A Genetically Modified, Enterovirus-Based Expression System in Human Induced Pluripotent Stem Cells for the Safe Study of Viral Pathomechanisms
Guiscard Seebohm1, Nathalie Strutz-Seebohm1, Jürgen Hescheler2, Stefan Peischard1*
1Institute for Genetics of Heart Diseases (IfGH), Department of Cardiovascular Medicine, University Hospital Münster, D-48149 Münster, Germany
2Institute for Neurophysiology, University of Cologne, 50931 Cologne, Germany
*Corresponding author: Stefan Peischard, Institute for Genetics of Heart Diseases (IfGH), Department of Cardiovascular Medicine, University Hospital Münster, D-48149 Münster, Germany.
Received: 23 July 2025; Accepted: 06 August 2025; Published: 22 Septemebr 2025
Article Information
Citation: Guiscard Seebohm, Nathalie Strutz- Seebohm, Jürgen Hescheler, Stefan Peischard. A Genetically Modified, Enterovirus-Based Expression System in Human Induced Pluripotent Stem Cells for the Safe Study of Viral Pathomechanisms. Archives of Clinical and Biomedical Research. 9 (2025): 364-375.
View / Download Pdf Share at FacebookAbstract
Coxsackievirus B3 (CVB3) is a major causative agent of viral myocarditis, a condition that can result in severe cardiac dysfunction and heart failure. Current research on CVB3 pathogenesis relies heavily on animal models or infectious virus systems, which are limited by biosafety requirements and translational challenges. To address these limitations, a novel human cell culture model based on induced pluripotent stem cells (hiPSCs) was developed, enabling controlled expression of viral genes without producing infectious particles. This system utilises a doxycycline-inducible Tet- On mechanism to regulate the expression of a modified CVB3 genome (CVB3ΔVP0-UTR), which lacks capsid protein functionality as well as 3´UTR and the 5´UTR, ensuring biosafety level 1 compliance. The results demonstrate that CVB3ΔVP0-UTR expression induces significant structural and functional changes in hiPSC-derived cardiomyocytes, including sarcomere disorganisation, reduced cell growth, impaired contractility, and extended contraction intervals. Furthermore, mitochondrial dysfunction was observed, characterised by fragmented mitochondrial networks, ROS accumulation, and decreased ATP production. These findings align with clinical features of CVB3-induced myocarditis and underscore mitochondrial dysregulation as a central pathological mechanism. This model provides a versatile platform for studying RNA virus pathogenesis and testing antiviral therapies. It offers significant advantages in terms of safety, controllability, and reproducibility while being adaptable to other RNA viruses. The present study underscores the potential of this model to further our understanding of viral myocarditis and to develop targeted therapeutic strategies.
Keywords
<p>CVB3; Disease model; HiPSC; Viral myocarditis</p>
Article Details
1. Introduction
RNA virus infections present a significant medical challenge, particularly due to limited therapeutic options and complex pathogenesis mechanisms [1]. Coxsackievirus B3 (CVB3) is a member of the family of enteroviruses and is one of the main viral causes of myocarditis, an inflammatory heart disease which can lead to dilated cardiomyopathy [2]. Furthermore, CVB3 infections have been linked to meningoencephalitis, pancreatitis and insulin-dependent diabetes mellitus type 1 [3,4].
The pathogenesis of CVB3-induced myocarditis involves direct cytopathic effects of the virus as well as a complex host immune response. Viral proteases such as 2A and 3C play a central role in this process by cleaving cellular structural proteins such as dystrophin, thereby impairing sarcomere structure and cell integrity [5,6]. Furthermore, CVB3 has been demonstrated to induce mitochondrial dysfunction, oxidative stress and energy metabolism disorders, which in turn lead to decreased cell viability and cardiac dysfunction [7-10].
Research on CVB3 has primarily utilised three methodological approaches: (1) the analysis of human tissue samples from patients with viral myocarditis [11], (2) the use of mouse or other animal models to study the pathogenesis of infection [10]. and (3) the in vitro infection of human cell lines with infectious virus particles [12,13]. However, it should be noted that all of these approaches have significant limitations: human samples are limited in availability and mostly represent advanced stages of disease; animal models do not fully reflect human physiology; and experiments with infectious viruses require biosafety level 2 or higher, which excludes many research laboratories from working with these viruses. Finally, infection with viable virus results in punctuated virus expression with variable virus loads in individual cells.
In order to overcome these limitations, an innovative cell culture model based on human induced pluripotent stem cells (hiPSCs) has been developed.
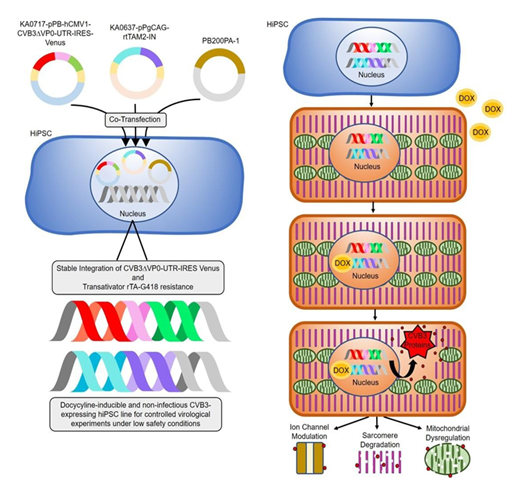
Figure 1 illustrates an innovative cell culture model for investigating CVB3-induced effects in a safe laboratory setting. On the left, the generation of transgenic hiPSCs by co-transfection of three vectors is depicted: a CVB3ΔVP0-UTR-IRES-Venus construct (KA0717-pPB-hCMV1-CVB3DVP0-UTR-IRES-Venus), an rtTA transactivator plasmid and resistance plasmid (KA0637-pPgCAG-rtTAM2-IN) [9]. Stable integration is achieved by co-transfection with the PiggyBac transposase vector (PB200PA-1). Stable integration of these components results in a controllable cell line in which viral gene expression can be induced by doxycycline application. The right figure (Figure 1, right) illustrates the functionality of the system. Transgenic hiPSC can be differentiated into several cell types e.g. hiPSC-derived cardiomyocytes. In the absence of doxycycline, viral gene expression remains inactive. However, upon doxycycline addition, the rtTA transactivator binds to the Tet-On promoter element, inducing the expression of the modified viral genome. This results in the production of viral proteins without the formation of infectious particles [9]. The expression of Venus serves as a visual marker of system activation. This model facilitates the study of viral effects on cellular structures, such as ion channels, sarcomeres and mitochondria, under biosafety level [1] conditions. The controlled expression of viral genes and the potential for differentiation into cardiomyocytes render this system a versatile platform for research into RNA virus pathogenic mechanisms and for the development of new therapeutic approaches.
Figure 1: Schematic representation of the innovative cell culture model for the controlled expression of viral genes. Left: co-transfection of hiPSCs with three plasmids (CVB3ΔVP0- UTR-IRES-Venus, rtTA transactivator and resistance plasmid, PiggyBac Transposase) for the stable integration of the modified CVB3 genome. Right: Transgenic hiPSC can be differentiated into cell types of interest e.g. cardiomyocytes. Depiction of the system's functionality. In the absence of doxycycline, viral gene expression remains inactive. However, the addition of doxycycline results in the induction of CVB3ΔVP0-UTR genome expression, thereby preventing the formation of infectious particles, while fluorescent Venus expression serves as a marker for system activation. Physiological effects of viral gene expression, including modulation of ion channels, sarcomere degradation and mitochondrial dysfunction can be studied under controlled conditions.
The regulation of viral gene expression is facilitated by a tetracycline-inducible system (Tet-On) that is activated by doxycycline [14,16]. The Tet-On system has firmly established itself as a highly effective tool for the precise control of gene expression, offering temporal and dose-dependent regulation [16]. The model offers several advantages: (1) biosafety, as no infectious particles are formed; (2) precise control of viral gene expression through doxycycline induction; (3) easy visualisation through Venus reporter expression; and (4) broad applicability for various investigations, from basic cellular mechanisms to drug testing. This approach provides a versatile platform for the study of RNA virus infections that overcomes the limitations of previous models and allows new insights into viral pathogenesis.
2. Materials and Methods
2.1 hiPSC culture
Sendai Foreskin 1 (SFS.1) human induced pluripotent stem cells were cultivated at 37°C and 5% CO2, in accordance with the protocol established by Zhang et al. [17] and Peischard et al. [18]. The SFS.1 cells were cultivated in FTDA medium (DMEM/F12) (Invitrogen #21331020) containing 5 µg/ml ITS (Becton Dickinson #354350), 0.1% human serum albumin (Biological Industries #05-720- 1B), 1X CD Lipid Concentrate (Invitrogen #1905031), 1X Penicillin/Streptomycin/Glutamine (Life Technologies #10378016), 10 ng/ml FGF2 (PeproTech #100-18B), 5 ng/ml Activin A (eBioscience #34-8993-85), 0.4 µg/ml TGFβ1 (eBioscience #34-8348-82) and 50 nM Dorsomorphin (Santa Cruz #sc-200689) [19,20]. The medium was changed on a daily basis. Once the cells had become fully confluent, they were washed with Phosphate Buffered Saline (PBS) and incubated with Accutase® (Sigma #A6964) that had been supplemented with 10 µM Y-27632 (Abcam Biochemicals #ab120129) for 10 minutes at 37°C.The enzymatic reaction was stopped by the addition of Fibroblast Growth Factor (FGF) Medium (FTDA) that had been supplemented with 10 µM Y-27632. The cells were then subjected to a centrifugation process at 200 x g for a duration of 2 minutes, after which they were resuspended in FTDA medium containing 10 µM Y- 27632. The seeding process was conducted at a cell density of 600,000 cells per well in a 6-well plate, which had been coated with a diluted Matrigel® solution (Becton Dickinson #354263) at a ratio of 1:75. The following day, the medium was changed to FTDA without Y-27632.
2.2 The generation of a transgenic coxsackievirus-inducible human iPS cell line
A three-vector system was used to generate the CVB3△VP0-UTR-inducible hiPS cell line SFS.1-CVB3△VP0-IRES-Venus. The vectors (PB200PA-1, KA0637-pPgCAG- rtTAM2-IN and KAO717-pPB-hCMV1-CVB3△VP0-IRES-Venus) were introduced into SFS.1wt cells at a ratio of 1:3:15 using FuGene6 lipofection [9]. The selection of FuGene6 was based on comparative preliminary experiments that demonstrated twice the transfection efficiency of FuGeneHD [9].
Following transfection and expression, a two-step selection process was initiated: (1) G418 selection for seven days to enrich cells containing KA0637-pPgCAG-rtTAM2-IN and (2) doxycycline-induced activation of rtTA, which triggers the expression of KAO717-pPB-hCMV1-CVB3△VP0-IRES-Venus. To mitigate potential for cytotoxic effects arising from the expression of CVB3△VP0-UTR, a protease inhibitor cocktail (Roche) was added to the culture medium. This strategy resulted in the emergence of Venus-positive colonies within three days of doxycycline treatment.
2.3 PCR for CVB3△VP0-UTR detection
PCR detection of CVB3△VPO-UTR in SFS.1 was carried out with a total reaction volume of 25 µL, comprising the following components: 1 µL template cDNA, 1.5 µL MgSO4 (25 mM), 0.5 µL KOD polymerase (1 U/µL), and 0.75 µL of primer P1491 (10 µM) (ATGCCCTCGGGATGTTCTGG), 0.75 μL of primer P1854 (10 μM) (GTCCGACTGGGACGCTTCTA), 2.5 μL dNTPs, 2.5 μL 10x KOD buffer and 15.5 μL nuclease-free water. Various colony-derived cDNAs from SFS.1-CVB3ΔVPO-UTRs (±DOX), as well as positive and negative controls, were used as templates. The PCR programme commenced with an initial denaturation step at 95°C for two minutes, followed by 40 cycles of denaturation at 95°C for 20 seconds, annealing at 63°C for 10 seconds, and extension at 70°C for 10 seconds. Subsequently, a final extension step at 70°C for one minute was performed, and the samples were stored at 4°C. For the purpose of analysis, 10 µL of the PCR product was mixed with 2 µL of 6x Loading Dye and separated by agarose gel electrophoresis. The expected fragment size was 472 bp.
2.4 qRT-PCR for virus load detection
The detection of CVB3△VPO-UTR utilising quantitative RT-PCR involved the preparation of a primer working solution with a concentration of 2.5 µM, which was achieved by combining 950 µl H2O with 25 µl of each forward and reverse primer. Two primer pairs were employed in this study: P5 (consisting of P1855 (GTGAAGGTCGGAGTCAACGG) and P1856 (TGAAGGGGTCATTGATGGCA)) and P1 (consisting of P1881 (ACGAATCCCAGTGTGTTTTGG) and P1882 (TGCTCAAAAACGGTATGGACAT)). The cDNA was used in a 1:10 dilution (20 µl cDNA + 80 µl H2O). CDNA samples from SFS.1 CVB3◻VP0-UTR, treated with different DOX concentrations (no Dox, 0.25 ng/ml, 0.5 ng/ml, 1 ng/ml, 5 ng/ml, 20 ng/ml, 50 ng/ml, 100 ng/ml, 500 ng/ml, 1 µg/ml and 2 µg/ml), were analysed. For the purpose of quality assurance, a number of controls were incorporated: firstly, a negative control (NC-cDNA) was included to check the reagents of the reverse transcription kit for contamination, secondly, a PCR negative control (NC_PCR) with water instead of cDNA was used to control the SYBR Green reagents, and thirdly, an RT control that contained all reagents of the RT kit except the reverse transcriptase. The qPCR reaction mixture was prepared as a master mix with 10 µl SYBR Green, 3 µl primer pair and 5 µl H2O, resulting in a volume of 18 µl, to which 2 µl of the respective cDNA sample was then added to yield a total volume of 20 µl per reaction.
2.5 Immunofluorescence staining and imaging
Cells were fixed with 4% PFA/PBS for 10 minutes and blocked for one hour in 2% BSA, 2% glycine and 0.2% Triton-X in PBS-T. Primary antibody against α-actinin (A7811, Sigma Aldrich) was incubated overnight at 4°C. Following this, the samples were washed with PBS-T and secondary antibody (SAB4600036, Sigma Aldrich) was applied at a ratio of 1:1000 for 1h at RT. The antibody solution was removed and cells were stained with WGA deep red (CF633 WGA, #29024-1, Biotium) in a 1:1000 ratio for 5 min. Staining solution was removed and cells were washed 3x with PBS. In the second washing step DAPI was added to the PBS in a ratio of 1:1000 and the cells were incubated for 5 min. Imaging was performed using a Leica DMI4000 confocal microscope.
2.6 MitoTracker™ CMXRos staining and analysis
Cardiomyocytes (4 weeks post-differentiation) were seeded at 100,000 cells per well on 12 mm cover slips and incubated overnight in KO-THAI medium with or without 2 µg/ml doxycycline. The cells were stained with 100 nM MitoTracker™ CMXRos for 15 min, fixed in 4% PFA/PBS and mounted for imaging. Analysis of network structure was performed using the MiNa plugin for imageJ [21,22].
2.7 The differentiation of SFS.1-CVB3△VP0-IRES-Venus into cardiomyocytes
SFS.1-CVB3△VP0-IRES-Venus were detached using Accutase® for a period of 8–10 min and treated in accordance with the aforementioned methodology. Following centrifugation, D0 differentiation medium was added, consisting of KO-DMEM (Life Technologies #10829018), 1X Penicillin/Streptomycin/Glutamine, 5 µg/ml ITS, 10 µM Y-27632, 20 ng/ ml FGF2, 1 nM CHIR-99021 (Axon Medchem #Axon1386), and 0.25 to 2.0 nM BMP-4 (R&D #314-BP-010) was used to resuspend the cells [23]. It is imperative to test the concentration of BMP-4 regularly in order to ensure optimal differentiation. The resuspended cells were seeded at 550,000 cells/24-well on Matrigel-coated wells. After 24 hours, the medium was changed, with the D0 medium being replaced with TS-ASC medium (KO-DMEM, 5.5 mg/l transferrin (Sigma #T8158- 100MG), 6.75 µg/l selenium). (Sigma #S5261-10G), 1X penicillin/streptomycin/glutamine, 250 µM ascorbate (Sigma #49,752-10G). The cells were then incubated in TS-ASC for a period of 24 hours, after which the medium was changed to TS-ASC Medium with 0.5 mM C59 (Tocris #5148). Thereafter, the cells were incubated in TS-ASC + C59 for 48 hours, with the medium being exchanged after 24 hours. Subsequently, the cells were maintained in TS-ASC medium for an additional 5 days until autonomous beating of the cells was observed [9,23]. The beating cells were then washed with phosphate-buffered saline (PBS) and incubated with TrypLE Select (1X) (Life Technologies #12563011) supplemented with 10 µM Y-27632 for 10 minutes at 37°C. Following this, the cells were isolated and subjected to a centrifugation process at 200 x g for a duration of 3 minutes. Thereafter, the cells were transferred into KO-THAI medium, comprising KO-DMEM, 1X penicillin/streptomycin/glutamine, 0.2 % human serum albumin, 250 µM ascorbate, 5 µg/ml ITS and 0.04 % (v/v) thioglycerol + 10 µM Y-27632 and seeded at a ratio of 1:5 onto 24-well plates that had been coated with 1:75 diluted Matrigel and 0.2 % gelatin in a 1:1 ratio. The following day, the medium was replaced with KO-THAI medium devoid of Y-27632.
2.8 Cardioexcyte CE96
HiPSC-derived cardiomyocytes, CVB3-induced and control cells, were seeded on 0.1% gelatin-coated NSP-96 Type Standard Stim plates (Nanion Technologies) at 30,000 cells/well in appropriate medium (KO-THAI) supplemented with 10 µM Y- 27632. After 24-hour incubation at 37°C and 5% CO2, medium was replaced without Y-27632. Cells were cultured until spontaneous contraction occurred (5 days post- seeding), then transferred to the CardioExcyte 96 system for baseline measurement of contraction via impedance for 2 hours.
2.9 Luciferase ATP-Assay
For the assessment of CVB3ΔVP0-UTR induced metabolic disruption in hiPSC cardiomyocytes, 96-well plates were prepared with a sequential coating procedure using 0.2% gelatin solution (1 hour, room temperature) followed by FBS (15 minutes, 37°C). Cardiomyocytes were thawed according to established protocols and seeded onto the pre-coated plates in a density of 30,000 cells/well. CVB3◻VP0-UTR expression was started the following day by DOX application at 2 mg/ml over-night. Subsequent luciferase reporter assays were performed using the Thermo Fisher A22066 kit following manufacturer's specifications.
3. Results
In order to investigate the effects of Coxsackievirus B3 (CVB3) gene expression in human cells, a doxycycline-inducible system was developed. This system was based on human induced pluripotent stem cells (hiPSCs) and allowed the controlled expression of a modified, non-infectious CVB3 genome (CVB3ΔVP0-UTR). The impact of this expression on cell morphology, growth, and function was then studied. The ensuing results furnish insights into the molecular and functional consequences of CVB3ΔVP0-UTR expression in hiPSCs and their differentiated cardiomyocytes.
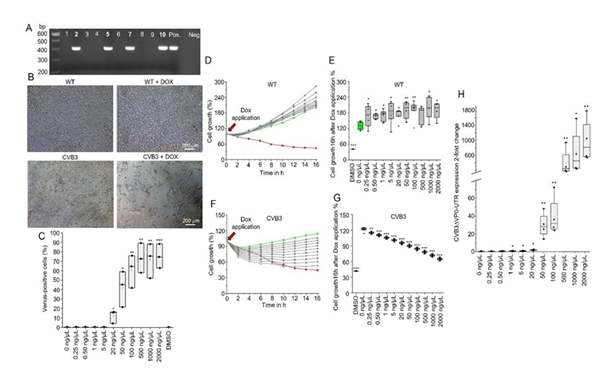
The PCR analysis (Figure 2A) confirms the successful integration of the CVB3ΔVP0- UTR construct into the hiPSCs, with specific bands (472 bp) visible in samples 2, 5, 7 and 10, indicating successful integration. In contrast, the negative control (Neg.) shows no amplification. These results confirm the stable transfection of the CVB3ΔVP0-UTR construct into the SFS.1WT cells, which forms the basis for an inducible expression system.
The microscopic images (Figure 2B) illustrate the effects of doxycycline-induced expression of CVB3ΔVP0-UTR on cell morphology and growth. SFS.1WT cells in the absence of doxycycline (DOX) and WT cells in the presence of DOX exhibit normal morphology, thereby confirming that doxycycline alone does not induce morphological changes. CVB3-transgenic cells without DOX also retain their normal morphology, demonstrating the strict regulation of the Tet-On system. In contrast, CVB3-transgenic cells with DOX manifest distinct morphological changes, including cell shrinkage and loss of integrity, indicative of the cytotoxic effects of viral protein expression.
FACS analysis (Figure 2C) demonstrates the percentage of Venus-positive cells as a function of DOX concentration, with a robust increase in the number of Venus-positive cells observed at concentrations higher than 50 µg/µL Dox (p < 0.01), thereby confirming a clear dose-dependence of the inducible system. These results are consistent with previous studies on the functionality of the Tet-On system.
Figure 2: Characterisation of the CVB3ΔVP0-UTR-inducible hiPSC system. A: Polymerase chain reaction (PCR) analysis to confirm the integration of the CVB3ΔVP0-UTR construct into the hiPSCs. The specific bands (472 base pairs (bp)) in samples 2, 5, 7 and 10 show successful integration. B: Microscopic images of cell morphology under different conditions. WT cells in the absence of doxycycline (DOX) and WT cells in the presence of DOX demonstrate normal morphology, while CVB3-transgenic cells in the absence of DOX remain unchanged and CVB3-transgenic cells in the presence of DOX show reduced cell count and density. C: FACS analysis of Venus-positive cells in relation to the DOX concentration, demonstrating that with increasing concentration, the number of Venus-positive cells increases significantly (p < 0.01). D: Cell growth analysis of SFS.1WT cells after 16 hours of DOX application. E: Effect of varying DOX concentrations on cell growth of SFS.1WT cells after 16 hours. F: Effect of varying DOX concentrations on cell growth of SFS.1 CVB3ΔVP0-UTR cells after 16 hours. G: Dose-dependent cell death of SFS.1 CVB3ΔVP0-UTR cells after 16 hours of DOX application. H: Quantitative RT-PCR for expression of the CVB3ΔVP0-UTR genome in dependence on the DOX concentration.
The cell growth analysis over a period of 16 hours (Figure 2D) demonstrates that control cells SFS.1WT (green line) exhibit continuous growth, while DMSO-treated cells (red line) demonstrate significant growth inhibition. DOX treatment with up to 2000 ng/µL Dox had no effect in cell growth in SFS.1WT. A similar observation is evident in the long-term growth analysis (Figure 2E), where control cells show no DOX induced effect after 16h of treatment. Application of DOX exhibits no significant effect at low concentrations (<5 ng/µL) exhibit no significant effect on cell growth, while higher concentrations (>20 ng/µL) significantly inhibit cell growth (p < 0.01) in SFS.1- CVB3ΔVP0-UTR (Figure 2F). This is corroborated by the dose-dependent growth inhibition in SFS.1-CVB3ΔVP0-UTR observed after 16 hours of DOX application (Figure 2G). These findings demonstrate a direct correlation between doxycycline dose, viral gene expression and cytotoxic effects. Furthermore, quantitative RT-PCR (Figure 2H) of the CVB3ΔVP0-UTR genome in SFS.1-CVB3ΔVP0-UTR demonstrates an exponential increase in gene expression with increasing DOX concentration (p < 0.01), thus confirming the effectiveness of the inducible system and the tight control of viral gene expression.
The differentiation of these cells into cardiomyocytes, along with the subsequent functional analyses, is a subject of considerable interest to study viral infection.
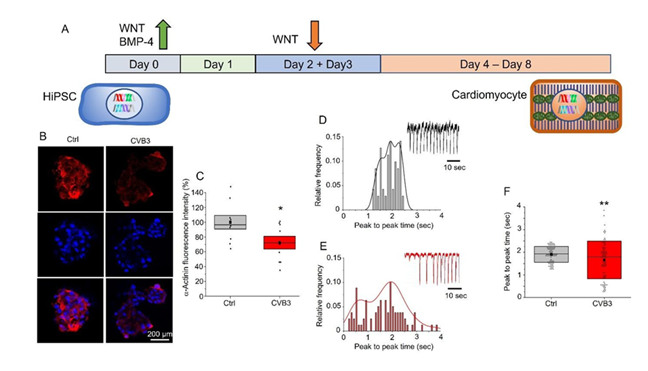
Figure 3: Functional analysis of CVB3ΔVP0-UTR-expressing cardiomyocytes. A: Schematic representation of the hiPSC-to-cardiomyocyte differentiation protocol utilising WNT and BMP-4 signalling modulators. B: Immunofluorescence images illustrating well-visible α-actinin staining in control cells (Ctrl) in comparison to a weak staining in CVB3ΔVP0-UTR -expressing cells. C: α -actinin staining intensity in a boxplot representation, demonstrating a significant reduction in CVB3-expressing cardiomyocytes (p < 0.05). D: Histogram of peak-to-peak time intervals in control cells, indicates regular contractions with a stable frequency. Regular activity is also depicted in the exemplary trace. E: Histogram for CVB3ΔVP0-UTR-expressing cardiomyocytes. Irregular contractions with a broader distribution of intervals. This can also be seen in the exemplary trace. F: Presents a boxplot that compares the peak-to-peak time intervals between control and CVB3-expressing cardiomyocytes.
The timeline protocol (Figure 3A) illustrates the differentiation process from hiPSCs to cardiomyocytes. On day 0, the differentiation process is initiated with the addition of WNT and BMP-4 signalling pathway activators, which result in mesodermal induction. Subsequently, on days 2 and 3, the WNT signalling pathway is inhibited by the WNT inhibitor C59 to promote cardiac specification. Thereafter, from day 4, the cells mature in KO-THAI medium into functional cardiomyocytes that show spontaneous contractions on day 8. This method follows established protocols for cardiac differentiation of hiPSCs and usually achieves differentiation efficiencies of about 90% [18,24].
Immunofluorescence images (Figure 3B) show an α -actinin staining in control cardiomyocytes (Ctrl) and CVB3-expressing cardiomyocytes (CVB3). A well-visible α- actinin staining is detectable in the control cells (red staining), whereas in CVB3- expressing cells, a clearly reduced α-actinin signal is evident. Quantification of this observation in control and CVB3-expressing cardiomyocytes show significant differences, which is illustrated by a boxplot (Figure 3C), whereas CVB3-expressing cells are significantly less intensely stained (p < 0.05), confirming a CVB3-induced reduction of sarcomeric structures. These observations are consistent with reports of structural damage in CVB3-induced myocarditis, in which viral proteases such as 2A cleave cellular structural proteins, thereby disrupting sarcomere organisation in mice and human [2,10].
The histograms (Figure 3D, E) and the contraction traces demonstrate the frequency of contraction behaviour in control and CVB3-expressing cardiomyocytes. While control cells exhibit regular contractions with a mean frequency of approximately 2 seconds per cycle, CVB3-expressing cells demonstrate a broader distribution of peak- to-peak time intervals, indicating impaired pace making and contraction. A comparison of the contraction intervals (Figure 3F) reveals significantly longer intervals in CVB3- expressing cells (p < 0.01), indicating a reduced contraction frequency and reduced pace maker activity, which is line with impaired pacemaker channel HCN4 activity observed in Peischard et al. [20].
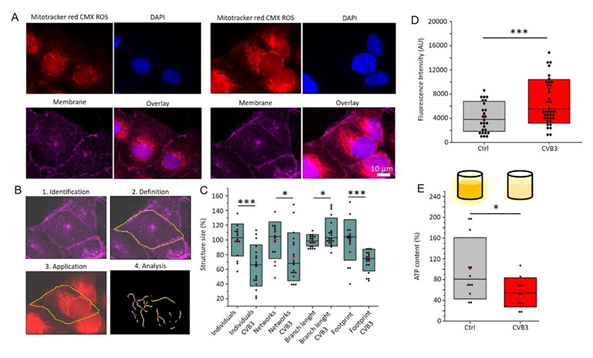
Mitochondrial networks (red) and cell nuclei (blue) in control and CVB3-expressing cardiomyocytes are revealed by MitoTracker™ and DAPI staining (Figure 4A). In control cells, well-organised, branched mitochondrial networks are visible, whereas in CVB3-expressing cells, fragmentation and disorganisation of mitochondria is evident. Overlay with membrane marker WGA deep red (magenta) shows that the mitochondrial structure in CVB3-expressing cells is severely affected. These observations are consistent with previous studies that have described mitochondrial disturbances in viral infections [10,25].
Figure 4B illustrates the workflow for the quantitative analysis of mitochondrial structure with MiNa, encompassing the identification, definition, application, and analysis of structural parameters such as size and branching. Quantitative analyses of mitochondrial structures (Figure 4C) reveals a significant reduction in network size (p < 0.001) and branch length (p < 0.05) and an increase in isolated mitochondria (p < 0.001) in CVB3-expressing cells. The results of fluorescence intensity analysis demonstrate a significant increase in the fluorescence intensity of MitoTracker™ CMXRos (Figure 4D) in CVB3-expressing cells (p < 0.001), supporting a disruption of mitochondrial membrane integrity and membrane cluster accumulation [9]. The ATP content is significantly reduced in CVB3-expressing cells compared to control cells (p < 0.05), indicating an impaired energy metabolism (Figure 4E). This observation is consistent with previous studies that have described a reduction in ATP production in mitochondrial dysfunction and viral infections.
Figure 4: Mitochondrial dysfunction in CVB3ΔVP0-UTR-expressing cardiomyocytes. A: Immunofluorescence images showing mitochondrial structures (MitoTracker™ CMXRos, red) and cell nuclei (DAPI, blue) in control (left) and CVB3-expressing cardiomyocytes (right). B: Workflow for quantitative analysis of mitochondrial morphology. Networks are identified and analysed by MiNa7ImageJ as described in the methods section. C: The results of the network analysis are presented as boxplots demonstrating a significant reduction in network size (p < 0.001) and branch length (p < 0.05), alongside an increase in isolated mitochondria (p < 0.001) in CVB3-expressing cells. D: The fluorescence intensity of MitoTracker™ CMXRos (p < 0.001) is significantly reduced in CVB3-expressing cells. E: The analysis of an ATP-luciferase assay demonstrates a statistically significant decrease in ATP content in CVB3-expressing cells compared to control cells (p < 0.05).
4. Discussion
The results of this study demonstrate that doxycycline-inducible expression of the modified non-infectious coxsackievirus B3 (CVB3ΔVP0-UTR) in human induced pluripotent stem cells and their derived cardiomyocytes induces key pathophysiological features of viral cardiomyopathies. This enables a precise investigation of the effects of viral proteins on human cells under highly controlled conditions.
4.1 The establishment of a controllable CVB3 expression system
The model presented is based on the stable integration of a modified CVB3 genome into the genome of human induced pluripotent stem cells. The CVB3ΔVP0-UTR genome has been integrated into the genome of SFS.1 cells (Figure 2A). Tet-On system has been established as an effective tool for controlled gene expression [26,27]. The results obtained demonstrate a dose-dependent induction of Venus fluorescence and expression of the CVB3ΔVP0-UTR genome subsequent to doxycycline application (Figure 2B, C, H). The observed exponential increase in Venus fluorescence and gene expression with increasing doxycycline concentration confirms the precision and efficiency of the system, as described in other studies.
In comparison with previous studies, the present model has the advantage that no infectious particles can be produced, thus allowing it to be operated under low biosafety level 1. This represents a substantial advancement, given that earlier studies on viral infections frequently necessitate stringent safety measures. Furthermore, the controlled and uniform expression of viral genes facilitates a precise temporal and dose-dependent analysis of viral effects, which is often challenging in conventional infection models [28].
The cytotoxic effects of CVB3ΔVP0-UTR expression, as demonstrated in the growth analyses (Figure 2D-G), are consistent with those reported in previous studies that described analogous effects during CVB3 infections [29,30]. The occurrence of these effects in the absence of infectious particles strongly suggests viral protein synthesis alone is sufficient to cause some severe cellular dysfunctions. This finding corroborates earlier hypotheses that viral non-structural proteins, such as proteases, can directly induce cellular damage [31,32]. The effects of these proteins on cardiomyocyte structure and function are clearly observable and can now be studied systematically in this very controlled, uniform, human model.
4.2 Analysis of cardiomyocyte physiology
The successful differentiation of the transgenic hiPSCs into functional cardiomyocytes (Figure 3A) enabled the investigation of the effects of viral gene expression on cardiac structure and function. The results obtained demonstrate a disorganisation of sarcomere structures in CVB3-expressing cardiomyocytes (Figure 3B), which is in line with previous reports of structural damage in viral myocarditis [5,33,34]. The observed reduction in α-actinin staining in CVB3-expressing cardiomyocytes (Figure 3C) could be due to impaired protein synthesis or increased protein degradation by viral proteases [35,36]. Previous studies have shown that the CVB3 protease 2A cleaves cellular proteins such as dystrophin, which leads to the impairment of the cell structure [32,37]. Furthermore, the activation of caspase signalling pathways by viral infection can lead to cell shrinkage and apoptosis [12].
The impaired contractility and prolonged repolarisation intervals observed in CVB3- expressing cardiomyocytes (Figure 3D-F) indicate an impairment of electromechanical coupling. These observations are consistent with clinical findings in patients with viral myocarditis, in whom reduced contractility and impaired electrical activity of the heart are observed. The molecular mechanisms may involve a disruption of calcium homeostasis by viral proteins, as previously described [10,30,34].
4.3 Mitochondrial dysfunction has been identified as a central pathogenic mechanism
Network analysis of mitochondrial structure (see Figure 4) demonstrates significant fragmentation of mitochondrial networks. Consistent with impaired structures a dysregulation of the membrane potential and reduced ATP production is detected in CVB3-expressing cardiomyocytes. These findings are consistent with a growing body of evidence identifying mitochondrial dysfunction as a central mechanism in viral infections and particularly in viral myocarditis [7,38,39].
The observed and quantified fragmentation of mitochondrial networks (Figure 4 A, B and C) indicates disrupted mitochondrial dynamics. Mitochondrial fusion and fission are essential processes for maintaining mitochondrial function and cell homeostasis [40,41]. Dysregulation of these processes, as observed in viral infections, can lead to mitochondrial fragmentation, dysfunction and ultimately cell damage [38].
The increased MitoTracker™ fluorescence intensity in CVB3-expressing cells (Figure 4D) indicates an altered mitochondrial membrane potential, which may be due to mitochondrial membrane hyperpolarisation, as observed in various forms of cell stress and viral infections [42,43]. Mitochondrial membrane hyperpolarisation can lead to impaired ATP synthesis and increased production of reactive oxygen species (ROS), which in turn contributes to oxidative stress and cell damage which is pathophysiologically relevant [44-46]. The reduced ATP production, verified by Luciferase ATP-Assay in CVB3-expressing cells (Figure 4E) confirms a functional impairment of the mitochondria. ATP is essential for contractile processes and cellular homeostasis in cardiomyocytes, and an energy deficiency can therefore lead to an impairment of cardiac function, as observed in viral myocarditis [16,47].
4.4 A comparison with other models and clinical relevance
In comparison with previous models, our approach offers several advantages: (1) the controlled expression of viral genes without the production of infectious particles, (2) the possibility to study specific viral proteins and their effects, and (3) the use of human cells that better represent human physiology than animal models.
Previous studies have described similar structural and functional changes in cardiomyocytes after CVB3 infection, such as sarcomere disorganisation, impaired contractility and mitochondrial dysfunction. The present model corroborates these findings and demonstrates that these effects can also occur in the absence of the production of infectious particles, thereby suggesting a direct effect of viral proteins [30,48,49]. The observed changes in CVB3-expressing cardiomyocytes reflect pathological features described in patients with viral myocarditis, such as sarcomere disorganisation, impaired contractility and mitochondrial dysfunction. This underscores the clinical significance of the model and its aptitude for investigating the pathogenesis of viral cardiomyopathies [30,50,51]. As this model is well-controllable in regard of virus expression level and duration it would be perfectly suited for antiviral drug testing approaches and drug development.
4.5 Therapeutic implications
The identification of mitochondrial dysfunction as a central pathomechanism in CVB3- induced cardiomyopathy opens up new therapeutic possibilities, with mitochondria- targeted antioxidants such as MitoQ or MitoTempo showing promise in reducing oxidative stress and improving mitochondrial function [38,52]. Indeed, a recent study demonstrated that treatment with MitoTempo can reduce viral load in chronic infections [52].
Modulators of mitochondrial dynamics may also have therapeutic potential, with compounds that promote mitochondrial fusion or inhibit fission having the capacity to reduce mitochondrial fragmentation and improve cellular energy production [38,53].
Furthermore, the inhibition of viral proteases such as 2A and 3C has been shown to reduce direct cellular damage [38,54]. Combination therapies that address both viral replication and mitochondrial dysfunction may prove to be particularly effective, as has recently been proposed for other RNA viruses [38,55].
More recent approaches, such as CRISPR/Cas13-based systems, hold considerable promise as well. These systems are capable of directly attacking and degrading viral RNA without having to target the tertiary structure of proteins [55]. The modular and customisable nature of such approaches could enable the faster development of tailored therapeutic interventions.
4.6 Outlook and future research
The model provides a versatile platform for studying viral pathogenesis and identifying potential therapeutic targets, and future studies could investigate the specific molecular mechanisms by which viral proteins cause mitochondrial dysfunction, e.g. by direct interaction with mitochondrial proteins or indirectly by modulating cellular signalling pathways.
Furthermore, the system could be used to study the role of specific viral proteins by generating targeted deletions or modifications of viral genes, which could lead to the identification of viral determinants responsible for particular pathological effects.
The integration of medium- to high-throughput screening methodologies with this model offers the potential to identify compounds that either inhibit viral protein function or enhance mitochondrial dysfunction. Consequently, this could result in the formulation of novel therapeutic strategies for viral diseases including viral myocarditis.
Finally, the system could be adapted to other RNA viruses to study their specific effects on human cells, which could lead to a better understanding of common and virus- specific pathomechanisms and enable broader therapeutic approaches.
5. Conclusion
The present study successfully establishes an inducible CVB3 expression system in human induced pluripotent stem cells and their derived cardiomyocytes. This system enables the controlled expression of viral genes without the production of infectious particles, thus providing a safe and versatile platform for studying viral pathogenesis.
The results demonstrate that the expression of CVB3ΔVP0-UTR genes causes significant structural and functional changes in human iPSC-cardiomyocytes, including sarcomere disorganisation, impaired contractility and mitochondrial dysfunction. These changes reflect pathological features observed in patients with viral myocarditis, underlining the clinical relevance of the model.
The identification of mitochondrial dysfunction as a central pathomechanism offers new therapeutic opportunities, particularly in the area of mitochondria-targeted antioxidants and modulators of mitochondrial dynamics. The model provides a versatile platform for future studies on molecular mechanisms of viral pathogenesis and for identifying potential therapeutic targets.
Funding:
The study was supported by the DFG SE1077 (Project funding number 437654872) and the Ministry of Culture and Science of the State of North Rhine- Westphalia MKW NRW (Project funding number 005-2310-0022).
Informed Consent Statement:
Not applicable.
Data Availability Statement:
The data presented in this study are available on request from the corresponding author. The presented gene array data are available in the supplementary material.
Acknowledgments:
We thank Anne Humberg and Thorsten König for the technical support. We also thank Cimin Kalali and Tabea Kretschmar for experimental support.
Conflicts of Interest:
The authors declare no conflict of interest.
References
- Mohanty P, Panda P, Acharya RK, et al. Emerging perspectives on RNA virus-mediated infections: from pathogenesis to therapeutic interventions. World J Virol 12 (2023): 242-255.
- Massilamany C, Gangaplara A, Reddy J. Intricacies of cardiac damage in coxsackievirus B3 infection: implications for therapy. Int J Cardiol 177 (2014): 330-339.
- Kanno T, Kim K, Kono K, et al. Group B coxsackievirus diabetogenic phenotype correlates with replication efficiency. J Virol 80 (2006): 5637-5643.
- Machado RS, Tavares FN, Sousa IP, Jr. Global landscape of coxsackieviruses in human health. Virus Res 344 (2024): 199367.
- Luo H, Wong J, Wong B. Protein degradation systems in viral myocarditis leading to dilated cardiomyopathy. Cardiovasc Res 85 (2010): 347-356.
- Zephyr J, Kurt Yilmaz N, Schiffer CA. Viral proteases: Structure, mechanism and inhibition. Enzymes 50 (2021): 301-333.
- Oh SJ, Lim BK, Yun J, et al. CVB3-Mediated Mitophagy Plays an Important Role in Viral Replication via Abrogation of Interferon Pathways. Front Cell Infect Microbiol 11 (2021): 704494.
- Pappritz K, Lin J, El-Shafeey M, et al. Colchicine prevents disease progression in viral myocarditis via modulating the NLRP3 inflammasome in the cardiosplenic axis. ESC Heart Fail 9 (2022): 925-941.
- Peischard, S. et al. The first versatile human iPSC-based model of ectopic virus induction allows new insights in RNA-virus disease. Sci Rep 10, 16804, doi:10.1038/s41598-020-72966-9 (2020).
- Rohrbeck, M. et al. Pathophysiological Mechanisms of Cardiac Dysfunction in Transgenic Mice with Viral Myocarditis. Cells 12, doi:10.3390/cells12040550 (2023).
- Pollack A, Kontorovich AR, Fuster V, et al. Viral myocarditis-diagnosis, treatment options, and current controversies. Nat Rev Cardiol 12 (2015): 670-680.
- Kraft L, Sauter M, Seebohm G, et al. In Vitro Model Systems of Coxsackievirus B3- Induced Myocarditis: Comparison of Commonly Used Cell Lines and Characterization of CVB3- Infected iCell((R)) Cardiomyocytes. Viruses 13 (2021).
- Mamana J, Humber GM, Espinal ER, et al. Coxsackievirus B3 infects and disrupts human induced-pluripotent stem cell derived brain-like endothelial cells. Front Cell Infect Microbiol 13 (2023): 1171275.
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline- responsive promoters. Proc Natl Acad Sci U S A 89 (1992): 5547-5551.
- Zhou X, Vink M, Klaver B, et al. Optimization of the Tet-On system for regulated gene expression through viral evolution. Gene Ther 13 (2006): 1382-1390.
- Fedorov LM, Tyrsin OY, Krenn V, et al. Tet-system for the regulation of gene expression during embryonic development. Transgenic Res 10 (2001): 247-258.
- Peischard S, Piccini I, Strutz-Seebohm N, et al. From iPSC towards cardiac tissue-a road under construction. Pflug Arch Eur J Phy 469 (2017): 1233-1243.
- Zhang M, Sebastian Schulte J, Heinick A, et al. Universal Cardiac Induction of Human Pluripotent Stem Cells in Two and Three- Dimensional Formats: Implications for In Vitro Maturation. Stem Cells 33 (2015): 1456-1469.
- Malan D, Zhang M, Stallmeyer B, et al. Human iPS cell model of type 3 long QT syndrome recapitulates drug-based phenotype correction. Basic Res Cardiol 111 (2016): 14.
- Peischard S, Möller M, Disse P, et al. Virus-induced inhibition of cardiac pacemaker channel HCN4 triggers bradycardia in human-induced stem cell system. Cell Mol Life Sci 79 (2022): 440.
- Chu CH, Tseng WW, Hsu CM, et al. Image Analysis of the Mitochondrial Network Morphology With Applications in Cancer Research. Front Phys-Lausanne 10 (2022).
- Valente AJ, Maddalena LA, Robb EL, et al. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem 119 (2017): 315-326.
- Disse P, Aymanns I, Mücher L , et al. Knockout of the Cardiac Transcription Factor NKX2-5 Results in Stem Cell- Derived Cardiac Cells with Typical Purkinje Cell-like Signal Transduction and Extracellular Matrix Formation. Int J Mol Sci 24 (2023).
- Rao J, Pfeiffer MJ, Frank S, et al. Stepwise Clearance of Repressive Roadblocks Drives Cardiac Induction in Human ESCs. Cell Stem Cell 18 (2016): 341-353.
- Peischard S, Ho HT, Theiss C, et al. A Kidnapping Story: How Coxsackievirus B3 and Its Host Cell Interact. Cellular physiology and biochemistry : International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology 53 (2019): 121-140.
- Das AT, Tenenbaum L, Berkhout B. Tet-On Systems for Doxycycline-inducible Gene Expression. Curr Gene Ther 16 (2016): 156-167.
- Ingole KD, Nagarajan N, Uhse S, et al. Tetracycline-controlled (TetON) gene expression system for the smut fungus Ustilago maydis. Front Fungal Biol 3 (2022): 1029114.
- Collins DE, Reuter JD, Rush HG, et al. Viral Vector Biosafety in Laboratory Animal Research. Comparative Med 67 (2017): 215-221.
- Tian L, Yang Y, Li C, et al. The cytotoxicity of coxsackievirus B3 is associated with a blockage of autophagic flux mediated by reduced syntaxin 17 expression. Cell Death Dis 9 (2018).
- Yang Y, Li W, You B, et al. Advances in cell death mechanisms involved in viral myocarditis. Front Cardiovasc Med 9 (2022): 968752.
- Chau DH, Yuan J, Zhang H, et al. Coxsackievirus B3 proteases 2A and 3C induce apoptotic cell death through mitochondrial injury and cleavage of eIF4GI but not DAP5/p97/NAT1. Apoptosis 12 (2007): 513-524.
- Vlok M, Solis N, Sadasivan J, et al. Identification of the proteolytic signature in CVB3-infected cells. J Virol 98 (2024): e0049824.
- Martin TG, Myers VD, Dubey P, et al. Cardiomyocyte contractile impairment in heart failure results from reduced BAG3-mediated sarcomeric protein turnover. Nat Commun 12 (2021).
- Sharma A, Marceau C, Hamaguchi R, et al. Human induced pluripotent stem cell-derived cardiomyocytes as an in vitro model for coxsackievirus B3-induced myocarditis and antiviral drug screening platform. Circ Res 115 (2014): 556-566.
- Luo H, Zhang J, Cheung C, et al. Proteasome inhibition reduces coxsackievirus B3 replication in murine cardiomyocytes. Am J Pathol 163 (2003): 381-385.
- Zhao G, Zhang HM, Qiu Y, et al. Cleavage of Desmosomal Cadherins Promotes gamma-Catenin Degradation and Benefits Wnt Signaling in Coxsackievirus B3-Induced Destruction of Cardiomyocytes. Front Microbiol 11 (2020): 767.
- Lim BK, Peter AK, Xiong D, et al. Inhibition of Coxsackievirus-associated dystrophin cleavage prevents cardiomyopathy. J Clin Invest 123 (2013): 5146-5151.
- Duan X, Liu R, Lan W, et al. The Essential Role of Mitochondrial Dynamics in Viral Infections. Int J Mol Sci 26 (2025).
- Oh SJ, Lim BK, Yun J, et al. Corrigendum: CVB3-Mediated Mitophagy Plays an Important Role in Viral Replication via Abrogation of Interferon Pathways. Front Cell Infect Microbiol 11 (2021): 757341.
- Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science 337 (2012): 1062-1065.
- Yu SB, Pekkurnaz G. Mechanisms Orchestrating Mitochondrial Dynamics for Energy Homeostasis. Journal of molecular biology 430 (2018): 3922-3941.
- Shang C, Liu Z, Zhu Y, et al. SARS-CoV-2 Causes Mitochondrial Dysfunction and Mitophagy Impairment. Front Microbiol 12 (2021): 780768.
- Xiao B, Deng X, Zhou W, et al. Flow Cytometry-Based Assessment of Mitophagy Using MitoTracker. Front Cell Neurosci 10 (2016): 76.
- Joshi DC, Bakowska JC. Determination of mitochondrial membrane potential and reactive oxygen species in live rat cortical neurons. J Vis Exp (2011).
- Park J, Lee J, Choi C. Mitochondrial network determines intracellular ROS dynamics and sensitivity to oxidative stress through switching inter-mitochondrial messengers. Plos One 6 (2011): e23211.
- Suski JM, Lebiedzinska M, Bonora M, et al. Relation between mitochondrial membrane potential and ROS formation. Methods Mol Biol 810 (2012): 183-205.
- Wang X, Zhang X, Wu D, et al. Mitochondrial flashes regulate ATP homeostasis in the heart. Elife 6 (2017).
- Yip F, Lai B, Yang D. Role of Coxsackievirus B3-Induced Immune Responses in the Transition from Myocarditis to Dilated Cardiomyopathy and Heart Failure. Int J Mol Sci 24 (2023).
- Zhang HM, Ye X, Su Y, et al. Coxsackievirus B3 infection activates the unfolded protein response and induces apoptosis through downregulation of p58IPK and activation of CHOP and SREBP1. J Virol 84 (2010): 8446-8459.
- Westermann D, Savvatis K, Lindner D, et al. Reduced degradation of the chemokine MCP-3 by matrix metalloproteinase-2 exacerbates myocardial inflammation in experimental viral cardiomyopathy. Circulation 124 (2011): 2082-2093.
- Yang D, Yu J, Luo Z, et al. Viral myocarditis: identification of five differentially expressed genes in coxsackievirus B3-infected mouse heart. Circ Res 84 (1999): 704-712.
- Mischke J, Klein S, Cornberg M, et al. MitoTempo treatment as an approach to cure persistent viral infections? Virology 600 (2024): 110280.
- Stasiak K, Dunowska M, Trewick S, et al. Genetic Variation in the Glycoprotein B Sequence of Equid Herpesvirus 5 among Horses of Various Breeds at Polish National Studs. Pathogens 10 (2021).
- Badorff C, Knowlton KU. Dystrophin disruption in enterovirus-induced myocarditis and dilated cardiomyopathy: from bench to bedside. Med Microbiol Immunol 193 (2004): 121-126.
- Tabata E, Kobayashi I, Morikawa T, et al. Evolutionary activation of acidic chitinase in herbivores through the H128R mutation in ruminant livestock. iScience 26 (2023): 107254.


 Impact Factor: * 5.8
Impact Factor: * 5.8 Acceptance Rate: 71.20%
Acceptance Rate: 71.20%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 