The Spermine Phosphate-Bound Cyclooctaoxygen Sodium Epigenetic Shell of Euchromatin DNA Is Destroyed by the Epigenetic Poison Glyphosate
Andreas J Kesel*
1Chammünsterstr. 47, D-81827 München, Bayern/Bavaria, Germany
*Corresponding author: Andreas J Kesel, Chammünsterstr. 47, D-81827 München, Bayern/Bavaria, Germany.
Received: 05 February 2023; Accepted: 15 February 2023; Published: 16 March 2023
Article Information
Citation:
Andreas J Kesel. The Spermine Phosphate-Bound Cyclooctaoxygen Sodium Epigenetic Shell of Euchromatin DNA Is Destroyed by the Epigenetic Poison Glyphosate. Archives of Clinical and Biomedical Research 7 (2023): 171-190.
View / Download Pdf Share at FacebookAbstract
Oxygen exists in two gaseous (dioxygen and ozone) and six solid allotropic modifications. An additional allotropic modification of oxygen, the cyclooctaoxygen, was predicted to exist in 1990. The first synthesis and characterization of cyclooctaoxygen as its sodium crown complex, isolated in the form of three cytosine nucleoside hydrochloride complexes, was reported in 2016. The sperminium hydrogen phosphate/cyclooctaoxygen sodium complex was calculated to cover the actively transcribed regions (2.6%) of bovine lymphocyte interphase genome. Cyclooctaoxygen seems to be naturally absent in hypoxia-induced highly condensed chromatin, taken as a model for eukaryotic metaphase/anaphase/early telophase mitotic chromatin. Hence, it is proposed that the cyclooctaoxygen sodium-bridged sperminium hydrogen phosphate and selenite coverage serves as an epigenetic shell of actively transcribed gene regions in eukaryotic ‘open’ euchromatin DNA. Cyclooctaoxygen sodium-bridged sperminium hydrogen selenite was calculated to serve as a marker shell component at ATG start codons in human euchromatin DNA mRNA genes, both at the translation initiation triplet and at 5′-untranslated region upstream ATGs. The total herbicide glyphosate (ROUNDUP®) and its metabolite (aminomethyl)phosphonic acid (AMPA) are proved to represent ‘epigenetic poisons’, since they both selectively destroy the cyclooctaoxygen sodium complex. This definition is of reason, since the destruction of cyclooctaoxygen is certainly sufficient to bring the protection shield of human euchromatin into collateral epigenetic collapse. The total herbicide glyphosate and its environmental metabolite (aminomethyl)phosphonic acid (AMPA) can be associated in vitro with catalytic detoriation of eukaryotic euchromatin genetic information. The epigenetic shell of eukaryotic euchromatin is susceptible to decay induced by catalytic epigenetic poisons threatening eukaryotic genomic heritage.
Keywords
AMPA; Cyclooctaoxygen; DNA; Glyphosate; Selenium; Spermine
AMPA articles; Cyclooctaoxygen articles; DNA articles; Glyphosate articles; Selenium articles; Spermine articles
Article Details
1. Introduction
In 1677 Antoni van Leeuwenhoek discovered [1] the characteristic crystals of spermine phosphate (spermine × 2 H3PO4 × 6 H2O) [2] in matured native human semen (Figure 1). Since then, there was collected conclusive evidence that human chromosomal DNA is closely associated with spermine phosphate [2]. The interaction of oxygen species with DNA has until recently only being focused on oxidative DNA damage and its pathophysiological consequences [3]. In 2015 Kirmes et al. reported an unprecedented interaction of eukaryotic chromatin DNA structure with atmospheric oxygen partial pressure [4]. Under switching to hypoxic conditions (1% O2, 5% CO2, 94% N2) the murine cardiomyocyte HL-1 cell chromatin rendered itself highly condensed, accompanied by redistribution of the polyamine pool (mainly spermine and spermidine) to the nucleus [4]. In 2016 Kesel et al. showed [5] that eukaryotic single-stranded DNA (ssDNA) binds a new allotropic form of oxygen, the cyclooctaoxygen (cyclo-O8), in form of its sodium (Na+) complex (cyclo-O8-Na+), especially when in coordination to spermine phosphate (sperminium hydrogen phosphate) [5]. A model for a logically resulting first epigenetic shell of eukaryotic in vivo DNA was proposed [5]. Also a partial substitution of the sperminium (C10H30N44+)-bound monohydrogen phosphate (HPO42–) anions by hydrogen selenite (HSeO3–) anions was postulated during these investigations [5], thereby providing an explanation for the well-known, but ‘mysterious’ [6], augmenting effects of the polyamine spermine [6] and the essential micronutrient selenium [5] on eukaryotic genome integrity and chromosomal DNA stability.

Figure 1: Parts of the original publication [1] from 1677 by Antoni van Leeuwenhoek with the description of the first light microscopic observation of crystalline spermine phosphate in human semen. (A) The title page 1040. (B) Page 1042 with fig. A showing the characteristic crystalline shape [2] of spermine × 2 H3PO4 × 6 H2O [2]. The last paragraph including fig. A is read in New Latin: “Et sum prædicta materia paucillum temporis steterat, in ea observabantur trilaterales figuræ ab utraque parte in aculeum desinentes, quibusdam longitudo minutissimæ arenæ, aliquæ aliquantulum majores, ut fig. A. Prætera, adeo nitidæ ac pellucidæ, ac si crystallinæ fuissent.”. English transcription: “And I mentioned the matter which stood for a short time, in which trilateral figures were observed from both sides ending in a sting, some in length of minute grains, some a little larger, as fig. A. Moreover, so sleek and translucent, as if it were crystalline.”.
2. Materials and Methods
2.1 Materials
The following materials were utilized: NC (C9H14ClN3O5 × 2 ClNaO8 × 2 H2O × ½ C3H6O) = [tetrakis(b-D-cytidin-N3-ium)(octoxocane-k4O1,O3,O5,O7)]sodium(5+) pentachloride dihydrate hemiacetonate (M = 1,370.22 g/mol) [5]; RC (C9H14ClN3O5 × 2 HClNa2O17 × ¼ C3H6O) = b-D-cytidine hydrochloride – m-chloro(m-hydroxy)bis(octoxocane-k4O1,O3,O5,O7)disodium (1:2) × 0.25 acetone (M = 1,003.06 g/mol) [5]; potassium iodide (KI) puriss. p.a., reag. ISO, reag. Ph.Eur., w (m/m) ≥ 99.5% (M = 166.00 g/mol) {Sigma-Aldrich Corp., St. Louis, MO, USA; pH 6.0–9.0 [J = 20 °C, 5% (m/m) in H2O], loss on drying ≤ 0.2% (J = 105 °C), total nitrogen (N) ≤ 0.001%, heavy metals (as Pb) ≤ 0.0005%, iodate (IO3–) ≤ 2 mg/kg}; starch puriss. p.a., from potato, reag. ISO, reag. Ph.Eur., soluble [Sigma-Aldrich Corp., St. Louis, MO, USA; pH 6.0–7.5, loss on drying ≤ 13% (J = 105 °C), sulfated ash ≤ 0.5%, substances reducing Fehling solution (as maltose) ≤ 0.7 %]; (aminomethyl)phosphonic acid (AMPA) (CH6NO3P, M = 111.04 g/mol) [Sigma-Aldrich Corp., St. Louis, MO, USA, Lot: MKBX8824V; w (m/m) = 98.5% (titration), carbon (C) 10.9%, nitrogen (N) 12.6%]; N-(phosphonomethyl)glycine (glyphosate) (C3H8NO5P, M = 169.07 g/mol) [Sigma-Aldrich Corp., St. Louis, MO, USA, Lot: MKBX1937V; w (n/n) > 99% (TLC), carbon (C) 21.3%, nitrogen (N) 8.2%, mp 230 °C (dec.)]; deuterated chloroform (CDCl3) [euriso-top®, Saint Aubin cedex, France, Lot: D007H, W2631; 99.80% D, H2O < 0.01%, stored over molecular sieve 3Å]; molecular sieve 3Å (0.3 nm, zeolithe, metal-aluminosilicate) [AppliChem GmbH, Darmstadt, Germany; Lot: 5H002478; water absorbency ≥ 20% (24 h, 80% humidity)]; elemental iodine I2 (M = 253.81 g/mol) (iodum resublimatum Ph.Eur. 7.0) [Caesar & Loretz (Caelo) GmbH, Hilden, Germany, Lot: 122890; w (m/m) = 99.9% (titration), non-volatile matter ≤ 0.1%, Br– and Cl– ≤ 250 ppm]. L-Ascorbic acid (vitamin C) Ph.Eur. 7.0 (M = 176.12 g/mol) was purchased from a local pharmacy store (Friedens-Apotheke, München-Trudering, Germany). Glyphosate monosodium salt (glyphosate-Na, C3H7NNaO5P, M = 191.05 g/mol) was purchased in form of ROUNDUP® GRAN 15,5 g granules [Monsanto Europe S.A., Antwerp, Belgium, Lot: T346 (prod. date 2011/12/12); contains 475 g/kg glyphosate-Na, wglyphosate-Na (m/m) = 47.5%, inert carrier material 33.5%, water and excipients 19%].
2.2 Calculation of the Genomic Coverage by the Cyclooctaoxygen Sodium-Bridged Spermine Phosphate Epigenetic Shell of Interphase DNA in Bovine Lymphocytes
The average volume of a human blood lymphocyte was taken as 206 f(emto)l(iter) [10-15 liter = (µm)3] [7, 8] after technical correction [8] of the published value (210 fl) [7]. The published average volume of a bovine blood lymphocyte (256 fl) [9] was taken as 214 fl after introducing a technical correction factor of f = 0.834 [8]. The reference genome size of Bos taurus (Hereford breed) was taken as 2,670,139,648 bp (RefSeq assembly accession number GCF_000003055.6) [10]. This genome showed 41.89% GC content [10]. The theoretical intracellular concentration of the sperminium hydrogen phosphate/cyclo-O8-Na+ complex required to cover all triplets of the dsDNA genome in a blood lymphocyte of B. taurus was calculated: [214 fl × 6.022140857 × 1023 mol-1]-1 × 2,670,139,648 × 2 × (3)-1 = 13.8127 mM. The coverage of B. taurus genome by the sperminium hydrogen phosphate/cyclo-O8-Na+ complex was calculated (mean ± s.d.: 2.6208 ± 0.4953%) from the published [11] fractions of spermine bound to dsDNA: 421 µM × [13.8127 mM]-1 = 3.0479% (in presence of 2 mM Mg2+ and 100 mM K+), 287 µM × [13.8127 mM]-1 = 2.0778% (2 mM Mg2+, 150 mM K+), and 378 µM × [13.8127 mM]-1 = 2.7366% (10 mM Mg2+, 100 mM K+). The number of base pairs for the number of protein-coding exons in B. taurus genome (49,107) [10] was calculated with the median of amino acid residues/gene (468) [10]: 49,107 × 3 bp × 468 = 68,946,228 bp (2.5821% of B. taurus genome).
2.3 Calculation of the Spermine Coverage of Highly Condensed Mitotic Metaphase DNA in HeLa S3 Cells
The average effective molecular mass of dGp/dCp was calculated as M = 309.19 g/mol, of dAp/dTp as M = 308.70 g/mol. The reference Homo sapiens genome size was taken as 3,099,441,038 bp (RefSeq assembly accession number GCF_000001405.40) [12]. This genome showed 40.5% GC content [12]. The molecular mass of this human genome dsDNA was calculated: [(0.405 × 309.19 g/mol) + (0.595 × 308.70 g/mol)] × [3,099,441,038 bases (b) × 2] = 1.191482507 × 1012 g/mol. The HeLa cell genome [13] was anticipated as 76 chromosomes (hypertriploid) + 22 abnormal chromosomes [13, 14]. The chromosomal DNA size of HeLa metaphase chromatin dsDNA was calculated as 19,539,129,390 bp with M = 1.207127414 × 1013 g/mol from published karyotyping [14]. The content of spermine in HeLa S3 cell metaphase chromatin was taken as 135.9 ± 16.1 pmol/µg DNA [15]. This was transformed into 135.9 ± 16.1 p(ico)mol (10-12 mol) spermine/82.84129651 z(epto)mol (10-21 mol) dsDNA = 1,640,486,155 (molecules spermine/dsDNA). Since one spermine molecule is assumed to cover six bp (in the pure spermine form of A-DNA duplex [16] and Z-DNA duplex [17]), this corresponds to a coverage of 1,640,486,155 × 6 × [19,539,129,390]-1 = 50.3754% of HeLa S3 cell metaphase chromatin dsDNA genome by spermine. The content of spermidine in HeLa S3 cell metaphase chromatin was taken as 116.1 ± 11.8 pmol/µg DNA [15]. This was transformed into 116.1 ± 11.8 p(ico)mol (10-12 mol) spermidine/82.84129651 z(epto)mol (10-21 mol) dsDNA = 1,401,474,927 (molecules spermidine/dsDNA). Since one spermidine molecule is assumed to cover six bp (in the pure spermidine form of Z-DNA duplex [18]), this corresponds to a coverage of 1,401,474,927 × 6 × [19,539,129,390]-1 = 43.0359% of HeLa S3 cell metaphase chromatin dsDNA genome by spermidine. Taken together, the polyamine (spermine/spermidine ratio 1.17) coverage of HeLa S3 cell metaphase chromatin dsDNA is 50.3754% + 43.0359% = 93.4113%.
2.4 Calculation of the Polyamine Coverage of Maximally Condensed Mitotic Late Anaphase/Early Telophase DNA in Murine Cryptal Enterocytes
The phosphorus (P) content of female Mus musculus strain C3H/HeJ cryptal enterocytic mitotic (late anaphase/early telophase) chromatin was taken as 298.5 ± 17.3 mmol (P)/kg [19]. The average effective molecular mass of one nucleotide unit (dGp/dCp and dAp/dTp) in M. musculus C3H/HeJ genome (2,701,131,316 bp) [20] was calculated with the GC content of 42.82% [20]: M = 308.91 g/mol. With the formula w (P) = (nP) × M-1 (nP, number of P atoms pro formula unit; M, molecular weight of formula unit) the phosphorus content w (P) of murine mitotic chromatin was calculated: 3,237.19 mmol (P)/kg (free dsDNA). The value for complete complexation with one spermine tetracation/six nucleotides is 2,912.86 mmol (P)/kg (spermine tetracation-complexed ssDNA), and for complete complexation with one spermidine trication/six nucleotides is 2,997.41 mmol (P)/kg (spermidine trication-complexed ssDNA). Taking the arithmetic mean of the published [11, 21] spermine/spermidine ratio 0.85, bound to rat liver DNA and RNA (2 mM Mg2+, 150 mM K+) [11], or in Sprague–Dawley rat liver nuclei [21], the theoretical value for complete complexation is {(2,912.86 × 0.85) + [(2,997.41 × (2 – 0.85)]} × (2)-1 = 2,961.48 (P)/kg (spermine tetracation/spermidine trication-complexed ssDNA). Phosphatidylcholine in its dipalmitoyl (C16:0) lipid composition (M = 734.04 g/mol) is the main constituent (60.8 ± 1.3%) of the phospholipid fraction in rat liver chromatin [22]. This molecular weight was corrected to an average effective molecular mass M = 765.03 g/mol, based on the fractions [22] and lipid compositions [22] of phospholipids (phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine, phosphatidylinositol, sphingomyelin) in Sprague–Dawley rat liver chromatin, to yield the reference value for the (P) content of phospholipid 1,307.13 mmol (P)/kg. The fractions of DNA (32.0 ± 4.1%), RNA (5.1 ± 1.6%), protein (62.6 ± 3.8%), and phospholipid (0.2 ± 0.1%) in Sprague–Dawley rat liver chromatin were taken as published [22], and applied on the murine mitotic chromatin nucleic acids (DNA + RNA). The published [19] value was corrected for the chromatin-bound cation (Na+, K+, Mg2+, Ca2+) [19], protein and phospholipid [22] content: {[841.0 mmol (Na+, K+, Mg2+, Ca2+)/kg + 298.5 mmol (P)/kg] × 100 × (37.1)-1} + [0.2 × (100)-1 × 1,307.13 mmol (P)/kg] = 3,074.04 mmol (P)/kg (ssDNA + ssRNA) in mitotic late anaphase/early telophase chromatin. The six nucleotide coverage (ssDNA + ssRNA) by the polyammonium cations was calculated: (3,237.19 – 3,074.04) × (3,237.19 – 2,961.48)-1 × 100% = 59.1745%. The genomic dsDNA six bp coverage by the polyammonium cations, corrected for the nuclear RNA content, was calculated: 59.1745% × [32.0 × (37.1)-1] × 2 = 102.0800%. This corresponds to a coverage of 5.8777 bp murine cryptal enterocytic mitotic (late anaphase/early telophase) chromatin dsDNA by one polyammonium cation (spermine/spermidine ratio 0.85). The six nucleotide coverage of nuclear RNA by the polyammonium cations was calculated: 59.1745% – [102.0800% × (2)-1] = 8.1345%, corresponding to a relative six nucleotide coverage of nuclear RNA: 8.1345 × (5.1)-1 × 100% = 159.5000%. This corresponds to a coverage of 3.7618 nucleotides of nuclear RNA by one polyammonium cation (spermine/spermidine ratio 0.85).
2.5 Calculation of the Hydrogen Selenite (HSeO3−) Coverage of Human Euchromatin DNA Specifically at ATG Start Codon Sequences
The reference H. sapiens genome size was taken as 3,099,441,038 bp (RefSeq assembly accession number GCF_000001405.40) [12]. The number of genes and pseudogenes in H. sapiens genome (59,265) was taken as published [12]. The median of the number of amino acid (aa) residues for a protein encoded by the actively transcribed part of the human genome (euchromatin) was calculated with the number of exon transcripts (394,894) [12] exhibiting the median length 143 b [12], and the number of single-exon (none intron to be spliced) transcripts (1,770) [12] exhibiting the median length 1,595 b [12]. The sum of the median exon transcript length and the median single-exon transcript length [(394,894 × 143 b) + (1,770 × 1,593 b)] = 59,289,452 b (number of bases coding for euchromatin mRNA) was divided by the number of genes (41,989) [12] to yield the median length of euchromatin mRNA transcripts [(394,894 × 143 b) + (1,770 × 1,593 b)] × (41,989)-1 = 1,412 b. This value was divided by three (as to the triplet nature of the genetic code) to give the median of aa residues/gene of human euchromatin [(394,894 × 143 b) + (1,770 × 1,593 b)] × (41,989)-1 × (3)-1 = 470.6745 aa. The percentage of actively transcribed regions in the human genome was calculated: 470.6745 aa × 3 × 59,265 × (3,099,441,038 bp)-1 = 2.7000% of H. sapiens genome. The theoretical intracellular concentration of the sperminium hydrogen phosphate/cyclo-O8-Na+ complex required to cover all triplets of the dsDNA genome in a blood lymphocyte of H. sapiens was calculated: [206 fl × 6.022140857 × 1023 mol-1]-1 × 3,099,441,038 b × 2 × (3)-1 = 16.6561 mM. The average volume of a human blood lymphocyte was taken as 206 fl [femtoliter = (µm)3] [7, 8] after technical correction [8] of the published value (210 fl) [7]. The optimal human blood serum selenium (as Se, M = 78.96 g/mol) level of 110 µg/l (110 ng/ml, this serum level corresponds to a concentration of 1.3931 µM) was taken from literature as concentration for optimal selenoprotein expression [23, 24]. The percentage of selenium (as selenite) concentration accumulated intracellularly in the cytosol from extracellular medium was taken from literature [25] [as 57.6% uptake into the cytosol of primary rat hepatocytes (1 min time, 10 µM Na275SeO3 extracellularly)]. The median number of aa-coding triplets required for one triplet to be covered with the sperminium selenite/cyclo-O8-Na+ complex in interphase euchromatin (in form of hydrogen selenite HSeO3−) was calculated by: (16.6561 mM × 2.7000% × 78.96 g/mol) × (110 µg/ml × 0.576)-1 = 560.4400 aa. The number of 5′-untranslated region (5′-UTR) upstream open reading frames (uORFs) upstream start codons (uATGs transcribed to uAUGs) in the 5′-UTR of human mRNA genes was calculated from literature [26] as the number of transcribed human genes with at least one uATG (11,670) divided by the number of human mRNA genes with annotated 5′-UTR (23,775): 11,670 × (23,775)-1 = 49.0852%. That means that 49% of human mRNA genes contain at least one uATG 5′-UTR start codon [26] [the downstream 3′-untranslated region (3′-UTR) contains none ATG start codon]. The total number of human uATGs/5′-UTR of human mRNA genes (16,504) was published (35,599) [27]. This gives the number of human uATGs/5′-UTR: 35,599 × (16,504)-1 = 2.1570 uATGs/5′-UTR (human). If this number is multiplied by the percentage of human mRNA genes containing at least one uATG 5′-UTR start codon [26], the number of hydrogen selenite HSeO3− molecules (within the sperminium hydrogen selenite/cyclo-O8-Na+ complex in interphase euchromatin) bound to human mRNA genes can be calculated: 2.1570 × 49.0852% = 1.0588 HSeO3−/5′-UTR of mRNA gene (human). Accordingly, together with the transcribed and translated gene open reading frame initiation codon ATG encrypted in the Kozak consensus sequence 5′-ACCATGG-3′ [28], the number of human mRNA gene-incorporated hydrogen selenite HSeO3− molecules (within the sperminium hydrogen selenite/cyclo-O8-Na+ complex in interphase euchromatin) can be calculated: 1 (HSeO3−/initiation ATG) + 1.0588 (HSeO3−/5′-UTR) = 2.0588 HSeO3−/mRNA gene (human). The median length of human mRNA transcripts (3,560 b) [12] can be transformed into the median aa length (triplets) of human translated genes (as to the triplet nature of the genetic code): 3,560 b × (3)-1 = 1,186.6667 aa. If divided by the number of human mRNA gene-incorporated hydrogen selenite HSeO3− molecules, the number of triplets required for the covering by one hydrogen selenite HSeO3− molecule (within the sperminium selenite/cyclo-O8-Na+ complex in interphase euchromatin) can be calculated: 1,186.6667 aa × {2.0588 [HSeO3−/mRNA gene (human)]}-1 = 576.3978 aa. From this value the optimal human blood serum selenium (as Se, M = 78.96 g/mol) level required for covering of all ATG codons in human mRNA genes (the downstream 3′-UTR contains none ATG start codon) was calculated: (16.6561 mM × 2.7000% × 78.96 g/mol) × (576.3978 aa × 0.576)-1 = 106.9546 µg/l (ng/ml; 1.3545 µM).
2.6 Calculation of the Apparent Acid Dissociation Constant of the Human Genome DNA
The intrinsic pKa of one isolated phosphodiester of DNA is 1.29, this is the pKa (J = 25 °C) of dimethyl phosphate [29]. The reference H. sapiens genome size was taken as 3,099,441,038 bp (RefSeq assembly accession number GCF_000001405.40) [12]. The apparent (effective) pK′a,HG = 2.5601 (J = 25 °C) of the haploid human genome was calculated (Figure 2A) according to the method of Katchalsky & Gillis [30], as based on the theoretical considerations of Kuhn & Kuhn [31]. The length rise of one B-DNA repeating unit (helix rise/bp) in solution (in the crystal: 0.336 nm [32]) was taken as: b = 0.334 × 10-7 cm (0.334 ± 0.01 nm [32]). The h0 was calculated by: 0.334 nm × 3,099,441,038 = 1.03521331 m (= theoretical length of H. sapiens dsDNA haploid genome as linear thread). The hn was calculated by: 0.5 × (0.334 nm + 0.5798 nm) × 3,099,441,038 = 1.41613461 m (theoretical length of half-neutralized H. sapiens dsDNA haploid genome as linear thread) (Figure 2A). The intermediate term (n − 1) was calculated as 0.4763 with l = 2.0321 and l′ = 0.0321 (Figure 2A). The 0.5798 nm pitch in one relaxed (fully neutralized) dsDNA repeating unit was calculated as statistic arithmetic mean dØ = 579.80 pm from the interphosphorus distance d (5′-P,5′-P) = 739.25 ± 14.37 pm (d ± s.d.; n = 4) in the fully ionized single-stranded hexanucleotide d(ApApApApApAp) (molecular modeling software: ACD/Chem Sketch version 12.01 with integrated ACD/3D Viewer, processed with Mercury 3.1 version 3.1.1), minus the typical phosphorus–oxygen distance d (P,O) = 159.45 pm [33], by the formula: dØ = 739.25 pm – 159.45 pm = 579.80 pm.
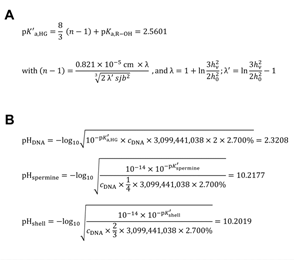
Figure 2: The calculation of the apparent acid dissociation constant of the haploid human genome, and of the interphase intranuclear micro-pH [41] values induced by human genome depending on its intrinsic epigenetic occupation status. (A) The calculation of the apparent (effective) pK′a,HG = 2.5601 (J = 25 °C) of the haploid human genome according to the method of Katchalsky & Gillis [30] and Kuhn & Kuhn [31] [pK′a,HG = apparent acid dissociation constant (J = 25 °C) of haploid H. sapiens genome B-DNA double helix; pKa,R–OH = 1.29 = theoretical pKa (J = 25 °C) [29] of one isolated internucleotide phosphodiester (R–OH) proton; s = 1 = number of statistical subunits on thread molecule [31]; j = 6 = number of spacing atoms (at least distance) in one dsDNA repeating unit [31]; b = 0.334 × 10-7 cm (0.334 ± 0.01 nm [32]) = length rise in cm of one B-DNA repeating unit (helix rise/bp) in solution; hn = end–to–end distance of dsDNA at half-neutralization; h0 = end–to–end distance of dsDNA]. (B) The theoretical micro-pH [41] values surrounding H. sapiens haploid (and diploid) interphase euchromatin were calculated as the intranuclear micro-pH induced by human haploid (and diploid) genome treated as a weak acid, from pHDNA, pHspermine, and pHshell, by applying the formula for pH induced by weak acids: pH = –log10 (Ka × ca)0.5, or the formula for pH induced by weak bases: pH = –log10 [(Kw × Kb) × (cb)-1]0.5 (Ka, acid dissociation constant; ca, concentration weak acid; Kw, dissociation constant of water, Kw = 10-14 (J = 25 °C); cb, concentration weak base). The single-stranded human genome intranuclear interphase concentration cDNA was calculated as: [1 genome × (6.022140857 × 1023 mol-1)-1] × (33.5268 fl)-1 = 49.5282 pM. The intranuclear micro-pH was calculated by the law for the calculation of the solution pH induced by salts of weak acids with weak bases: pHsalt = 0.5 × (pHacid + pHbase).
2.7 Calculation of the Hypothetical Intranuclear Micro-pH Mediated by Single Spermine Occupation of Human Interphase Euchromatin
The reference H. sapiens genome size was taken as 3,099,441,038 bp (RefSeq assembly accession number GCF_000001405.40) [12]. The number of base pairs for the number of protein-coding exons in H. sapiens genome (59,265) [12] was calculated with the median of amino acid residues/gene (470.6745 aa) (see Section 2.5): 470.6745 aa × 59,265 × 3 = 83,683,573 bp (2.7000% of H. sapiens genome). The intrinsic pH of H. sapiens haploid interphase genome was calculated with the volume of a human blood lymphocyte interphase nucleus taken as 33.5268 fl [femtoliter = (µm)3] [8, 9] after technical correction (f = 0.834) [8] of the published value (40.2 ± 2.0 fl) [9]. Because H. sapiens dsDNA haploid genome is to be treated as a weak acid [pK′a,HG = 2.5601 (J = 25 °C)] (Figure 2A), the hypothetical nuclear pHDNA = 2.3208, mediated by H. sapiens haploid interphase genome dsDNA, can be calculated (Figure 2B). The concentration cDNA was calculated as: [1 genome × (6.022140857 × 1023 mol-1)-1] × (33.5268 fl)-1 = 49.5282 pM. This concentration was multiplied by the number of phosphodiester moieties in dsDNA (3,099,441,038 × 2) and corrected for actively transcribed gene regions of H. sapiens genome (2.7000%). The four pKa values (± s.d.) of spermine pKa1 = 10.86 ± 0.06, pKa2 = 10.05 ± 0.01, pKa3 = 8.82 ± 0.01, pKa4 = 7.95 ± 0.01 [potentiometric titration in H2O, J = 25 °C, ionic strength (NaCl) 0.1] [34] give the apparent (effective) pK′spermine = 9.42 ± 0.02 (mean ± s.d.) (J = 25 °C). Because the hypothetical nuclear concentration of spermine tetracation covering B-DNA is one quarter of cDNA, if we assume one spermine molecule covering four base pairs (single occupation) of B-DNA [35], and correcting for actively transcribed gene regions of H. sapiens genome (2.7000%), the pHspermine = 10.2177 can be calculated (Figure 2B). Therefore, the hypothetical intranuclear micro-pH surrounding H. sapiens haploid interphase euchromatin when covered (single quartet occupation) by the spermine tetracation can be calculated as: pHDNA/spermine = 0.5 × (2.3208 + 10.2177) = 6.2693. For the diploid dsDNA genome, after completed S phase during interphase, the hypothetical micro-pH surrounding H. sapiens diploid interphase euchromatin when covered (single quartet occupation) by the spermine tetracation is, again, pHDNA/spermine = 6.2693.
2.8 Calculation of the Theoretical Intranuclear Micro-pH Mediated by Sperminium Hydrogen Phosphate/Cyclooctaoxygen Sodium Complex Occupation of Human Interphase Euchromatin
The apparent pK′spermine was corrected for the monohydrogen phosphate complexation to get the apparent (effective) pK′shell of the sperminium hydrogen phosphate/cyclooctaoxygen sodium complex epigenetic shell. Ortho-phosphoric acid H3PO4 shows the pKa1 = 2.161, pKa2 = 7.207, and pKa3 = 12.325 (J = 25 °C) [36]. Therefore, the pK′spermine is to be corrected for the monohydrogen phosphate of the epigenetic shell: pK′shell = [9.42 + ({[(2.161 + 7.207) × 0.5] + 12.325} × 0.5)] × 0.5 = 8.9623. Because the theoretical nuclear concentration of the sperminium hydrogen phosphate/cyclooctaoxygen sodium complex covering dsDNA is two-third of cDNA, when we assume one epigenetic complex molecule covering three base pairs on both strands of dsDNA (double occupation), and correcting for actively transcribed gene regions of H. sapiens genome (2.7000%), the pHshell = 10.2019 (Figure 2B). Therefore, the theoretical intranuclear micro-pH surrounding H. sapiens haploid interphase euchromatin when covered (double triplet occupation) by the sperminium hydrogen phosphate/cyclooctaoxygen sodium complex can be calculated as: pHDNA/shell = 0.5 × (2.3208 + 10.2019) = 6.2614. For the diploid dsDNA genome, after completed S phase during interphase, the theoretical micro-pH surrounding H. sapiens diploid interphase euchromatin when covered (double triplet occupation) by the sperminium hydrogen phosphate/cyclooctaoxygen sodium complex is, again, pHDNA/shell = 6.2614.
2.9 Color Assay for Cyclo-O8-Na+ Contained in RC – Destruction of Cyclo-O8-Na+ by the Glyphosate Metabolite (Aminomethyl)Phosphonic Acid
Stock preparations were: KI (52 mg KI in 2,000 µl H2O) (1), KI + starch (52 mg KI + 18 mg starch in 2,000 µl H2O) (2), RC + KI (34 mg RC + 52 mg KI in 2,000 µl H2O) (3), RC + KI + AMPA (34 mg RC + 52 mg KI + 22 mg AMPA in 2,000 µl H2O) (4), RC + KI + starch (34 mg RC + 52 mg KI + 18 mg starch in 2,000 µl H2O) (5), RC + KI + starch + AMPA (34 mg RC + 52 mg KI + 18 mg starch + 22 mg AMPA in 2,000 µl H2O) (6). The concentrations in solution were: RC, 16.95 mM (with cyclo-O8-Na+, 67.79 mM); KI, 156.63 mM; AMPA, 99.06 mM. The stock solutions were incubated at room temperature (RT, J = 14.0 °C) for 30 min, then at elevated temperature (J = 25.2 °C) for 2 min, after which time the first photograph series (Figure 3) was taken. The solutions were then incubated at elevated temperature for 48 min, after which time the second photograph (Figure 3) series was taken. The solutions were further incubated at RT for 14 h. Afterwards, both RC + KI (3) and RC + KI + AMPA (4) were mixed with 1,000 µl deuterated chloroform (CDCl3), and extracted by shaking. Concomitantly, RC + KI + starch (5) and RC + KI + starch + AMPA (6) were both mixed with 8 mg of solid L-ascorbic acid (concentration of L-ascorbic acid in solution 22.71 mM). The solutions were succeedingly incubated at elevated temperature for 15 min, after which time the third photograph series (Figure 3) was taken.
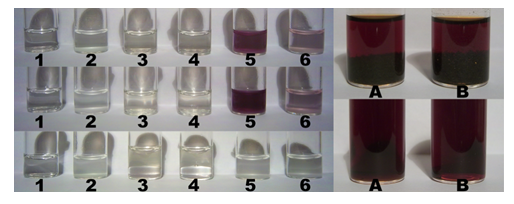
Figure 3: Color assays for cyclo-O8-Na+ contained in RC, for the destruction of cyclo-O8-Na+ by the glyphosate metabolite (aminomethyl)phosphonic acid (AMPA) (left, 1–6), and for the potential reduction of elemental iodine by AMPA (right, A and B). Solutions (left, 1–6) were: KI (1), KI + starch (2), RC + KI (3), RC + KI + AMPA (4), RC + KI + starch (5), and RC + KI + starch + AMPA (6). The concentrations in solution were: RC, 16.95 mM (with cyclo-O8-Na+, 67.79 mM); KI, 156.63 mM; AMPA, 99.06 mM. The solutions were incubated at two room temperatures for prolonged time. Afterwards (left, bottom row), both RC + KI (3) and RC + KI + AMPA (4) were extracted with deuterated chloroform (bottom phase), and (left, bottom row) both RC + KI + starch (5) and RC + KI + starch + AMPA (6) were treated with L-ascorbic acid. Legend: left, top row (1–6) = first photograph series; left, middle row (1–6) = second photograph series; left, bottom row (1–6) = third photograph series; right, top row (first photograph series): (A) AMPA (76.55 mM) + iodine (as I2, 78.80 mM), (B) iodine (as I2, 78.80 mM); right, bottom row (second photograph series): (A) AMPA (51.03 mM) + iodine (as I2, 52.53 mM), (B) iodine (as I2, 52.53 mM).
2.10 Control Color Assay for Potential Reduction of Elemental Iodine by the Glyphosate Metabolite (Aminomethyl)Phosphonic Acid
Stock preparations were: blank without AMPA (1,000 µl H2O), AMPA stock solution (34 mg AMPA in 1,000 µl H2O), and two iodine stock solutions [each 80 mg elemental iodine in 2,000 µl 90% (v/v) aqueous ethanol]. The stock preparations were incubated at room temperature (RT, J = 14.0 °C) for 15 min with occasional shaking, then at elevated temperature (J = 25.2 °C) for 15 min, until the stock preparations were nearly dissolved (AMPA full, iodine not fully). Then the AMPA stock solution was injected into the first iodine stock solution (it results solution A), no decoloration resulted. The H2O blank was injected into the second iodine stock solution (it results solution B), no decoloration resulted. The concentrations in solution were now: AMPA, 102.07 mM; iodine as I2, 105.07 mM. After incubation at elevated temperature for 15 min, into both solutions 1,000 µl of water were injected, crystallization of elemental iodine followed, and no decoloration resulted. The first photograph series (Figure 3) was taken after 25 min incubation at elevated temperature. After incubation at elevated temperature for 15 min, 2,000 µl of 90% (v/v) aqueous ethanol were injected in both A and B. The mixtures were shaken, the iodine dissolved to give clear deep brown solutions, and no decoloration resulted. After 5 min incubation at elevated temperature the second photograph (Figure 3) series was taken. Both A and B did not show any further change at RT during 24 h observation.
2.11 Color Assay for Cyclo-O8-Na+ Contained in RC – Destruction of Cyclo-O8-Na+ by Glyphosate and ROUNDUP®
Stock preparations were: RC + KI + starch (34 mg RC + 57 mg KI + 18 mg starch in 2,000 µl H2O) (1), RC + KI + starch + glyphosate (34 mg RC + 57 mg KI + 18 mg starch + 34 mg glyphosate in 2,000 µl H2O) (2), RC + KI + starch + glyphosate-Na (34 mg RC + 57 mg KI + 18 mg starch + 87 mg ROUNDUP® GRAN in 2,000 µl H2O) (3), RC + KI + glyphosate (34 mg RC + 57 mg KI + 34 mg glyphosate in 2,000 µl H2O) (4), RC + KI + glyphosate-Na (34 mg RC + 57 mg KI + 87 mg ROUNDUP® GRAN in 2,000 µl H2O) (5) [a saturated solution of 18 mg glyphosate (free acid) in 1,000 µl of H2O showed pH 1.5 (J = 14.0 °C)]. The concentrations in solution were: RC, 16.95 mM (with cyclo-O8-Na+, 67.79 mM); KI, 171.69 mM; glyphosate, 100.55 mM; glyphosate-Na, 108.15 mM. The stock solutions were incubated at room temperature (RT, J = 14.7 °C) for 30 min, then at elevated temperature (J = 25.2 °C) for 70 min, after which time the photograph series (Figure 4) was taken. A control color assay for potential reduction of elemental iodine by glyphosate and ROUNDUP® GRAN was performed. Stock preparations were: blank without glyphosate [75 mg elemental iodine in 2,000 µl 45% (v/v) aqueous ethanol], glyphosate stock solution [52 mg glyphosate in 2,000 µl 45% (v/v) aqueous ethanol], and ROUNDUP® GRAN stock solution [124 mg ROUNDUP® GRAN in 2,000 µl 45% (v/v) aqueous ethanol]. The stock preparations were incubated at room temperature (RT, J = 14.1 °C) for 45 min with occasional shaking. Then 75 mg solid elemental iodine were added to both the glyphosate and ROUNDUP® GRAN stock preparation. After incubation at elevated temperature for 1.5 h, the stock preparations were nearly dissolved (ROUNDUP® GRAN full, glyphosate and iodine not fully). Then 1,000 µl of 90% (v/v) aqueous ethanol were injected into each preparation [resulting in solution A1 (glyphosate), A2 (glyphosate-Na), and B (blank)], no decoloration resulted. The concentrations in solution were now: glyphosate, 102.52 mM; glyphosate-Na, 102.77 mM; iodine as I2, 98.50 mM. The photograph series (Figure 4) was taken after 6 h incubation at elevated temperature, no decoloration resulted. Both A1, A2, and B did not show any further change at RT during 24 h observation.
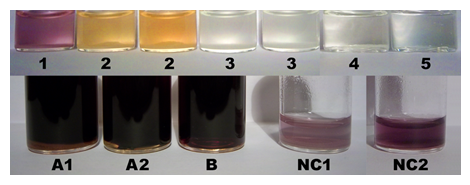
Figure 4: Color assays for the destruction of cyclo-O8-Na+ contained in RC by glyphosate and ROUNDUP® GRAN (top, 1–5), for the potential reduction of elemental iodine by glyphosate and ROUNDUP® GRAN (bottom, A1, A2, and B), and for cyclo-O8-Na+ contained in NC (bottom, NC1 and NC2). Solutions (top, 1–5) were: RC + KI + starch (1), RC + KI + starch + glyphosate (free acid) (2), RC + KI + starch + ROUNDUP® GRAN (3), RC + KI + glyphosate (free acid) (4), and RC + KI + ROUNDUP® GRAN (5). The concentrations in solution were: RC, 16.95 mM (with cyclo-O8-Na+, 67.79 mM); KI, 171.69 mM; glyphosate, 100.55 mM; glyphosate-Na, 108.15 mM. The solutions were incubated at two room temperatures for prolonged time. Legend (bottom): (A1) glyphosate (free acid, 102.52 mM) + iodine (as I2, 98.50 mM), (A2) ROUNDUP® GRAN (glyphosate-Na, 102.77 mM) + iodine (as I2, 98.50 mM), (B) iodine (as I2, 98.50 mM), (NC1, NC2) NC (18.25 mM, with cyclo-O8-Na+, 18.25 mM) + KI (259.04 mM) after 10 h (NC1) and 50 h (NC2) incubation.
A color assay for cyclo-O8-Na+ contained in NC was accordingly performed. The preparation was: NC + KI + starch (25 mg NC + 43 mg KI + 18 mg starch in 1,000 µl H2O). The concentrations in solution were: NC, 18.25 mM (with cyclo-O8-Na+, 18.25 mM); KI, 259.04 mM. The preparation was incubated at elevated temperature for 10 h, after which time the first photograph (Figure 4) series was taken. The preparation was further incubated at room temperature (RT, J = 13.7 °C) for 40 h, after which time the second photograph series (Figure 4) was taken.
2.12 Enzyme Assay of the Glyphosate Metabolite (Aminomethyl)Phosphonic Acid with Human Mitochondrial g-Aminobutyric Acid Transaminase
Recombinant human (16p13.2) mitochondrial 4-aminobutyrate aminotransferase (ABAT) (E.C. 2.6.1.19), mature full length protein aa 29–500 with N-terminal His-SUMO-tag and C-terminal Myc-tag, was purchased from MyBioSource, Inc. (San Diego, CA, USA). The sequence (472 aa) is:
sqaaakvdvefdydgplmktevpgprsqelmkqlniiqnaeavhffcnyeesrgnylvdvdgnrmldlysqissvpigyshpa
llkliqqpqnasmfvnrpalgilppenfveklrqsllsvapkgmsqlitmacgscsnenalktifmwyrskergqrgfsqeeletc
minqapgcpdysilsfmgafhgrtmgclatthskaihkidipsfdwpiapfprlkypleefvkenqqeearcleevedlivkyrk
kkktvagiivepiqseggdnhasddffrklrdiarkhgcaflvdevqtgggctgkfwahehwglddpadvmtfskkmmtggf
fhkeefrpnapyrifntwlgdpsknlllaeviniikredllnnaahagkalltglldlqarypqfisrvrgrgtfcsfdtpddsirnklili
arnkgvvlggcgdksirfrptlvfrdhhahlflnifsdiladfk
(PLP-binding K329 active site in bold). The 500 aa ABAT precursor protein (4-aminobutyrate aminotransferase, mitochondrial isoform 1 precursor, H. sapiens, NCBI Reference Sequence NP_000654.2) additionally bears the N-terminal peptide 1–28. The mature ABAT (aa 29–500, 53.27 kDa) is responsible for the catabolism of g-aminobutyric acid (GABA), an important, mostly inhibitory neurotransmitter in the central nervous system, into succinic semialdehyde. The active enzyme is a homodimer of 53 kDa subunits, each condensed to one pyridoxal 5′-phosphate (PLP). The human ABAT deficiency phenotype includes psychomotor retardation, hypotonia, hyperreflexia, lethargy, refractory seizures, and electroencephalographic (EEG) abnormalities. To investigate whether AMPA is a substrate for human mitochondrial GABA transaminase, experiments were performed with commercially available recombinant human GABA transaminase according to the procedure of Schor et al. [37] with some modifications. Incubations with 500 nmol AMPA, in an assay volume of 120 µl did not show activity of GABA transaminase towards AMPA, while the control assay using 500 nmol 15N-GABA as substrate did resulted in the formation of the expected enzyme product. Subsequent inhibition experiments, with co-incubations of fixed amounts (500 nmol) of 15N-GABA with increasing amounts of AMPA (0–2,000 nmol), revealed that AMPA did not act as an inhibitor of the GABA transaminase-catalysed reaction of 15N-GABA to succinic semialdehyde. These combined results strongly suggest that AMPA is not a substrate for human GABA transaminase.
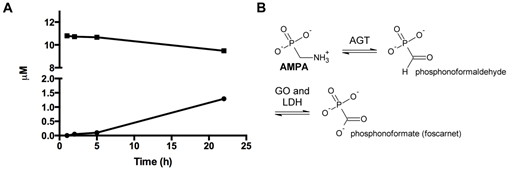
Figure 5: The catabolism of AMPA by human peroxisomal alanine:glyoxylate aminotransferase (AGT). (A) Time course of the AMPA half-transamination reaction of human AGT. The enzyme at a concentration of 5 µM was incubated at J = 25 °C in 100 mM potassium phosphate buffer (pH 7.4). At the indicated times, aliquots were withdrawn and denatured. After removal of the precipitated protein by centrifugation, the supernatants were subjected to HPLC analysis (squares, PLP; circles, PMP). (B) The generation of phosphonoformate (foscarnet) from AMPA over the intermediate phosphonoformaldehyde by the rate-limiting transamination of the major environmental glyphosate metabolite AMPA. Phosphonoformaldehyde, seen as a glyoxylate analogue, could be oxidized by glycolate oxidase (GO) and lactate dehydrogenase (LDH) [66]. Foscarnet is a potent inhibitor of eukaryotic DNA polymerase a [67, 68], an enzyme crucially involved in maintaining chromosomal integrity and telomere length [69].
2.13 Enzyme Assay of the Glyphosate Metabolite (Aminomethyl)Phosphonic Acid with Human Wild-Type Alanine:Glyoxylate Aminotransferase
Recombinant human alanine:glyoxylate aminotransferase (AGT) was expressed in E. coli and purified as described [38]. The enzyme at 5 µM concentration was incubated with 100 mM AMPA at J = 25 °C in 100 mM potassium phosphate buffer pH 7.4. At various times (1, 2, 5, 22 h), aliquots were withdrawn and the reaction was stopped by adding trichloroacetic acid 10% (v/v). The total amount of PLP and pyridoxamine 5′-phosphate (PMP) was determined by HPLC analysis as previously described (Figure 5A) [39]. The product of the half-transamination of AMPA by AGT is phosphonoformaldehyde which can be further oxidized to phosphonoformic acid (phosphonoformate, foscarnet) (Figure 5B).
2.14 Software
Molecular modeling was performed with ACD/Chem Sketch version 2022.1.0 with integrated ACD/3D Viewer (Advanced Chemistry Development, Inc., Toronto, Ontario, Canada) and processed with Mercury 2022.3.0 [The Cambridge Crystallographic Data Centre (CCDC), Cambridge, United Kingdom]. Additional molecular modeling was performed with ACD/Chem Sketch version 12.01 with integrated ACD/3D Viewer (Advanced Chemistry Development, Inc., Toronto, Ontario, Canada) and processed with Mercury 3.1 version 3.1.1 [The Cambridge Crystallographic Data Centre (CCDC), Cambridge, United Kingdom].
2.15 Photographing
Original photographs of the color reactions (Figure 3, Figure 4) were taken with the Kodak EasyShare M550 digital camera (Eastman Kodak Company, Rochester, NY, USA) and transferred to computer with the software Adobe PhotoDeluxe 2.0 Business Edition 1.0 (Adobe Systems Incorporation, San Jose, CA, USA).
3. Results
3.1 Calculation of the Genomic Coverage by the Cyclooctaoxygen Sodium-Bridged Spermine Phosphate Epigenetic Shell of Interphase DNA in Bovine Lymphocytes
The theoretical intracellular concentration of the sperminium hydrogen phosphate/cyclo-O8-Na+ complex required to cover all triplets of the dsDNA genome in a blood lymphocyte of Bos taurus was calculated as 13.81 mM. The actual concentration of spermine was measured as 1.57 ± 0.12 (mM ± s.d.) [11]. Therefore, the genomic dsDNA coverage of B. taurus genome can be calculated as 2.62 ± 0.50 (% ± s.d.), since one unit of sperminium hydrogen phosphate/cyclo-O8-Na+ complex is assumed to cover three nucleotides. A good correlation was obtained when this value was compared to the proportion of protein-coding exons in B. taurus genome which was calculated as 2.58%. For comparison, the human genome contains 2.70% protein-coding exons (see Section 2.5). This pointed to complete coverage of actively transcribed gene regions in B. taurus interphase genome by the sperminium hydrogen phosphate/cyclo-O8-Na+ complex.
3.2 Calculation of the Spermine Coverage of Highly Condensed Mitotic Metaphase DNA in HeLa S3 cells
The content of spermine in HeLa S3 cell metaphase chromatin was calculated as 135.9 ± 16.1 pmol spermine/82.84 zmol dsDNA, and 116.1 ± 11.8 pmol spermidine/82.84 zmol dsDNA [15]. This corresponds to 1.64 × 109 molecules spermine pro one HeLa S3 cell dsDNA genome, and 1.40 × 109 molecules spermidine pro one HeLa S3 cell dsDNA genome. Since one spermine molecule is assumed to cover six base pairs (in the pure spermine form of A-DNA duplex [16] and Z-DNA duplex [17]), and one spermidine molecule is assumed to cover six base pairs (in the pure spermidine form of Z-DNA duplex [18]), this corresponds to a genomic coverage of 50.4% by the spermine tetracation, and of 43.0% by the spermidine trication. This accounts for 93.4% polyamine occupation of HeLa S3 cell dsDNA highly condensed metaphase chromatin by spermine and spermidine, calculated for six base pairs/polyamine unit. As one spermine molecule, in one special occasion, was found to cover four base pairs of a unique B-DNA [35], these values could be anticipated as being lower, since chromosomal DNA is predominantly in the B-DNA form.
3.3 Calculation of the Polyamine Coverage of Maximally Condensed Mitotic Late Anaphase/Early Telophase DNA in Murine Cryptal Enterocytes
The elemental phosphorus content [w (P) in mmol/kg dry weight] in female Mus musculus strain C3H/HeJ cryptal enterocytic mitotic (late anaphase/early telophase) chromatin was published [19]. The obtained in vivo value corresponds to a genomic dsDNA coverage (calculated for 6 bp/polyamine) of 102.1% (100% coverage is 5.88 bp/polyamine), and a nuclear RNA coverage (calculated for 6 nucleotides/polyamine) of 159.5%, by the spermine tetracation and spermidine trication (spermine/spermidine ratio 0.85). This strongly points to a function of polyamine occupation for nuclear RNA, assuming 100% coverage as 3.76 nucleotides/polyamine molecule. These results, both for dsDNA and nuclear RNA [hnRNA with pre-mRNAs, snRNA, snoRNA, RNase P, RNase MRP, various ncRNAs (lncRNA) and other nuclear RNAs], are a logic consequence of the maximal condensation grade peaking in late anaphase/early telophase mammalian chromatin [40].
3.4 Calculation of the Hydrogen Selenite (HSeO3−) Coverage of Human Euchromatin DNA Specifically at ATG Start Codon Sequences
A perfect correlation was obtained when the hydrogen selenite substitution content of the cyclooctaoxygen sodium-bridged sperminium hydrogen phosphate protection shell of human euchromatin was calculated from the optimal human blood serum level for the essential human micronutrient selenium (see Section 2.5). The optimal human blood serum level of selenium (as Se) was taken as 110 µg/l (ng/ml) from literature [23, 24]. The median number of aa-coding triplets required for one triplet to be covered with the sperminium hydrogen selenite/cyclo-O8-Na+ complex in interphase euchromatin (in form of hydrogen selenite HSeO3−) at the published [23, 24] optimal human blood serum level of selenium [as Se, 110 µg/l (ng/ml)] was calculated to be 560.44 aa. The optimal human blood serum level to cover all ATG triplet codons in human euchromatin mRNA genes, both at the mRNA gene translation initiation triplet ATG codon and at the 5′-UTR uORFs upstream start codons (uATGs transcribed to uAUGs), was calculated to be 107 µg/l (ng/ml). The number of 5′-UTR uORFs upstream start codons (uATGs transcribed to uAUGs) in the 5′-UTR of mRNA genes was calculated from literature [26] as the number of transcribed human genes with at least one uATG divided by the number of human mRNA genes with annotated 5′-UTR. 49% of human mRNA genes contain at least one uATG start codon in their 5′-UTR [26] [the downstream 3′-untranslated region (3′-UTR) contains none ATG start codon]. The number of triplets required for the covering of all ATG triplet codons in human interphase euchromatin mRNA genes by one hydrogen selenite HSeO3− molecule (within the sperminium hydrogen selenite/cyclo-O8-Na+ complex) at the calculated optimal human blood serum level of selenium 107 µg/l (ng/ml) (576.40 aa) correlated with the determined heuristic value (560.44 aa). Consequently, a model of the postulated [5] first epigenetic shell of euchromatic in vivo DNA, as exemplified for a single-stranded hexanucleotide d(ApCpCpApTpGp), part of the transcribed and translated human gene open reading frame initiation codon ATG encrypted in the Kozak consensus sequence 5′-ACCATGG-3′ [28], introducing a molecular biological model for sperminium hydrogen phosphate/cyclo-O8-Na+/ssDNA and sperminium hydrogen selenite/cyclo-O8-Na+/ssDNA interactions, was deduced (Figure 6).
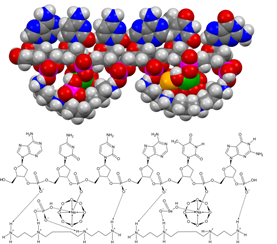
Figure 6: Molecular modeling of the postulated [5] first epigenetic shell of euchromatic in vivo DNA, as exemplified for a single-stranded hexanucleotide d(ApCpCpApTpGp), part of the transcribed and translated human gene open reading frame initiation codon ATG encrypted in the Kozak consensus sequence 5′-ACCATGG-3′ [28], introducing a molecular biological model for sperminium hydrogen phosphate/cyclo-O8-Na+/ssDNA (left) and sperminium hydrogen selenite/cyclo-O8-Na+/ssDNA (right) interactions. Element color codings: grey, carbon; white, hydrogen; blue, nitrogen; red, oxygen; purple, phosphorus; green, sodium; yellow, selenium.
3.5 Calculation of the Apparent Acid Dissociation Constant of the Human Genome DNA, and the Intranuclear Micro-pH Mediated by Single Spermine Occupation, in Comparison to Spermine Phosphate/Cyclooctaoxygen Sodium Complex Occupation, of Human Interphase Euchromatin
The apparent (effective) pK′a,HG = 2.56 (J = 25 °C) of the haploid human genome was calculated (Figure 2A) according to the method of Katchalsky & Gillis [30], as based on the theoretical considerations of Kuhn & Kuhn [31]. The hypothetical intranuclear pHDNA = 2.32, mediated by H. sapiens haploid interphase genome dsDNA without any neutralizing shell, can be calculated (Figure 2B). Assuming one spermine molecule covering four base pairs (single occupation) of B-DNA [35], and correcting for actively transcribed gene regions of H. sapiens genome, the hypothetical intranuclear micro-pH [41] surrounding H. sapiens haploid interphase euchromatin when covered (single quartet occupation) by the spermine tetracation alone can be calculated pHDNA/spermine = 6.27. For the diploid dsDNA genome, after completed S phase during interphase, the pHDNA/spermine is identical. The theoretical intranuclear micro-pH surrounding H. sapiens haploid interphase euchromatin when covered (double triplet occupation) by the sperminium hydrogen phosphate/cyclooctaoxygen sodium complex can be calculated pHDNA/shell = 6.26. For the diploid dsDNA genome, after completed S phase during interphase, the pHDNA/shell is identical.
3.6 Color Assay for Cyclo-O8-Na+ Contained in RC – Destruction Of Cyclo-O8-Na+ by the Glyphosate Metabolite (Aminomethyl)Phosphonic Acid
The cyclo-O8-Na+ complex [m-chloro(m-hydroxy)bis(octoxocane-k4O1,O3,O5,O7)disodium] contained in RC (4 mol cyclo-O8-Na+ pro mol cytidine × HCl) reacted with potassium iodide and potato starch to an intensely colored (reddish violet) [(cyclo-O8-Na+)2(I42–)]–amylose complex [RC + KI + starch (5)] (Figure 3) which was destroyed by the glyphosate metabolite (aminomethyl)phosphonic acid (AMPA) [RC + KI + starch + AMPA (6)]. The nature of this complex is based on the starch-catalysed formation of tetraiodide I42– [(I–I–I–I)2–], which is known to be of ruby red color in crystalline form [42], was frequently observed in crystals [43], and was theoretically predicted to exist in solution [44]. The tetraiodide I42– is in turn complexed to cyclo-O8-Na+ and inserted into the amylose helix (Figure 7A and 7B). Multiple controls excluded that the [(cyclo-O8-Na+)2(I42–)]–amylose complex is formed (i) in blanks [KI (1), and KI + starch (2)], (ii) without starch [RC + KI (3)], and (iii) from AMPA and RC + KI [RC + KI + AMPA (4)] (Figure 3). That the reddish violet complex involved triiodide I3– [(I–I–I)–] or pentaiodide I5– [(I–I–I–I–I)–] anions could be excluded by the observed color. Triiodide I3– is deep brown [45] and pentaiodide I5– is deep blue in color [46]. Pentaiodide I5– is also deep blue in the well-known complex with starch [47]. Furthermore, the [(cyclo-O8-Na+)2(I42–)]–amylose complex was (i) reduced (decolorized) by L-ascorbic acid (vitamin C), and (ii) the residual color after AMPA-catalysed destruction of cyclo-O8 was pale pink (rosé) in color, not intense blue. This proved that (i) the reddish violet complex contained reducible iodine units, and (ii) did not contain the reducible iodine units as triiodide, pentaiodide and/or other higher polyiodides [48]. Without starch no [(cyclo-O8-Na+)2(I42–)] and/or iodine was formed, as was proved by deuterated chloroform (CDCl3) extraction of the incubated RC + KI (3) and RC + KI + AMPA (4) solutions (Figure 3). Therefore, the iodine in tetraiodide I42– must have being formed by starch catalysis.
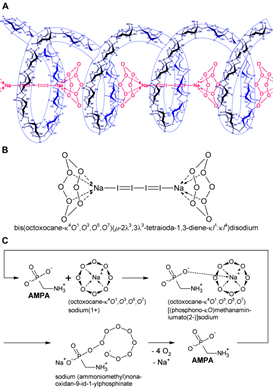
Figure 7: Explanation of the color reaction for cyclo-O8-Na+ contained in RC, and the destruction of cyclo-O8-Na+ by the glyphosate metabolite (aminomethyl)phosphonic acid (AMPA). (A) The cyclo-O8-Na+ complex contained in RC reacted with potassium iodide under catalysis by potato starch to an intensely colored (reddish violet) [(cyclo-O8-Na+)2(I42–)] complex stabilized within the starch-contained amylose helix. (B) The proposed chemical formula for the amylose-complexed [(cyclo-O8-Na+)2(I42–)]: bis(octoxocane-k4O1,O3,O5,O7)( m-2l3,3l3-tetraioda-1,3-diene-kI1: kI4)disodium. (C) A logically deduced catalytic ‘rolling-circle’ mechanism for the AMPA-catalysed degradation of cyclo-O8-Na+. The cyclooctaoxygen ring is split to a phosphonate-esterified nonaoxidanide which is stabilized by ionic binding to the primary ammonium cation of AMPA. The phosphonate-esterified nonaoxidanide eliminates four oxygen O2 molecules by a ‘rolling-circle’ cascade, regenerating AMPA.
A logically deduced catalytic ‘rolling-circle’ mechanism for the AMPA-catalysed degradation of cyclo-O8 is hence proposed (Figure 7C). AMPA exhibits three acid dissociation constants: pKa1 = 0.9 (phosphonic acid, 1st), pKa2 = 5.6 (phosphonic acid, 2nd), pKa3 = 10.2 (primary ammonium R–NH3+) [49]. Therefore, AMPA is fully (zwitter)ionized at physiological pH 7.4. One anionic oxygen of the phosphonate group binds to the sodium cation in cyclo-O8-Na+, the other anionic phosphonate oxygen splits the cyclooctaoxygen ring creating a phosphonate-esterified nonaoxidanide which is stabilized by ionic binding to the primary ammonium cation of AMPA. The phosphonate-esterified nonaoxidanide eliminates four oxygen O2 molecules by a ‘rolling-circle’ cascade, in reversal of the proposed [5] synthesis of cyclooctaoxygen, yielding AMPA and Na+. This would be clearly a catalytic mechanism, since AMPA is regenerated in the reaction cycle. Hence AMPA is able to destroy many cyclo-O8-Na+ complexes without being consumed itself. To exclude that the destruction of cyclo-O8-Na+ by AMPA is an artifact, it was tested if AMPA reduces (decolorizes) iodine in near equimolar mixture (Figure 3). In all variations tested, including a blank control, AMPA was not oxidized by iodine, and, in turn, did not reduce (decolorize) iodine molecules. Therefore, the AMPA-catalysed destruction of cyclo-O8 was selective, and not a mere reduction of the tetraiodide I42– [(I–I–I–I)2–]-contained iodine unit in the [(cyclo-O8-Na+)2(I42–)]–amylose complex.
3.7 Color Assay for Cyclo-O8-Na+ Contained in RC – Destruction of Cyclo-O8-Na+ by Glyphosate and ROUNDUP®
The developed color assay was applied onto the free acid of glyphosate and the monosodium salt of glyphosate contained in ROUNDUP® GRAN granules. The colored [(cyclo-O8-Na+)2(I42–)]–amylose complex [RC + KI + starch (1)] (Figure 4) was destroyed by glyphosate [RC + KI + starch + glyphosate (2)] and ROUNDUP® GRAN [RC + KI + starch + glyphosate-Na (3)]. Without starch no [(cyclo-O8-Na+)2(I42–)] and/or other color complex was formed, as was proved by the controls RC + KI + glyphosate (4) and RC + KI + glyphosate-Na (5) (Figure 4). The RC + KI + starch + glyphosate (2) solution was colored yellow by the strong acid glyphosate (Figure 4), through H+ action onto starch producing the characteristic yellow dextrins [50]. Glyphosate exhibits four acid dissociation constants: pKa1 = 0.78 (phosphonic acid, 1st), pKa2 = 2.29 (carboxylic acid), pKa3 = 5.96 (phosphonic acid, 2nd), pKa4 = 10.98 (secondary ammonium R1–NH2+−R2) [49]. Therefore, glyphosate (free acid) represents a strong acid. A logically deduced catalytic ‘rolling-circle’ mechanism for the glyphosate-catalysed degradation of cyclo-O8 is hence proposed (Figure 8). One anionic oxygen of the phosphonate group binds to the sodium cation in cyclo-O8-Na+, the other anionic phosphonate oxygen splits the cyclooctaoxygen ring creating a phosphonate-esterified nonaoxidanide which is stabilized by ionic binding to the secondary ammonium cation of glyphosate. The phosphonate-esterified nonaoxidanide eliminates four oxygen O2 molecules by a ‘rolling-circle’ cascade, yielding glyphosate and Na+ in a catalytic mechanism. Glyphosate is regenerated in the catalytic cycle. Glyphosate is able to destroy many cyclo-O8-Na+ complexes without being consumed itself (Figure 8).
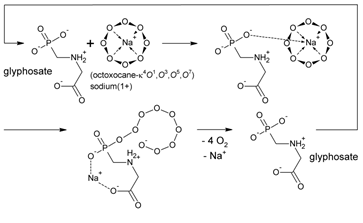
Figure 8: A logically deduced catalytic ‘rolling-circle’ mechanism for the (fully ionized) glyphosate-catalysed degradation of cyclo-O8-Na+. The cyclooctaoxygen ring is split to a phosphonate-esterified nonaoxidanide which is stabilized by ionic binding to the secondary ammonium cation of glyphosate (and complexation of the sodium cation). The phosphonate-esterified nonaoxidanide eliminates four oxygen O2 molecules by a ‘rolling-circle’ cascade, regenerating glyphosate.
To exclude that the destruction of cyclo-O8-Na+ by glyphosate is an artifact, it was tested if glyphosate, or glyphosate-Na, reduces (decolorizes) iodine in near equimolar mixture (Figure 4). Glyphosate was not oxidized by iodine, and, in turn, did not reduce (decolorize) iodine molecules. Therefore, the glyphosate-catalysed destruction of cyclo-O8 was selective. To confirm the general nature of the color assay, it was extended to the cyclo-O8-Na+ contained in NC (1 mol cyclo-O8-Na+ pro 4 mol cytidine × HCl) [5]. The reddish violet [(cyclo-O8-Na+)2(I42–)]–amylose complex was indeed formed from NC (Figure 4), but much more slowly (> 10 h) than from RC.
3.8 Enzymatic Investigations with the Glyphosate Metabolite (Aminomethyl)Phosphonic Acid
AMPA was tested for being accepted as an enzymatic substrate for human mitochondrial g-aminobutyric acid transaminase (ABAT) [37], and wild-type human liver peroxisomal alanine:glyoxylate aminotransferase (AGT) [38]. Both enzymes were selected because of the chemical analogy between AMPA and b-alanine/L-alanine. ABAT represents also a b-alanine transaminase [37]. ABAT did not catalyse any AMPA half-transamination. Data obtained with human AGT indicate that the enzyme is slowly able to catalyse the half-transamination of AMPA (Figure 5B), with a rate of kcat = –0.0108 ± 0.0009 µM PLP/h/µM AGT consumed, or kcat = –0.0104 ± 0.0009 µM PMP/h/µM AGT formed, respectively (PLP, pyridoxal 5′-phosphate; PMP, pyridoxamine 5′-phosphate) (Figure 5A). This catalytic constant is approximately 16.2 million-fold lower than that of the physiological transamination of L-alanine (human wild-type AGT: kcat = 45 ± 2 s-1; kcat = 162,000 h-1) [51].
4. Discussion
One reason why the cyclooctaoxygen sodium-bridged spermine phosphate epigenetic shell of in vivo DNA was overlooked until now should be its destruction during DNA purification by the classical phenol extraction method of Schuster, Schramm & Zillig [52]. This original phenol extraction, although variously modified for nowadays use, consistently precipitates the spermine as sperminediium (terminal NH3+) di(phenolate) at pH 7.9–8.0, since the pKa value of phenol is 9.97 [potentiometric titration in H2O, J = 25 °C, ionic strength (NaCl) 0.1], and the four pKa values of spermine are: pKa1 = 10.86 (terminal [53] NH3+), pKa2 = 10.05 (terminal NH3+), pKa3 = 8.82 (inner [53] NH2+), pKa4 = 7.95 (inner NH2+) [potentiometric titration in H2O, J = 25 °C, ionic strength (NaCl) 0.1] [34]. The cyclo-O8-Na+ could react with alkaline (pH 7.9–8.0) buffered phenol to disodium rhodizonate (C6Na2O6), a known oxidation product of p-benzoquinone, which in turn is an oxidation product of phenol. Regardless of the chemical details, the commercial salmon sperm DNA (utilized in [5]) and calf thymus DNA preparations are devoid of cyclo-O8-Na+ and spermine phosphate complexation, since the methods utilized for calf thymus and salmon sperm sodium deoxyribonucleate preparation [treatment with sodium dodecyl sulfate, high salt (NaCl) treatment, repeated ethanol precipitation] certainly remove the cyclooctaoxygen sodium-bridged spermine phosphate epigenetic shell. Our findings have important consequences for the epigenetics [54] of eukaryotic in vivo DNA. Kesel et al. suggested [5] a model for a first epigenetic shell of in vivo DNA (Figure 6), based on the observed complexation of cyclo-O8-Na+ and spermine phosphate to ssDNA. In this model [5] the phosphate backbone of ssDNA binds one cyclo-O8-Na+ pro three nucleotides, and this binary complex binds one spermine monophosphate to form a ternary epigenetic core of DNA. The monohydrogen phosphate bridges the cyclo-O8-Na+ with the sperminium cation, and the cyclo-O8-Na+ has an inverted alternating orientation [5]. Interestingly, the sperminium tetracation cannot bind alone to DNA in this model, since the distances [d (N1,N4) = 490 pm; d (N4,N9) = 620 pm; d (N1,N12) = 1,600 pm] between the four ammonium nitrogens do not fit the intrastrand phosphate–phosphate distance of dsDNA (B-DNA: dØ = 700 pm [55]; A-DNA: dØ = 590–600 pm [55]; Z-DNA: dØ = 590 pm (step pCp), dØ = 600 pm (step pGp) [56]). Therefore, it is quite remarkable that in this model for the first epigenetic shell of in vivo DNA [5] a repeating unit is formed from cyclo-O8-Na+ and spermine phosphate that perfectly fits both the triplet nature of the genetic code and the repeating distance of the phosphate anion backbone of DNA. In view of the important findings of Kirmes et al. [4], that an interaction of eukaryotic chromatin DNA structure with atmospheric oxygen partial pressure takes place, previous postulations [5] have to be defined more precisely. Since under switching to hypoxic conditions eukaryotic cell chromatin gets highly condensed [4], accompanied by redistribution of the polyamine pool to the nucleus [4], the cyclooctaoxygen sodium-bridged spermine phosphate epigenetic shell can only be restricted to actively transcribed gene regions of eukaryotic ‘open’ euchromatin, excluding occupation of condensed chromatin. Hypoxia should largely prevent metabolic formation of cyclooctaoxygen. Both under hypoxic conditions and in the metaphase of mitosis, where spermine synthesis is highest [57], coincident with an extraordinary high condensation grade (15,000–20,000-fold) of metaphase chromatin [58], none cyclooctaoxygen should be involved in covering the highly condensed chromatin DNA. Here no discrimination between eu- and heterochromatin is made in metaphase, and all mitotic eukaryotic chromatin DNA is complexed with spermine tetracation and spermidine trication (and, at small proportions, with putrescine and cadaverine dications). In summary, this reflects the high mitotic chromatin condensation grade and is confirming the results with hypoxia-induced chromatin condensation under coinciding polyamine pool nuclear translocation [4]. All these considerations point to the correctness of the model that the cyclooctaoxygen sodium-bridged spermine phosphate epigenetic shell is restricted to actively transcribed ‘hot spot’ gene regions of eukaryotic ‘open’ euchromatin. Importantly, this epigenetic shell of eukaryotic ‘open’ euchromatin covers each strand of dsDNA separately, one at the positive strand, one at the negative strand (double occupation), whereas the highly condensed dsDNA structures bind one polyamine molecule directly at the double strand (single occupation) [16–18, 35]. This is substantiated by the precise calculation of the apparent acid dissociation constant of the human genome DNA. Kesel et al. also elaborated a model for selenium (as hydrogen selenite, HSeO3–, at physiological pH 7.4) protection of DNA [5]. Selenium, the element of the moon [59], was discovered by Jöns Jacob Berzelius (1779–1848) in 1817 and was named by him in honor of the Greek goddess of the moon Selene (σεληνη) [60]. Selenium is essential to mammalian physiology at nutritional levels, but supraphysiological intake of selenium is known to be toxic for mammals [5]. Sodium selenite (Na2SeO3), as hydrogen selenite HSeO3– at pH 7.0 (selenious acid H2SeO3: pKa1 = 2.62, pKa2 = 8.32 [61]), binds to calf thymus genomic B-DNA at pH 7.0 [62], and to Saccharomyces cerevisiae A-RNA at pH 7.0 [63]. Selenium has the ability to protect DNA from noxious influences (oxidative stress, radiation, cytotoxic agents) [5], and is essential to genomic stability [5], but the exact molecular biological basis for these phenomena is unknown. If in the model of a first epigenetic shell of in vivo DNA [5] the monohydrogen phosphate is replaced by hydrogen selenite, an epigenetic explanation for the interaction of selenium with eukaryotic in vivo DNA could be given. A coinciding correlation was obtained when the hydrogen selenite substitution content of the cyclooctaoxygen sodium-bridged sperminium hydrogen phosphate protection shell of human euchromatin was calculated from the optimal human blood serum level for the essential human micronutrient selenium (see Section 2.5). The optimal human blood serum level of selenium (as Se), given as 110 µg/l (ng/ml) from literature [23, 24], coincided with the calculated optimal selenium human blood serum level, 107 µg/l (ng/ml), required to cover all ATG triplet codons in human euchromatin mRNA genes, both at the mRNA gene translation initiation triplet ATG codon and the 5′-UTR uORFs upstream start codons (uATGs transcribed to uAUGs). 49% of human mRNA genes contain at least one uATG start codon in their 5′-UTR [26] [the downstream 3′-untranslated region (3′-UTR) contains none ATG start codon]. The median number of aa-coding triplets required for one triplet to be covered with the sperminium hydrogen selenite/cyclo-O8-Na+ complex in interphase euchromatin (in form of hydrogen selenite HSeO3−) at the published [23, 24] optimal human blood serum level of selenium [as Se, 110 µg/l (ng/ml)] was calculated as to be 560.44 aa. This result correlated well with the calculated number of aa triplets (576.40) required for the covering of all ATG triplet codons in human interphase euchromatin mRNA genes by one hydrogen selenite HSeO3− molecule (within the sperminium hydrogen selenite/cyclo-O8-Na+ complex) at the calculated optimal human blood serum level of selenium [as Se, 107 µg/l (ng/ml)]. A model of the postulated [5] first epigenetic shell of euchromatic in vivo DNA was deduced, as exemplified for a single-stranded hexanucleotide d(ApCpCpApTpGp), part of the transcribed and translated human gene open reading frame initiation codon ATG encoded in the Kozak consensus sequence [28] (Figure 6). This model accounts for, at least some of, the well-known bimodal, protective and toxic, in vivo effects exerted by selenium onto mammalian physiology. A complete specific substitution pattern of hydrogen selenite for monohydrogen phosphate at ATG open reading frame initiation codons would be essential for eukaryotic gene regulation, but if the displacement ratio HSeO3–/HPO42– exceeds a certain physiological tolerance level, and hydrogen selenite is randomly incorporated into the eukaryotic protection shell of human euchromatin, the epigenetic equilibrium should collapse. The high, both acute and chronic, mammalian toxicity of sodium selenite (Na2SeO3) should be due, at least in part, to direct detrimental effects of supraphysiological levels of hydrogen selenite HSeO3– on mammalian chromosomal DNA integrity and regulation of genome expression. Assuming an essential biological function for the cyclooctaoxygen sodium-bridged spermine phosphate and selenite epigenetic shell, it was searched for substances able to selectively destroy this epigenetic protection structure, and the total herbicide glyphosate, N-(phosphonomethyl)glycine (ROUNDUP®, Monsanto), and its major environmental metabolite (aminomethyl)phosphonic acid (AMPA) [64] were tested on the cyclo-O8-Na+ complex contained in RC. Glyphosate was chosen because it represents the top-selling total herbicide worldwide [65], and RC was selected because of its highest molar cyclo-O8-Na+ content in the complex series NC, dNC, and RC [5]. Glyphosate and AMPA show chemical properties which might predispose them for destruction of cyclooctaoxygen in general. Glyphosate and AMPA are very hydrophilic and amphoteric, and their phosphonate moieties could be suitable to interact with cyclo-O8-Na+. It was unequivocally shown that glyphosate and AMPA indeed selectively destroy the cyclo-O8-Na+ complex contained in RC (Figure 3, Figure 4, Figure 7). I therefore conclude that glyphosate and the major environmental glyphosate metabolite AMPA [64] also destroy the cyclooctaoxygen sodium-bridged spermine phosphate and selenite epigenetic shell of human euchromatin, because destruction of cyclooctaoxygen is sufficient to bring this essential protection shield of human euchromatin into collateral epigenetic collapse. To get support for the selectivity of AMPA as an epigenetic poison, the affinity of AMPA towards human mitochondrial g-aminobutyric acid transaminase (ABAT) [37], and to wild-type human liver peroxisomal alanine:glyoxylate aminotransferase (AGT) [51], was determined. ABAT represents also a b-alanine transaminase [37], and both enzymes were selected because of the structural similarity between AMPA and b-alanine/L-alanine. AMPA showed essentially no affinity to ABAT, but was very showly catabolized by AGT (Figure 5A). This latter result is of interest, since the product of the half-transamination of AMPA by AGT is phosphonoformaldehyde (Figure 5B) which can be oxidized (peroxisomal glycolate oxidase, cytoplasmic lactate dehydrogenase [66]) to phosphonoformic acid (phosphonoformate, foscarnet) (Figure 5B). Foscarnet represents a well-known inhibitor of mammalian [67, 68] and viral [67, 68] DNA-dependent DNA polymerases. Eukaryotic DNA polymerase a is crucially involved in chromosome maintenance, DNA repair and recombination, transcriptional silencing, checkpoint activation, and telomere length maintenance [69]. Mammalian DNA polymerase a is potently inhibited by foscarnet [67, 68]. Therefore, the low-affinity half-transamination of AMPA by AGT, the rate-limiting step leading to foscarnet, could negatively influence human chromosome maintenance, DNA damage repair, and telomere length preservation, mediated by the AMPA catabolite foscarnet inhibition of DNA polymerase a. This enzymatic catabolism provides an additional, minor mechanism of destabilization and impairment of eukaryotic chromosomal DNA indirectly induced by the environmental glyphosate metabolite AMPA.
5. Conclusions
An improved and corrected molecular biological model is proposed for a first epigenetic shell of eukaryotic euchromatin (Figure 6, Figure 9). This model incorporates an epigenetic explanation for the interactions of the essential micronutrient selenium (as selenite) with eukaryotic euchromatin (Figure 6). The sperminium hydrogen phosphate/cyclooctaoxygen sodium complex was calculated to cover the actively transcribed regions (2.6%) of bovine lymphocyte interphase genome dsDNA (double occupation). The polyamine (spermine/spermidine ratio 1.17) coverage of HeLa S3 cell metaphase chromatin dsDNA was calculated as 93.4% (single occupation). In murine cryptal enterocytic mitotic (late anaphase/early telophase) chromatin the obtained in vivo value corresponds to complete genomic coverage (single occupation), and to comprehensive and extensive nuclear RNA coverage, by the spermine tetracation and spermidine trication (spermine/spermidine ratio 0.85). Because cyclooctaoxygen seems to be naturally absent in hypoxia-induced highly condensed chromatin [4], a model [70] is proposed for the cyclooctaoxygen sodium-bridged spermine phosphate (and selenite) epigenetic shell of actively transcribed gene regions in eukaryotic ‘open’ chromatin DNA (Figure 9). Furthermore, a working model is tabulated in summary for the selective cell cycle-dependent epigenetic occupation of eukaryotic DNA (Table 1).
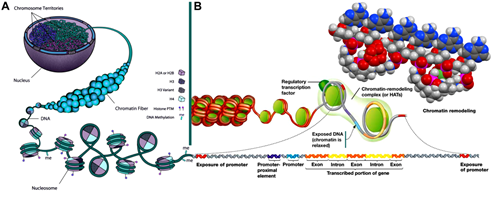
Figure 9: An improved and corrected model for the cyclooctaoxygen sodium-bridged spermine phosphate (and selenite) epigenetic shell [5] of actively transcribed gene regions in eukaryotic interphase ‘open’ chromatin DNA. (A) The nucleus of an eukaryotic cell with chromosome territories, chromatin fiber (10 nm ‘beads–on–a–string’ fiber), nucleosome structure, nucleosome octamer core histone proteins [H2A/H2B, H3/H3 variant, H4] with posttranslational histone protein modifications (histone PTM), and decondensing DNA with regulative cytosine nucleobase 5-methylation sites (me). Adapted and modified in part from [70]. (B) The decondensation of chromatin enabling gene transcription in eukaryotic interphase ‘open’ chromatin DNA. The relaxed chromatin, regulatory transcription factor, chromatin-remodeling complex/histone acetyl transferases (HATs), and chromatin remodeling are indicated. The ‘open’ DNA with the transcription-prone gene is generally structured in promoter, promoter-proximal genetic elements, transcribed/expressed gene exons and non-expressed gene introns. The cyclooctaoxygen sodium-bridged spermine phosphate (and selenite) epigenetic coverage of this actively transcribed gene region is symbolized.
What may be the evolutionary conserved biological significance, and human pathophysiological implication, of this selective epigenetic shell? Obviously, the selective cyclooctaoxygen sodium-bridged spermine phosphate (and selenite) epigenetic shell serves to discriminate euchromatin from heterochromatin (Table 1). Interestingly, a supraphysiological concentration of selenite induces S−G2/M cell cycle arrest, locked in interphase [71], and inhibits DNA and RNA synthesis [72], again pointing on a crucial determination of the optimally hydrogen selenite-marked ATG codons in eukaryotic DNA mRNA genes. Both a deficiency (not all ATG codons marked by HSeO3−) and an excess (randomly distribution of HSeO3−-tagging in genome, no more restricted to only ATG codons) of selenium concentration as hydrogen selenite exerts detrimental effects on eukaryotic gene regulation and expression. This reflects the specific life-essential role of selenium in eukaryotic gene processing and maintenance, a long-sought function of selenium in human nutritional physiology, previously defined as ‘mysterious’ essentiality of selenium for human life [59, 71].
Furthermore, during transcription of actively transcribed gene regions in eukaryotic ‘open’ chromatin the double helix must be unwound by DNA helicases and the strands must be separated to enable access to DNA-dependent RNA polymerases I, II, and III. This creates intermediate DNA single-strand regions which are prone to chemical structure damage by multiple noxious impacts like reactive oxygen species (ROS), mutagens, and UV light. The selective cyclooctaoxygen sodium-bridged spermine phosphate (and selenite) epigenetic occupation of these sensitive single-stranded stretches could serve as an intrinsic protection against structural damage. This would be a logic explanation for the selective nature of the separate occupation of both DNA strands, consequently retained when strands are separated for transcription of mRNA. Also intriguing seem to be pH effects, since spermine is a strong base, and the major pH-related damage to DNA is depurination [73] creating apurinic sites at low pH [73]. The precisely calculated intranuclear micro-pH gain, obtained by sperminium hydrogen phosphate/cyclooctaoxygen sodium complexation of B-DNA individual strands (essentially the same as the intranuclear micro-pH gain for condensed B-DNA strand-overarchingly covered by sperminium tetracations) should protect against low pH-depurination creating apurinic DNA lesions leading to DNA single-strand breaks [73]. In this context the formation of kinetin (N6-furfuryl-9H-adenine) from DNA is well known (Figure 10) [74]. It should be emphasized that kinetin is not contained in native mammalian DNA, contrary to misleading claims [75, 76], but is formed only during non-oxidative DNA damage. A mechanism for the kinetin formation in, or from, DNA was proposed [75, 76], but it seems not to be conclusive in chemical reason, since furfural does not react with the adenine 6-NH2 group under condensation to a Schiff base under in vivo conditions [74].
|
Cell cycle phase |
Heterochromatin |
Euchromatin |
|||
|
Interphase |
Function (concise) |
Spermine-occupation? |
Cyclo-O8-Na+-occupation? |
Spermine-occupation? |
Cyclo-O8-Na+-occupation? |
|
G0 (Gap 0) |
Resting and quiescence |
No |
No |
Yes (2 ×) – With cyclo-O8-Na+ |
Yes (2 ×) – On ‘open’ Chr |
|
G1 (Gap 1) |
Transcription and histone synthesis |
No |
No |
Yes (2 ×) – With cyclo-O8-Na+ |
Yes (2 ×) – On ‘open’ Chr |
|
S (Synthesis) |
DNA synthesis |
No |
No |
Yes (2 ×) – With cyclo-O8-Na+ |
Yes (2 ×) – On ‘open’ Chr |
|
G2 (Gap 2) |
Translation |
No |
No |
Yes (2 ×) – With cyclo-O8-Na+ |
Yes (2 ×) – On ‘open’ Chr |
|
Mitosis |
Function (concise) |
Spermine-occupation? |
Cyclo-O8-Na+-occupation? |
Spermine-occupation? |
Cyclo-O8-Na+-occupation? |
|
Prophase |
Chr condenses into chromosomes, nucleolus disappears |
Yes (1 ×) – Condensing Chr |
No |
Yes (1 ×) – Condensing Chr |
No |
|
Prometaphase |
Kinetochore and polar microtubules attach, mitotic spindle formed, nucleus disappears |
Yes (1 ×) – Condensed Chr |
No |
Yes (1 ×) – Condensed Chr |
No |
|
Metaphase |
Centrosomes pull chromosomes, chromosome centromeres line up at metaphase plate |
Yes (1 ×) – Highly condensed Chr |
No |
Yes (1 ×) – Highly condensed Chr |
No |
|
Anaphase |
Chromosomes break at centromeres, sister chromatids separated by microtubules |
Yes (1 ×) – Maximally condensed Chr in late anaphase |
No |
Yes (1 ×) – Maximally condensed Chr in late anaphase |
No |
|
Telophase |
Chr reformed from chromosomes, nucleus and nucleolus reappear |
Yes (1 ×) – Maximally condensed Chr in early telophase |
No |
Yes (1 ×) – Maximally condensed Chr in early telophase |
No |
Table 1: Tabulation of the selective cell cycle-dependent occupation of eukaryotic DNA by epigenetic polyamine shells. 1 ×, single quartet occupation (one polyamine pro both strands); 2 ×, double triplet occupation (one polyamine pro one strand); Chr, chromatin.
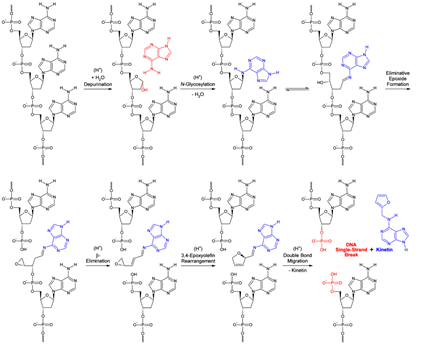
Figure 10: A chemical mechanistic deduced logical scheme for the generation of kinetin (N6-furfuryl-9H-adenine) [74] from DNA by proton catalysis (kinetin-generating “base flip”, KGBF). Protonation of the adenine nucleobase of the shown d(pApApAp) sequence at purine N-7 induces depurination [73], creating an apurinic site [73]. Proton-catalysed N-glycosylation [77, 78] at the 6-NH2 of adenine regenerates a nucleobase-inverted 2′-deoxynucleotide which is in furanose ring-opening equilibrium with its ald(os)imine form. Two eliminations simultaneously follow, the first (auto-catalysed by 3′-O-phosphate conjugate base proton reception) by SN2-nucleophilic eliminative epoxide formation, the second by proton-catalysed (auto-catalysed by the previously liberated 3'-O-monohydrogen phosphate conjugate acid) b-elimination (E1 unimolecular elimination), which leads to an 3,4-epoxyolefin: N-{(1E,2E)-3-[(2S)-oxiran-2-yl]prop-2-en-1-ylidene}-9H-purin-6-amine. The chiral (S)-2-vinyloxirane structure rearranges to a chiral (R)-2-alkyl-2,5-dihydrofuran due to 3,4-epoxyolefin rearrangement [79, 80]: N-[(2R)-2,5-dihydrofuran-2-ylmethyl]-9H-purin-6-amine. The latter compound rearranges to kinetin through facile double bond migration driven by aromatization to a furan. A DNA single-strand break is left back.
I therefore propose a chemical mechanistic deduced logical scheme for the generation of kinetin from DNA by proton catalysis (kinetin-generating “base flip”, KGBF) (Figure 10), based on proton-catalysed depurination [73] and subsequent inverted adenine 6-NH2 N-glycosylation [77, 78], in consequence [79, 80] leaving back a DNA single-strand break. It is therefore concluded that the sperminium hydrogen phosphate/cyclooctaoxygen sodium complex serves to protect ssDNA from nucleic acid-mediated intrinsic low intranuclear micro-pH-induced depurination, including KGBF, creating apurinic sites and concomitant DNA single-strand breaks at eukaryotic genome regions engaged in active transcription. In conclusion, it is logically obvious that any chemical agent, biochemical precursor (selenium) deficiency, and/or physical circumstance compromising the sperminium hydrogen phosphate/selenite–cyclooctaoxygen sodium complexation will inevitably lead to a severe disturbation of eukaryotic genome integrity, to an increased mutation rate, and to genomic DNA single-strand breaks. This is, in part, proved by the Snyder–Robinson X-linked mental retardation syndrome [5, 81], characterized by a defect in spermine synthesis, leading to nearly complete loss of the polyamine spermine. Therefore, it was initiated a search for chemical agents selectively destroying the epigenetic shell of eukaryotic euchromatin. A candidate molecule was found, and, hence, it was defined as an ‘epigenetic poison’. The total herbicide glyphosate, N-(phosphonomethyl)glycine (ROUNDUP®, Monsanto), and its major environmental metabolite (aminomethyl)phosphonic acid (AMPA) [64] were found, rather unequivocally, to selectively destroy the cyclo-O8-Na+ complex contained in RC. Glyphosate and AMPA came into focus because (i) glyphosate represents the top-selling total herbicide worldwide [65], (ii) their chemical structure (phosphonate + amine) and properties (strongly hydrophilic and acidic) seemed to enable them to interact with cyclooctaoxygen sodium, (iii) glyphosate and ROUNDUP® are suspected to damage DNA and cause cancer in humans [82], and (iv) AMPA is already widely distributed in global ecosystems like (surface) water [83]. I allow me to conclude on basis of my findings that glyphosate, ROUNDUP® and AMPA are major examples of slow-acting, insidious ‘epigenetic poisons’, (i) slowly eroding and detoriating human, animal and plant genomic integrity, (ii) rattening human, animal and plant inborne protection of hereditary information against mutation, and (iii) disturbing the processing of human, animal and plant genetic information by transcription. It is hence inevitable for me to define glyphosate, ROUNDUP® and AMPA as a significant threat for human, animal and plant genomic stability, especially for future human generations forced to live under the glyphosate-, ROUNDUP®- and AMPA-induced radiomimetic effects.
Acknowledgments
The author is deeply indepted to T. Westfeld, E.-M. May, D. Wiegel, W. Wübbolt, O. Meier, R. Sachs, A. Karbach, W. Bergmeier, J. Moldenhauer and H.-J. Hühn (Currenta GmbH & Co. OHG, Leverkusen, Germany) for analytical services. I heartily thank H.J. Jodl for helpful discussions, E.A. Struys (University Hospital Vrije Universiteit, Amsterdam, The Netherlands) for performing the ABAT assays, and B. Cellini (University of Perugia, Department of Experimental Medicine, Italy) for conducting the AGT assays. I am obliged to K. Hecker, K. Meuser and K. Hecker (HEKAtech GmbH, Wegberg, Germany) for expert elemental analyses.
References
- van Leeuwenhoek A. Observationes D. Anthonii Lewenhoeck, de natis è semine genitali animalculis. Philos Trans R Soc Lond 12 (1677-1678): 1040-1046.
- Rosenheim O. The isolation of spermine phosphate from semen and testis. Biochem J 18 (1924): 1253-1262.
- Cooke MS, Evans MD, Dizdaroglu M, et al. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J 17 (2003): 1195-1214.
- Kirmes I, Szczurek A, Prakash K, et al. A transient ischemic environment induces reversible compaction of chromatin. Genome Biol 16 (2015): 246.
- Kesel AJ, Day CW, Montero CM, et al. A new oxygen modification cyclooctaoxygen binds to nucleic acids as sodium crown complex. Biochim Biophys Acta 1860 (2016): 785-794.
- Igarashi K, Kashiwagi K. Polyamines: mysterious modulators of cellular functions. Biochem Biophys Res Commun 271 (2000): 559-564.
- Segel GB, Cokelet GR, Lichtman MA. The measurement of lymphocyte volume: importance of reference particle deformability and counting solution tonicity. Blood 57 (1981): 894-899.
- Kuse R, Schuster S, Schübbe H, et al. Blood lymphocyte volumes and diameters in patients with chronic lymphocytic leukemia and normal controls. Blut (Berlin) 50 (1985): 243-248.
- Sipe CR, Chanana AD, Cronkite EP, et al. Studies on lymphocytes XIII. Nuclear volume measurement as a rapid approach to estimate proliferative fraction. Scand J Haematol 16 (1976): 196-201.
- National Center for Biotechnology Information (NCBI). Bos_taurus_UMD_3.1.1 (2016).
- Watanabe S, Kusama-Eguchi K, Kobayashi H, et al. Estimation of polyamine binding to macromolecules and ATP in bovine lymphocytes and rat liver. J Biol Chem 266 (1991): 20803-20809.
- National Center for Biotechnology Information (NCBI). GRCh38.p14 (2022).
- Adey A, Burton JN, Kitzman JO, et al. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature 500 (2013): 207-211.
- Macville M, Schröck E, Padilla-Nash H, et al. Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Res 59 (1999): 141-150.
- Goyns MH. Polyamine content of a non-aqueously isolated chromosome preparation. Exp Cell Res 122 (1979): 377-380.
- Jain S, Zon G, Sundaralingam M. Base only binding of spermine in the deep groove of the A-DNA octamer d(GTGTACAC). Biochemistry 28 (1989): 2360-2364.
- Egli M, Williams LD, Gao Q, et al. Structure of the pure-spermine form of Z-DNA (magnesium free) at 1-Å resolution. Biochemistry 30 (1991): 11388-11402.
- Ohishi H, Tozuka Y, Da-Yang Z, et al. The rare crystallographic structure of d(CGCGCG)2: the natural spermidine molecule bound to the minor groove of left-handed Z-DNA d(CGCGCG)2 at 10 °C. Biochem Biophys Res Commun 358 (2007): 24-28.
- Cameron IL, Smith NKR, Pool TB. Element concentration changes in mitotically active and postmitotic enterocytes. An X-ray microanalysis study. J Cell Biol 80 (1979): 444-450.
- National Center for Biotechnology Information (NCBI). C3H_HeJ_v1 (2016).
- Sarhan S, Seiler N. On the subcellular localization of the polyamines. Biol Chem Hoppe-Seyler 370 (1989): 1279-1284.
- Viola-Magni MP, Gahan PB, Pacy J. Phospholipids in plant and animal chromatin. Cell Biochem Funct 3 (1985): 71-78.
- Hurst R, Armah CN, Dainty JR, et al. Establishing optimal selenium status: results of a randomized, double-blind, placebo-controlled trial. Am J Clin Nutr 91 (2010): 923-931.
- Muecke R, Waldschock K, Schomburg L, et al. Whole blood selenium levels and selenium supplementation in patients treated in a family doctor practice in Golßen (state of Brandenburg, Germany): a laboratory study. Integr Cancer Ther 17 (2018): 1132-1136.
- Park YC, Whanger PD. Toxicity, metabolism and absorption of selenite by isolated rat hepatocytes. Toxicology 100 (1995): 151-162.
- Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci USA 106 (2009): 7507-7512.
- Crowe ML, Wang XQ, Rothnagel JA. Evidence for conservation and selection of upstream open reading frames suggests probable encoding of bioactive peptides. BMC Genomics 7 (2006): 16.
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res 12 (1984): 857-872.
- Kumler WD, Eiler JJ. The acid strength of mono and diesters of phosphoric acid. The n-alkyl esters from methyl to butyl, the esters of biological importance, and the natural guanidine phosphoric acids. J Am Chem Soc 65 (1943): 2355-2361.
- Katchalsky A, Gillis J. Theory of the potentiometric titration of polymeric acids. Recl Trav Chim Pays-Bas 68 (1949): 879-897.
- Kuhn W, Kuhn H. Die Frage nach der Aufrollung von Fadenmolekülen in strömenden Lösungen. Helv Chim Acta 26 (1943): 1394-1465.
- Mandelkern M, Elias JG, Eden D, et al. The dimensions of DNA in solution. J Mol Biol 152 (1981): 153-161.
- Mermer A, Starynowicz P. Charge-density distribution in potassium dihydrogen phosphoglycolate – a comparison of phosphate and phosphonate groups. Acta Crystallogr B 68 (2012): 625-635.
- Aikens D, Bunce S, Onasch F, et al. The interactions between nucleic acids and polyamines. II. Protonation constants and 13C-NMR chemical shift assignments of spermidine, spermine, and homologs. Biophys Chem 17 (1983): 67-74.
- Tari LW, Secco AS. Base-pair opening and spermine binding—B-DNA features displayed in the crystal structure of a gal operon fragment: implications for protein–DNA recognition. Nucleic Acids Res 23 (1995): 2065-2073.
- Wiberg N (Ed). Holleman–Wiberg, Lehrbuch der Anorganischen Chemie, 101. ed, Walter de Gruyter, Berlin, New York (1995): 771.
- Shor DSM, Struys EA, Hogema BM, et al. Development of a stable-isotope dilution assay for g-aminobutyric acid (GABA) transaminase in isolated leukocytes and evidence that GABA and b-alanine transaminases are identical. Clin Chem 47 (2001): 525-531.
- Cellini B, Montioli R, Bianconi S, et al. Construction, purification and characterization of untagged human liver alanine-glyoxylate aminotransferase expressed in Escherichia coli. Protein Pept Lett 15 (2008): 153-159.
- Oppici E, Fodor K, Paiardini A, et al. Crystal structure of the S187F variant of human liver alanine: glyoxylate aminotransferase associated with primary hyperoxaluria type I and its functional implications. Proteins 81 (2013): 1457-1465.
- Llères D, James J, Swift S, et al. Quantitative analysis of chromatin compaction in living cells using FLIM–FRET. J Cell Biol 187 (2009): 481-496.
- Rhodes CT. Determination of micro-pH in solid drug delivery systems. Drug Dev Ind Pharm 25 (1999): 1221-1222.
- Herbstein FH, Kapon M, Schwotzer W. Crystal structure of tetrakis(phenacetin) dihydrogentetraiodide dihydrate {[H5C2OC6H4N(H)C(CH3)=O] 4 • H2I4 • 2 H2O}. Helv Chim Acta 66 (1983): 35-43.
- Hitchcock PB, Hughes DL, Leigh GJ, et al. Preparation of new vanadium(II) iodides and crystal structure of hexakis(acetonitrile)vanadium(II) (tetraiodide). J Chem Soc, Dalton Trans (1994): 3683-3687.
- Genser EE, Connick RE. Exchange of iodide ion with triiodide ion studied by nuclear magnetic resonance. J Chem Phys 58 (1973): 990-996.
- Schenck H-U, Simak P, Haedicke E. Structure of polyvinylpyrrolidone-iodine (povidone-iodine). J Pharm Sci 68 (1979): 1505-1509.
- Herbstein FH, Kapon M. Zigzag chains of alternating iodine molecules and triiodide ions in crystalline (phenacetin)2 • HI5. Nature Phys Sci 239 (1972): 153-154.
- Saenger W. The structure of the blue starch–iodine complex. Naturwissenschaften 71 (1984): 31-36.
- Svensson PH, Kloo L. Synthesis, structure, and bonding in polyiodide and metal iodide–iodine systems. Chem Rev 103 (2003): 1649-1684.
- Tomlin CDS (Ed). The Pesticide Manual: A World Compendium, Incorporating the Agrochemicals Handbook, eleventh ed, British Crop Protection Council, Farnham, Surrey, UK (1997).
- Terpstra KR, Woortman AJJ, Hopman JCP. Yellow dextrins: evaluating changes in structure and colour during processing. Starch/Stärke 62 (2010): 449-457.
- Cellini B, Bertoldi M, Montioli R, et al. Human wild-type alanine:glyoxylate aminotransferase and its naturally occurring G82E variant: functional properties and physiological implications. Biochem J 408 (2007): 39-50.
- Schuster H, Schramm G, Zillig W. Die Struktur der Ribonucleinsäure aus Tabakmosaikvirus. Z Naturforsch B 11 (1956): 339-345.
- Takeda Y, Samejima K, Nagano K, et al. Determination of protonation sites in thermospermine and in some other polyamines by 15N and 13C nuclear magnetic resonance spectroscopy. Eur J Biochem 130 (1983): 383-389.
- Waddington CH. The epigenotype. Endeavour (Engl Ed London) 1 (1942): 18-20.
- Crick FHC, Watson JD. The complementary structure of deoxyribonucleic acid. Proc R Soc Lond A Math Phys Sci 223 (1954): 80-96.
- Saenger W, Hunter WN, Kennard O. DNA conformation is determined by economics in the hydration of phosphate groups. Nature 324 (1986): 385-388.
- Sunkara PS, Ramakrishna S, Nishioka K, et al. The relationship between levels and rates of synthesis of polyamines during mammalian cell cycle. Life Sci 28 (1981): 1497-1506.
- Li G, Sudlow G, Belmont AS. Interphase cell cycle dynamics of a late-replicating, heterochromatic homogeneously staining region: precise choreography of condensation/decondensation and nuclear positioning. J Cell Biol 140 (1998): 975-989.
- Flohé L, Andreesen JR, Brigelius-Flohé R, et al. Selenium, the element of the moon, in life on earth. IUBMB Life 49 (2000): 411-420.
- Berzelius JJ. Lettre de M. Berzelius à M. Berthollet sur deux Métaux nouveaux. Ann Chim Phys (Paris) 7 (1817): 199-206.
- Borah S, Kumar PP. Ab initio molecular dynamics study of Se (IV) species in aqueous environment. Phys Chem Chem Phys 18 (2016): 26755-26763.
- Nafisi S, Montazeri M, Manouchehri F. The effect of Se salts on DNA structure. J Photochem Photobiol B 113 (2012): 36-41.
- Nafisi S, Manouchehri F, Montazeri M. RNA adducts with Na2SeO4 and Na2SeO3 – Stability and structural features. J Mol Struct 1006 (2011): 547-552.
- Rueppel ML, Brightwell BB, Schaefer J, et al. Metabolism and degradation of glyphosate in soil and water. J Agric Food Chem 25 (1977): 517-528.
- Benbrook CM. Trends in glyphosate herbicide use in the United States and globally. Environ Sci Eur 28 (2016): 3.
- Holmes RP, Assimos DG. Glyoxylate synthesis, and its modulation and influence on oxalate synthesis. J Urol 160 (1998): 1617-1624.
- Helgstrand E, Eriksson B, Johansson NG, et al. Trisodium phosphonoformate, a new antiviral compound. Science 201 (1978): 819-821.
- Sabourin CLK, Reno JM, Boezi JA. Inhibition of eucaryotic DNA polymerases by phosphonoacetate and phosphonoformate. Arch Biochem Biophys 187 (1978): 96-101.
- Khan S, Ahmed S. Role of swi7H4 mutant allele of DNA polymerase a in the DNA damage checkpoint response. PLoS One 10 (2015): e0124063.
- Rosa S, Shaw P. Insights into chromatin structure and dynamics in plants. Biology 2 (2013): 1378-1410.
- Zeng H. Selenium as an essential micronutrient: roles in cell cycle and apoptosis. Molecules 14 (2009): 1263-1278.
- Frenkel GD. Effects of sodium selenite and selenate on DNA and RNA synthesis in vitro. Toxicol Lett 25 (1985): 219-223.
- Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Ann Rev Genet 20 (1986): 201-230.
- Miller CO, Skoog F, Okumura FS, et al. Isolation, structure and synthesis of kinetin, a substance promoting cell division. J Am Chem Soc 78 (1956): 1375-1380.
- Barciszewski J, Siboska GE, Pedersen BO, et al. A mechanism for the in vivo formation of N6-furfuryladenine, kinetin, as a secondary oxidative damage product of DNA. FEBS Lett 414 (1997): 457-460.
- Barciszewski J, Massino F, Clark BFC. Kinetin—a multiactive molecule. Int J Biol Macromol 40 (2007): 182-192.
- Fuller WD, Sanchez RA, Orgel LE. Studies in prebiotic synthesis. VI. Synthesis of purine nucleosides. J Mol Biol 67 (1972): 25-33.
- Maurel MC, Convert O. Chemical structure of a prebiotic analog of adenosine. Orig Life Evol Biosph 20 (1990): 43-48.
- Mustard TJL, Mack DJ, Njardarson JT, et al. Mechanism and the origins of stereospecificity in copper-catalyzed ring expansion of vinyl oxiranes: a traceless dual transition-metal-mediated process. J Am Chem Soc 135 (2013): 1471-1475.
- Ilardi EA, Njardarson JT. Ring expansions of vinyloxiranes, -thiiranes, and -aziridines: synthetic approaches, challenges, and catalytic success stories. J Org Chem 78 (2013): 9533-9540.
- de Alencastro G, McCloskey DE, Kliemann SE, et al. New SMS mutation leads to a striking reduction in spermine synthase protein function and a severe form of Snyder-Robinson X-linked recessive mental retardation syndrome. J Med Genet 45 (2008): 539-543.
- Koller VJ, Fürhacker M, Nersesyan A, et al. Cytotoxic and DNA-damaging properties of glyphosate and Roundup in human-derived buccal epithelial cells. Arch Toxicol 86 (2012): 805-813.
- Coupe RH, Kalkhoff SJ, Capel PD, et al. Fate and transport of glyphosate and aminomethylphosphonic acid in surface waters of agricultural basins. Pest Manag Sci 68 (2012): 16-30.


 Impact Factor: * 5.8
Impact Factor: * 5.8 Acceptance Rate: 71.20%
Acceptance Rate: 71.20%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 