least 30 min. Then, cells were washed with PBS and stained with 10 μg/ml Propidium Iodide (Roche), 100µg/ml RNase (Sigma-Aldrich) in PBS, and incubated at 37°C for 30 min in the dark. Cell cycle analysis was then performed with a FACS Calibur flow cytometer (Becton- Dickinson, Franklin Lakes, NJ, USA), while FlowJo software (Ashland, OR, USA) was used for data analysis. Analysis of each sample was performed on greater than 10,000 events, and gating on forward scatter versus side scatter was used to exclude cell debris and doublets. Apoptotic cells are represented by a sub G0 population seen to the left of the G1 peak.
Caspase 3/7 Activity
GSC-23 cells were seeded in quadruplicate (1.5 × 104 cells) into 96-well plate 1 day after transduction with the indicated miRNAs and 50 μL of Caspase-Glo 3/7 reagent was added in quadruplicate the days of measure (7 and 10 days after transduction) and incubated during 1h at 37ºC. Bioluminiscent fluorescence was detected using spectrofluorometer (SpectraMAX Gemini XS, Molecular devices). Luminescence is proportional to the amount of caspase activity present, and so the proportional fluorescence intensity of the different treatment cell groups enables determination of differences in caspase activity.
Annexin V Assay
GSC-23 cells were seeded in a density of cells into 6-well plate and after 7 days of infection cells were washed and resuspended in 1x Binding Buffer. APC Annexin V and SYTOX Blue were added to cells and incubated during 15 min in the dark and analyzed by flow cytometry (Becton-Dickinson, Franklin Lakes, NJ, USA).
Transmission Electron Microscopy
GSC-23 cells were collected 7 days after transduction with miR-425-5p or Sc sponge lentivirus and fixed with a solution containing 4% glutaraldehyde in 0.1M cacodylate buffer, pH 7.4 during 1h at 4ºC. Cells were then centrifuged, treated with 0.25M of sacarose in 0.1M cacodylate buffer, fixed with 1% buffered osmium tetroxide for 1h at 4ºC and stained in block with 1% Millipore-filtered uranyl acetate. The samples were dehydrated with increasing concentrations of ethanol, infiltrated and embedded in LX-112 medium. After this, cells were polymerized in an oven at 60ºC for 2 days. Ultrathin section (65nm) were cut in a Leica Ultracut microtome, stained with uranyl acetate and lead citrate in a Leica EM Stainer, and examined in a Jeol 1210 transmission electron microscope (Jeol Ltd., Herts, UK).
In silico prediction of SOX2 binding sites and miRNA target analysis
SOX2 binding sites 2 kb upstream of the miR-425-5p translation start site were identified via PROMO algorithm [13] using search term ‘SOX2’ (Transcription factor identifier: T01836). In the present study miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/), miRDB (http://mirdb.org/miRDB/) and PicTar (http://pictar.mdc-berlin.de/) platforms were used to predict miR-425-5p target genes.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation assays were performed in GBM1A neurosphere cells expressing transgenic SOX2 using the MAGnify Chromatin Immunoprecipitation system (Life Technologies, Grand Island, NY, USA). Immunoprecipitation was performed with anti-SOX2 (R&D Systems, Minneapolis, MN, USA) or anti-IgG (Life Technologies). We select a region of the promoter of miR-425-5p containing the highest density of predicted SOX2 binding sites for qRT-PCR analysis. Primer sequences for SOX2 (region 1) in the miR-425-5p promoter used were: forward primer (5´ to 3´) CCTGCCCCACGGATCTAA and reverse primer (5´ to 3´) AGCAGGGGACGAAATCCAA.
Protein Extraction and Immunoblotting Assays
For the immunoblotting assays, cells were lysed in (3-[(3-cholamidopropyl)-dimethylammonio]-1-propane sulfonate) buffer for 30 minutes on ice. Samples containing identical amounts of protein (20 µg) were resolved by NuPAGE 4% to 20% Tris-glycine gradient gel (Invitrogen), transferred to nitrocellulose membrane (Bio-Rad Laboratories; Hercules; CA), and blocked in 5% non-fat milk in phosphate-buffered saline/Tween-20. Membranes were probed with antibody against full-length Caspase 3 (Cell Signaling). Membranes were developed according to the protocol for enhanced chemiluminiscence from Amersham (Piscataway, NJ). Equal loading was confirmed using an α-Tubulin antibody (Sigma-Aldrich).
Animal Studies
Ethical approval for animal studies was granted by the Animal Ethical Committee of the University of Navarra (CEEA) under the protocol number CEEA/093-14. All animal studies were done in the veterinary facilities of the Center for Applied Medical Research in accordance with institutional, regional, and national laws and ethical guidelines for experimental animal care. Athymic nude female mice, which were between 6- to 8-week-old, were obtained from Harlan Laboratories (Barcelona, SP) and maintained at The Center for the Applied Medical Research (CIMA, Pamplona, SP) in specific pathogen-free conditions and fed standard laboratory chow. Female nude mice are commonly used in preclinical studies involving brain tumor xenografts, due to their lower aggressiveness relative to that of male nude mice. Less aggressive behavior is preferred in group-housed mice because it reduces the risk of injuries when randomizing or pooling mice; such injuries might represent a confounding factor in the studies. A total of 8 female mice per group were used in this study. GSC-11 cells were transduced with Scramble-sponge or miR-425- 5p sponge and 2 days after transduction equal numbers of viable GSC-11 cells (5.0x105 cells) were engrafted into the caudate nucleus of athymic mice by means of the guide-screw system previously described [14]. Mice were monitored for anesthetic recovery and postsurgical pain during anesthetic recovery until regaining the ability to maintain an upright posture and walk normally, every 1h for the following 4 h, and daily thereafter. After surgery animals were assigned into the cages in a randomized manner. Mice were monitored daily for wellbeing assessment, which included hydration status, signs of pain or distress, and signs of tumor growth, including cranial protuberance and neurologic deficits manifested as seizures and abnormal gait or posture. Animals were euthanized by asphyxia in a CO2 chamber at the first signs of neurologic deficit or weight loss greater than 20%.
Statistical Analysis
Experimental data are represented as the mean ± standard deviation of three technical replicates and were compared using Student's t-test. One-way ANOVA test and Tukey or Bonferroni correction, depending on type of comparison, were used to analyze experiments with multiple simultaneous comparisons. Significant P-values are indicated with asterisks as follows: *P < 0.05, **P < 0.01, ***P < 0.001. Survival data was compiled using the Kaplan- Meier methodology and compared across arms using the log-rank test (Mantel-Cox), with p values lower than 0.05 considered statistically significant. All statistical analyses were performed using the GraphPad Prism Software (GraphPad Software, San Diego, CA).
Results
Molecular profiling identified a subset of SOX2-regulated miRNAs in glioma stem cells
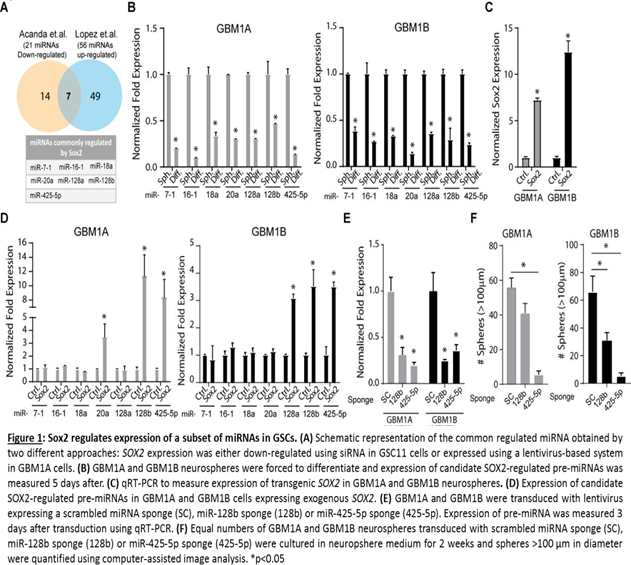
To uncover SOX2 regulated miRNAs we repressed SOX2 expression in GSC-11 cells using siRNA technology and examined its effects on the cell transcriptome using miRNA microarray analysis. Hierarchical clustering analysis identified 21 down- regulated and 35 up-regulated miRNAs (p < 0.01) in GSCs after SOX2 repression [11]. To provide robustness to this miRNA profile, we cross-referenced our miRNA array results to a set of miRNAs identified by Lopez-Bertoni and colleagues to be regulated by exogenous expression of SOX2 in GSCs [15]. Lopez et. al. identified 21 down-regulated and 56 up-regulated miRNAs in GSCs expressing exogenous SOX2. We determined high-confidence candidate SOX2-regulated miRNAs by combining the list of 21 down- regulated miRNAs after SOX2 knock-down with the 56 miRNAs up-regulated by exogenous SOX2 expression (i.e. miR-7-1, miR-16-1, miR18a, miR-20a, miR-128a, miR-128b and miR-425-5p) (Figure 1A). To validate this subset of miRNAs as bonafide SOX2 targets we first measured their response to GSC forced differentiation, a condition that decreases endogenous levels of SOX2 [15]. We observed in two distinct GSC lines that all 7 miRNAs interrogated were repressed under differentiating conditions compared to neurospheres (Figure 1B). To validate the results from the PCR miRNA array, we next measured the expression of this same set of miRNAs using qRT-PCR in GSCs expressing transgenic SOX2 and found that forced expression of SOX2 consistently increased levels of miR- 128b and miR-425-5p in each GSC line (Figure 1C and 1D).
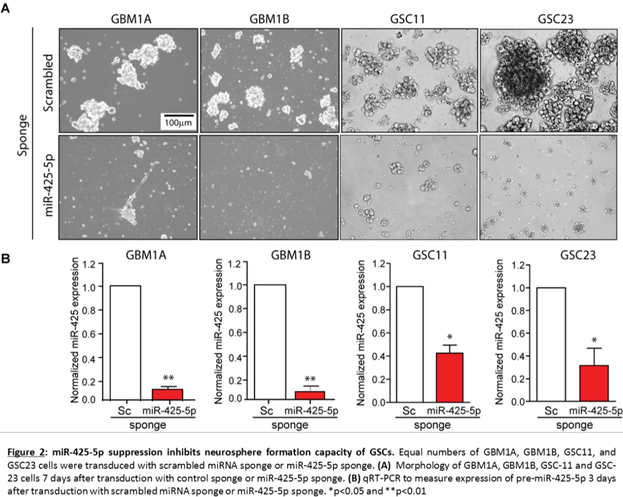
These results suggest that miR-128b and miR-425-5p are common downstream targets of SOX2 and may regulate GSC growth and survival. To test this hypothesis, we inhibited endogenous miR-128b and miR-425-5p using lentivirus expressing inhibitors (sponges) specifically targeting these miRNAs and measured sphere-forming capacity (>100 μm diameter) of two distinct GSC isolates. A lentivirus expressing a scrambled miRNA sequence (Sc.) was used as control. Using this approach, we observe 70-80% inhibition of endogenous miR-128b and miR-425-5p (Figure 1E). Interestingly, miR-425-5p inhibition repressed neurosphere formation capacity in two distinct GSC lines but miR-128b inhibition repressed this phenotype in only one of these lines (Figure 1F). Inhibition of miR-425-5p in two additional GSC lines resulted in strong inhibition of sphere formation capacity (Figure 2). Taken together, these results demonstrate that miR-425-5p is regulated by SOX2 and functions as a potent determinant of sphere-formation capacity in GSCs.
miR-425-5p regulates survival of GSCs
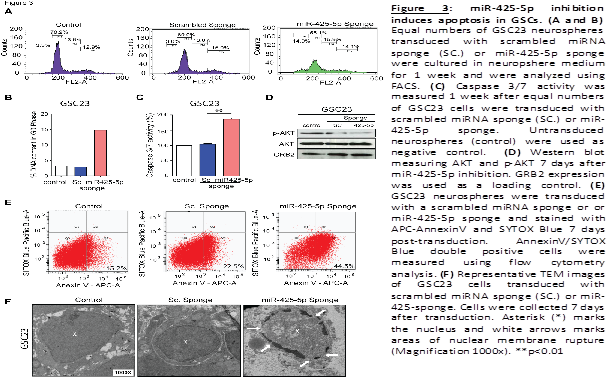
Multiple pathways, including dysregulation of cell growth and survival, can affect neurosphere formation capacity. To explore whether miR-425-5p impacts cell-cycle progression, miR-425-5p expression was inhibited in GSCs as described above and cell-cycle analysis was performed. We observed that inhibition of endogenous miR-425-5p expression leads to a significant increase in sub-G0 fraction compared to cells transduced with the control sponge (Sc.) or untransduced GSCs (control) (Figure 3A and 3B). No significant changes in S phase or G2M distribution were observed. This increase in the sub-G0 phase led us to postulate that miR- 425-5p may be involved in the regulation of cell death mechanisms in GSCs. To further analyze a possible apoptotic cell death activation, we use a fluorometric assay and detected a 2-fold increase in Caspase activation in GSCs transduced with the miR-425-5p sponge compared to control sponge and untransduced cells (Figure 3C). The PI3K- AKT pathway is a signal transduction pathway that promotes survival and growth in response to different stimuli (Dudek et al., 1997). Therefore, we also assessed the expression of AKT and its phosphorylated active form in GSCs. Western blot analysis showed a decrease in the expression of the phosphorylated form after miR-425-5p inhibition as compared with control sponge and parental cells (Figure 3D). Inhibition of miR-425-5p also increased the proportion of cells staining positive for Annexin-V (44.5%) as compared with the control sponge transduced cells (22.5%) and untransduced cells (15.2%) (Figue 3E). These results indicated that inhibition of miR-425-5p activates apoptosis process and suppress survival pathways in GSCs.
To further dissect the morphological changes produced in cells after activation of apoptosis, we performed transmission electron microscopy (TEM) in GSCs transduced with miR-425-5p sponge compared with control sponge and untransduced cells. TEM revealed marked condensation of nuclear chromatin and disintegration of the nuclear membrane after miR-425-5p inhibition (Figue 3F). Untransduced GSCs and control sponge transduced cells displayed normal cell and nuclear morphology. Collectively, these data show that miR-425-5p controls survival of GSCs by regulating apoptotic mechanisms.
miR-425-5p expression is enriched in GBM
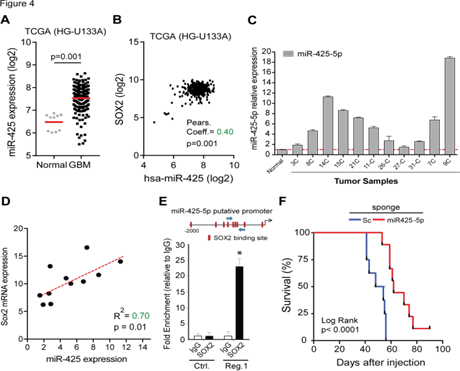
To examine the potential clinical relevance of this newfound SOX2:miR-425-5p axis we examined gene expression profiles of GBM patients. Analysis of TCGA datasets (HG- U133A) shows that miR-425-5p expression is increased in GBM compared to normal brain (Figue 4A). Consistent with our in vitro results, further analysis of this dataset revealed a significant positive correlation between SOX2 and miR- 425-5p expression in clinical specimens (Figure 4B). Additionally, we found a significant increase in miR-425-5p expression in 11 human surgical GBM specimens obtained by us (Figure 4C) and a significant correlation between SOX2 and miR-425-5p expression (R2 = 0.7, P<0.01) in these GBM samples (Figure 4D). This strong positive correlation between SOX2 and miR-425-5p suggested that SOX2 exerts transcriptional control of miR-425-5p expression through promoter DNA binding. In silico analysis of the miR-425-5p promoter region 5’ from the transcription start site identified multiple binding sites for SOX2 (Figure 4E, top panel). To validate the predicted SOX2 binding sites on the miR-425-5p promoter we selected the region with the greatest concentration of SOX2 binding sites and performed quantitative chromatin immunoprecipitation. A region of DNA containing no SOX2 binding sites was used as negative control. Our results show 23-fold SOX2 enrichment at the promoter region of miR-425-5p when compared with the IgG control (Figure 4E, bottom panel).
miR-425-5p expression and survival data was retrieved from the TCGA database using the BETASTASIS portal (http://www.betastasis.com). (A) miR-425-5p levels are enriched in GBM compared to normal brain. (B) Positive correlation between miR-425-5p and SOX2 expression in GBM. Pearson coefficient analysis was applied to establish correlations from gene expression data. (C) Evaluation of the expression of miR-425-5p in 11 glioblastoma samples using qRT-PCR analysis. (D) Correlation between SOX2 mRNA expression and miR- 425-5p expression in 11 glioblastoma samples. Scatter plot illustrate the correlation between SOX2 and miR- 425-5p expression levels. Linear regression analysis was used to establish correlation. (E) The putative promoter region of miR-425-5p has multiple SOX2 binding sites (red rectangles) predicted by PROMO search tool. Blue arrows indicate the region for which primer were designed for PCR analysis (Reg. 1) (top panel). DNA purified from chromatin immunoprecipitation was analyzed by qRT-PCR using primer pairs designed to amplify fragments containing SOX2 binding sites (Reg. 1) and primers targeting promoter region lacking SOX2 binding sites with the putative miR-425-5p promoter (Ctrl.) in GBM1A neurospheres expressing exogenous SOX2. (F) Kaplan–Meier survival analysis in nude mice after orthotopic implantation of GSC11 tumor cells transduced with miR-425-5p sponge or scrambled miRNA sponge (SC.). P values were determined using a log-rank test. *p<0.05
The results presented so far predict that in vivo inhibition of miR-425-5p will inhibit tumor growth. To investigate the effects of miR-425-5p on GBM tumorigenicity in vivo, GSCs stably transduced in vitro with miR-425- 5p sponge or control sponge were implanted in the putamen of nude mice (8 mouse/group). GSCs were transduced and selected before engraftment in mice. The median survival of mice receiving GSCs transduced with miR-425-5p sponge was 65 days compared to a median survival of 51 days in mice that received GSCs transduced with control sponge (Figure 4F). All subjects were euthanized when symptoms of terminal tumor burden, such as greater than 20% weight loss, hunched posture, cranial protrusion, and significant neurologic symptoms, were observed. Together, these results show that inhibition of miR-425-5p results in a survival benefit in an aggressive GBM model in vivo.
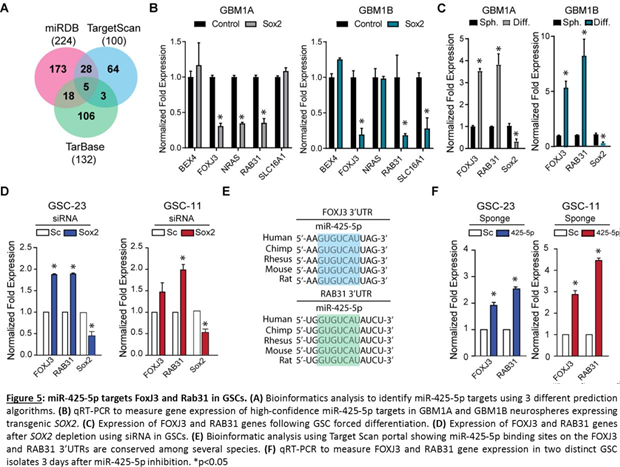
miR-425-5p targets FOXJ3 and RAB31 in GSCs
miRNA function is highly dependent on the targetome they regulate [16]. To identify the potential down-stream targets of miR-425-5p we interrogated three different target prediction algorithms and focused on the gene targets common to all three platforms (Figure 5A). This analysis predicted BEX4, FOXJ3, NRAS, RAB31, and SLC16A1 genes as high-probability miR-425-5p targets. To determine if any of these predicted targets are regulated by the SOX2:miR-425-5p axis, we measured the expression of these genes in GSCs expressing exogenous SOX2 that induces miR-425-5p expression (Figur 1C and 1D). miR-425-5p induction under these conditions was expected to repress its downstream targets. Consistent with this prediction, we observed that FOXJ3 and RAB31 mRNA levels were consistently down-regulated in two different GSC lines expressing transgenic SOX2 (Figure 5B). Additionally, FOXJ3 and RAB31 gene expression was up-regulated after forced differentiation of GSCs, conditions that repress SOX2 and miR-425-5p expression (Figure 5C and Figure 1B). Moreover, siRNA mediated repression of SOX2 also increased gene expression levels of FOXJ3 and RAB31 (Figure 5D). Bioinformatics analysis shows that the binding site for miR-425-5p in the FOXJ3 and RAB31 3’UTR is conserved among several species (Figure 5E). Inhibition of endogenous miR-425-5p significantly increased FOXJ3 and RAB31 gene expression compared to control neurospheres. Taken together, these results show that the SOX2:miR-425-5p axis regulates FOXJ3 and RAB31 gene expression in GSCs.
Discussion
SOX2 is a key member of the transcriptional regulatory network that controls pluripotency and self-renewal of stem cells [10,17]. SOX2 is enriched in undifferentiated glioma cell populations and plays an important role in maintaining the GSC phenotype [10,18,19]. Studies using mouse models of high-grade brain tumors show that SOX2 is required to maintain stem cell subpopulation in vivo, strongly suggesting that SOX2 drives tumorigenesis by maintaining the cancer stem cell population within high-grade brain tumors [20]. Importantly, high levels of SOX2 are associated with GBM aggressiveness and poor prognosis [21,22].
SOX2 regulates a complex network of downstream targets that include coding and non-coding genes, but which of these targets are functional and amenable to therapeutic intervention remains to be elucidated. We recently reported that SOX2 controls a network of non-coding RNAs in GSCs, including differentially expressed miRNAs (Acanda de la Rocha, A M et al. 2016). By combining results from two distinct gene-expression platforms we identified miR-7-1, miR-16-1, miR18a, miR-20a, miR-128a, miR-128b and miR-425-5p as high-confidence miRNAs directly regulated by SOX2 (Fig. 1A). Of these candidate miRNAs the SOX2:miR-425-5p axis was considered most likely to have an important role in GBM tumorigenesis based on its enrichment and correlation with SOX2 expression in clinical GBM specimens (Figure 4) in addition to loss- and gain-of-function responses in multiple GSC isolates including low-passage patient-derived GSCs.
Consistent with these observations, recent studies show that miR-425-5p enhances proliferation, controls cell death mechanisms, and promotes migration and metastasis in different cancer models [23-27] demonstrating that miR-425-5p can promote malignancy in multiple contexts. Recent findings show that miR-425-5p inhibition suppresses gastric cancer cell proliferation [28]. Additionally, Ahir and colleagues observed that treatment of breast cancer cells with CuO-Nw-FA resulted in down-regulation of miR-425-5p levels leading to induction of apoptosis and reduction of metastasis, through up-regulation of PTEN and inhibition of PI3/AKT/CDK pathway [29]. In line with these results, we show that down-modulation of miR-425-5p expression potently induces apoptosis in GSCs and suppress survival pathway demonstrating it’s critical role in glioma stem cell survival. Additionally, in vivo studies show that in vivo inhibition of miR-425-5p significantly extends animal survival in a GBM orthotopic mouse model (Figure 4F). Taken together, our results demonstrate that miR-425-5p is involved in regulating cell proliferation and apoptosis in GSCs.
Using multiple in vitro and in silico endpoints we identified RAB31 and FOXJ3 as miR-425-5p downstream targets in GSCs (Figure 5). FOXJ3 is a transcription factor that is expressed in neuroectoderm, neural crest, and in myotome [30] and is an important regulator of myofiber identity and muscle regeneration [31]. In line with our observations, FOXJ3 gene levels are induced during myogenic differentiation, consistent with a role in lineage restriction during muscle development [32,33]. Interestingly, it has been reported that another miRNA, miR-517a-3p, accelerates cell proliferation, migration and invasion by inhibiting FOXJ3 expression in human lung cancer [34]. Thus, FOXJ3 appears to be down- regulated in cancer by different context dependent miRNAs. RAB31, a member of the Ras superfamily, is enriched in oligodendrocytes and plays a critical role regulating the trans-Golgi network [35,36]. Consistent with our results, RAB31 is expressed in GFAP-positive fibers in the adult mouse brain and transgenic RAB31 expression enhances the differentiation of NPCs to astrocytes [37]. However, little is known about FOXJ3 and RAB31 as they pertain to maintenance of survival and phenotypic regulation of GSCs. Nevertheless, more experiments are needed to elucidate whether RAB31 and FOXJ3 are direct or indirect targets of miR-425-5p and their functional role in GSCs.
Our findings identify miR-425-5p as a potential therapeutic tool for inhibiting stem cell populations in GBM. Although the role of miRNAs as an adjuvant therapeutic option in the clinic has yet to be determined, there are recent studies that explore the use of nanoparticles, peptide carriers, and EVs to deliver miRNAs [38,39]. The study conducted by Bhaskaran et al. [40] demonstrated the feasibility of a miRNA- based gene therapy for GBM, and provide a roadmap for the development of future miRNA-based therapies. Therefore, our study shows that miR-425-5p is an interesting candidate to be further used for miRNA-therapy in GBM, for which novel therapeutics are desperately needed.
In conclusion, we add miR-425-5p to the network of SOX2-induced miRNAs that support GSC self-renewal, survival, and GBM growth in vitro and in vivo. Identifying how miR-425-5p functions in conjunction with other epigenetic oncogenic mechanisms should broaden our understanding of GBM and potentially lead to new therapeutic opportunities.
Acknowledgments
We thank to Ms. Eukene Vélaz Lizarraga at University of Navarra for her valuable help during sample preparation for Transmission Electron Microscopy.
Funding
The authors would like to thank the following organizations for financial support: American Brain Tumor Association (HLB); the United States NIH grants R01NS073611 and RO1NS096754 (JL); The Instituto de Salud Carlos III and Fondos Feder Europeos (PI13/125 and PI16/00066 MMA), the Spanish Ministry of Science and Innovation (IEDI-2015-00638 to MMA), and the Basque Foundation for Health Research (BIOEF, BIO13/CI/005). Foundation LA CAIXA (MMA), Foundation “El sueño de Vicky”, Asociation Pablo Ugarte-Fuerza Julen (MMA), DOD team science award (MMA, JF, and CG-M), Brain Cancer SPORE P50 CA127001 (JF and C-GM). AAR is supported by a fellowship from Friends of the University of Navarra (ADA) and EMBO short term stay (EMBO- ASTF 307-2015).
Conflict of Interest Disclosure Statement
The authors declare that they have no conflict of interest.
References
- Stupp R, Mason WP, van den Bent, Martin J, Weller M, Fisher B, Taphoorn MJB et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352 (2005): 987-996.
- Tran B, Rosenthal MA. Survival comparison between glioblastoma multiforme and other incurable cancers. J Clin Neurosci 17 (2010): 417-421.
- Stewart LA. Chemotherapy in adult high-grade glioma: A systematic review and meta- analysis of individual patient data from 12 randomised trials. Lancet 359 (2002): 1011-1018.
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T et al. Identification of human brain tumour initiating cells. Nature 432 (2004): 396-401.
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J et al. Identification of a cancer stem cell in human brain tumors. Cancer Res 63 (2003): 5821-5828.
- Sarkar A, Hochedlinger K. The sox family of transcription factors: Versatile regulators of stem and progenitor cell fate. Cell Stem Cell 12 (2013): 15-30.
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126 (2006): 663-676.
- Alonso MM, Diez-Valle R, Manterola L, Rubio A, Liu D, Cortes-Santiago N et al. Genetic and epigenetic modifications of Sox2 contribute to the invasive phenotype of malignant gliomas. PLoS One 6 (2011): e26740.
- Berezovsky AD, Poisson LM, Cherba D, Webb CP, Transou AD, Lemke NW et al. Sox2 promotes malignancy in glioblastoma by regulating plasticity and astrocytic Neoplasia 16 (2014): 19-25.
- Gangemi RMR, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P et al. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells 27 (2009): 40-48.
- Acanda de la Rocha, A M, Lopez-Bertoni H, Guruceaga E, Gonzalez-Huarriz M, Martinez-Velez N, Xipell E et al. Analysis of SOX2-regulated transcriptome in glioma stem cells. PLoS One 11 (2016): e0163155.
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 64 (2004): 7011-7021.
- Messeguer X, Escudero R, Farré D, Núñez O, Martínez J, Albà MM. PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18 (2002): 333-334.
- Lal S, Lacroix M, Tofilon P, Fuller GN, Sawaya R, Lang FF. An implantable guide-screw system for brain tumor studies in small animals. J Neurosurg 92 (2000): 326-333.
- Lopez-Bertoni H, Lal B, Li A, Caplan M, Guerrero-Cázares H, Eberhart CG et al. DNMT- dependent suppression of microRNA regulates the induction of GBM tumor-propagating phenotype by Oct4 and Sox2. Oncogene 34 (2015): 3994-4004.
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 16 (2017): 203-222.
- Fong H, Hohenstein KA, Donovan PJ. Regulation of self-renewal and pluripotency by Sox2 in human embryonic stem cells. Stem Cells 26 (2008): 1931-1938.
- Annovazzi L, Mellai M, Caldera V, Valente G, Schiffer D. SOX2 expression and amplification in gliomas and glioma cell lines. Cancer Genomics Proteomics 8 (2011): 139-147.
- Ikushima H, Todo T, Ino Y, Takahashi M, Saito N, Miyazawa K et al. Glioma-initiating cells retain their tumorigenicity through integration of the sox axis and Oct4 protein. J Biol Chem 286 (2011): 41434-41441.
- Favaro R, Appolloni I, Pellegatta S, Sanga AB, Pagella P, Gambini E et al. Sox2 is required to maintain cancer stem cells in a mouse model of high-grade oligodendroglioma. Cancer Res 74 (2014): 1833-1844.
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40 (2008): 499-507.
- Sathyan P, Zinn PO, Marisetty AL, Liu B, Kamal MM, Singh SK et al. Mir-21-Sox2 axis delineates glioblastoma subtypes with prognostic impact. J Neurosci 35 (2015): 15097-15112.
- Fang F, Song T, Zhang T, Cui Y, Zhang G, Xiong Q. MiR-425-5p promotes invasion and metastasis of hepatocellular carcinoma cells through SCAI-mediated dysregulation of multiple signaling pathways. Oncotarget 8 (2017): 31745-31757.
- Zhang Z, Wen M, Guo J, Shi J, Wang Z, Tan B et al. Clinical value of miR-425-5p detection and its association with cell proliferation and apoptosis of gastric cancer. Pathol Res Pract 213 (2017): 929-937.
- Zhang Y, Hu X, Miao X, Zhu K, Cui S, Meng Q et al. MicroRNA-425-5p regulates chemoresistance in colorectal cancer cells via regulation of programmed cell death 10. J Cell Mol Med 20 (2016): 360-369.
- Yan Y-, Gong F-, Wang B-, Zheng W. MiR-425-5p promotes tumor progression via modulation of CYLD in gastric cancer. Eur Rev Med Pharmacol Sci 21 (2017): 2130-2136.
- Sun L, Jiang R, Li J, Wang B, Ma C, Lv Y et al. MicoRNA-425-5p is a potential prognostic biomarker for cervical cancer. Ann Clin Biochem 54 (2017): 127-133.
- Zhang Z, Li Y, Fan L, Zhao Q, Tan B, Li Z et al. microRNA-425-5p is upregulated in human gastric cancer and contributes to invasion and metastasis in vitro and in vivo. Exp Ther Med 9 (2015): 1617-1622.
- Ahir M, Bhattacharya S, Karmakar S, Mukhopadhyay A, Mukherjee S, Ghosh S et al. Tailored-CuO-nanowire decorated with folic acid mediated coupling of the mitochondrial- ROS generation and miR425-PTEN axis in furnishing potent anti-cancer activity in human triple negative breast carcinoma cells. Biomaterials 76 (2016): 115-132.
- Landgren H, Carlsson P. FoxJ3, a novel mammalian forkhead gene expressed in neuroectoderm, neural crest, and myotome. Dev Dyn 231 (2004): 396-401.
- Alexander MS, Shi X, Voelker KA, Grange RW, Garcia JA, Hammer RE et al. Foxj3 transcriptionally activates Mef2c and regulates adult skeletal muscle fiber type identity. Dev Biol 337 (2010): 396-404.
- Shen L, Chen L, Zhang S, Du J, Bai L, Zhang Y et al. MicroRNA-27b regulates mitochondria biogenesis in myocytes. PLoS One 11 (2016): e0148532.
- Yamamoto H, Morino K, Nishio Y, Ugi S, Yoshizaki T, Kashiwagi A et al. MicroRNA- 494 regulates mitochondrial biogenesis in skeletal muscle through mitochondrial transcription factor A and forkhead box j3. Am J Physiol Endocrinol Metab 303 (2012): 1419.
- Jin J, Zhou S, Li C, Xu R, Zu L, You J et al. MiR-517a-3p accelerates lung cancer cell proliferation and invasion through inhibiting FOXJ3 expression. Life Sci 108 (2014): 48-53.
- Ng EL, Wang Y, Tang BL. Rab22B's role in trans-golgi network membrane dynamics. Biochem Biophys Res Commun 361 (2007): 751-757.
- Rodriguez-Gabin AG, Cammer M, Almazan G, Charron M, Larocca JN. Role of rRAB22b, an oligodendrocyte protein, in regulation of transport of vesicles from trans golgi to endocytic compartments. J Neurosci Res 66 (2001): 1149-1160.
- Chua CEL, Goh ELK, Tang BL. Rab31 is expressed in neural progenitor cells and plays a role in their differentiation. FEBS Lett 588 (2014): 3186-3194.
- Zeng A, Wei Z, Yan W, Yin J, Huang X, Zhou X et al. Exosomal transfer of miR-151a enhances chemosensitivity to temozolomide in drug-resistant glioblastoma. Cancer Lett 436 (2018): 10-21.
- Lee TJ, Yoo JY, Shu D, Li H, Zhang J, Yu JG et al. RNA nanoparticle-based targeted therapy for glioblastoma through inhibition of oncogenic miR-21. Mol Ther 25 (2017): 1544- 1555.
- Bhaskaran V, Nowicki MO, Idriss M, Jimenez MA, Lugli G, Hayes JL, et al. The functional synergism of microRNA clustering provides therapeutically relevant epigenetic interference in glioblastoma. Nat Commun 10 (2019): 44-z.


 Impact Factor: * 5.8
Impact Factor: * 5.8 Acceptance Rate: 71.20%
Acceptance Rate: 71.20%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 