Hair - A Good Source of DNA to Solve the Crime
Vaishali B. Mahajan1*, Vijaya Padale2, Deepak Y. Kudekar3, Bhausaheb P. More4, Krishna V. Kulkarni5
1Regional Forensic Science Laboratory, Government of Maharashtra, Home Department, Nashik, Maharashtra, India.
2Directorate of Forensic Science Laboratories, Government of Maharashtra, Home Department, Kalina, Santacruz East, Maharashtra, India.
*Corresponding Author: Vaishali B. Mahajan, Regional Forensic Science Laboratory, Government of Maharashtra, Home Department, Dindori Road, Nashik - 440002, Maharashtra, India
Received: 18 July 2019; Accepted: 01 August 2019; Published: 05 August 2019
Article Information
Citation: Vaishali B. Mahajan, Vijaya Padale, Deepak Y. Kudekar, Bhausaheb P. More, Krishna V. Kulkarni. Hair - A Good Source of DNA to Solve the Crime. Archives of Clinical and Biomedical Research 3 (2019): 287-295.
View / Download Pdf Share at FacebookAbstract
Most often hairs are picked up at the crime scenes and are used as contributing biological evidences in the crime cases. This can be helpful in determining the perpetrators of the crime and providing more information about what happened during the actual incidence. Hairs are commonly encountered in the forensic investigations and can be the good source of DNA. The optimal quantity of template DNA required for PCR based STR Profiling is about 1 ng and sufficient quantity may be present in the root sheath to prove the crime. In the present case, hairs were found in the fist of the deceased and they proved to be an important evidence that linked to the murderer. DNA was extracted from hair roots and profile was generated using PCR based STR Genotyping technology. The DNA profile obtained from the hair root matched with the DNA profile obtained from reference blood of the accused and proved his involvement in the crime.
Keywords
<p>Polymerase Chain Reaction; DNA; Short Tandem Repeat; Genotype; Allele</p>
Article Details
Abbreviations
DNA: Deoxy Ribonucleic Acid
PCR: Polymerase Chain Reaction
STR: Short Tandem Repeat
Introduction
When biological material is transferred between a perpetrator and a victim in violent crimes such as murder or rape, the DNA recovered from the crime scene as well as the biological material recovered from the bodies of the victim or accused has the power to potentially identify the perpetrator. Use of sufficient genetic markers enables individualization of the collected samples. The best identification including a high power of discrimination and rapid analysis speed has been achieved with STR (short tandem repeat) DNA markers. STR DNA profiling technology has emerged as the most accurate and trusted scientific method to prove the crime and to get justice for the victims. During analysis of sensitive cases such as murder, rape, theft or burglary, the presence of any biological evidence having a source of DNA helps the investigators to confirm the presence of the perpetrator at the crime scene.
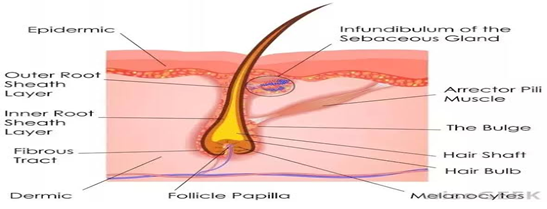
In the crime investigation, hairs are commonly encountered and have proven as an important source of DNA which is helpful in establishing the scope of the crime [1]. Some of the hairs include the hair bulb which is composed of living cells that generate the keratin shaft. The bulb cells contain DNA (Figure 1). This DNA can help connecting the suspect to the weapon and the crime scene. Also, it provides a strong evidence if found over the body of the deceased or the accused. There is major role of crime scene investigator to check the crime scene thoroughly and collect all the evidences that can be used to solve the crime. Medical officers conducting postmortem also have important role of collecting all evidences over or inside the body of the deceased while performing the autopsy. Proper collection, preservation and timely submission of evidences are of utmost important while solving any crime case. Failure to comply with any of these parameters may cause the loss of important evidence and degradation of samples. Moreover, it might pose challenges while proving the case in the court. DNA profiling of the collected evidences provides the gender and genetic information of an individual. The study of hair morphology provides information as to whether it has been cut or pulled out in a certain way. The amount of DNA within a single hair is often less than the optimal amount required for nuclear DNA markers. Most of the DNA is within a root sheath i.e. bulb with trace amounts of mitochondrial DNA in the shaft. PCR based STR genotyping using nuclear DNA targets highly repetitive DNA regions called microsatellites [2]. The quantity of starting template required for nuclear DNA profiling is 1 ng which may be present in the root sheath but not within the hair shaft. Mitochondrial loci are the preferred targets for hair shafts due to their higher copy number [3]. Analysis of DNA obtained from intact hair roots is now routinely performed in the area of forensic science [4]. Short tandem repeat (STR) analysis is regularly performed for generating DNA profiles from blood, saliva and hair samples encountered as evidence to solve the crime [5-8]. Traditional workflow for generating DNA profiles includes below steps:
- Extraction of DNA from body samples found on different substrates
- Quantification of extracted DNA
- Amplification of DNA using polymerase chain reaction (PCR) based STR reactions
- Denaturation of amplified DNA
- Genotyping using short tandem repeat (STR) technique
- Analysis and comparison of generated DNA profiles.
Now-a-days, advanced instruments such as AutoMate Express and EZ1 Advanced which work on robotic principle are helpful to minimize the analysis time. Commercially available kits for amplification of DNA allow the faster turn-around time [9, 10].
In the case received in our laboratory (Forensic Science Laboratory, Nashik, Maharashtra), 6 to 7 hair strands were collected from the fist of the deceased male. Our laboratory is routinely performing nuclear DNA profiling on the hair roots. In this case, only two hairs were found with the hair bulbs which were used for DNA profiling and connected the involvement of the female accused in the crime.
Materials and Methods
PrepFiler Express DNA extraction kit. Lot No. 1807201.
AmpFlSTR® Identifiler kit. Lot No. 1807261
HiDi Formamide.
Liz 600 Size standard.
Quantifiler Duo DNA kit. Lot No. 1710101.
AutoMate ExpressTM Forensic DNA Extraction System. Catlog number: 4441763
PCR thermal cycler GeneAmp 9700. Catlog number: 4375786
3500 Genetic Analyzer. Catlog number: 4406017
- Isolation of DNA
- Among the 6 to 7 hair strands, only two hairs were having the hair bulbs. Those bulbs were placed in 2ml micro centrifuge tube after cutting 5 mm from the base of the strand.
- For the reference profile, 40 µl blood of the accused woman was taken into another micro centrifuge tube.
- 500 µl Lysis buffer from PrepFiler Express F DNA extraction kit [11] was added to both the sample tubes.
- The sample tubes were kept on thermo shaker at 750 rpm at 70?C for 40 min.
- The tubes were then centrifuged at 10,000 rpm for 2 min.
- Cartridges from PrepFiler Express F DNA extraction kit were loaded to the cartridge rack in AutoMate Express DNA extraction system [12],. Sample tubes, elution tubes and tips were loaded as per machine guidelines and the machine program was run as per the recommended machine protocol (Table 1).
- After completion of program, elution tubes containing extracted DNA in highly pure form was stored at 4?C till the next PCR amplification process.
A wide variety of methods based on different principles are available for extraction of DNA from the forensic samples. The phenol/chloroform extraction is one of the sensitive methods for recovery of DNA from wide variety of forensic samples. However, this process requires usage of large volumes of toxic phenol and chloroform solutions. Also it is tedious and time consuming [13]. The PrepFilerTM Forensic DNA extraction Kit (Applied Biosystems, Foster City, CA) enables the isolation of DNA from biological samples that contain small quantities of biological material in such a way that it removes the substances interfering with PCR. Additionally, the extracted DNA is having sufficiently high concentration due to which the volume of extract for downstream analysis is minimal [14].
|
For use with (Equipment) |
Automate Express Forensic DNA Extraction System |
|
Capacity |
1 to 13 samples per run |
|
Dimensions |
50.8 cm (20 inch)(wrapped in paper) x 55.9 cm (22 inches) (D) x 57.2 cm (22.5 inch)(H) |
|
Input power |
240VA, 50/60 Hz Grounding is necessary, Required input power AC 100-240 V |
|
Kits designed for this instrument |
PrepFiler Express and PrepFiler Express BTA |
|
Pipetting range |
20-250 µl |
|
Program Features |
Protocol card pre-programmed for purifying nucleic acids from forensic samples |
|
Reaction time |
30 min (up to 13 samples) |
|
Weight |
55kg (121 pounds) |
Table 1: AutoMate Express Forensic DNA Extraction System Parameters
Quantification of the extracted DNADNA Extracted from both the hair and reference samples was quantified using Quantifiler® Duo DNA Quantification Kit [15,16] on an Applied Biosystems 7500 Real-Time PCR System according to manufacturer recommended protocols. Quantified DNA was taken for downstream application.
PCR based STR AnalysisThe quantified DNA extracted from hair roots and reference blood sample of the accused was processed for Polymerase Chain Reaction using the AmpFlSTR® Identifiler PCR Amplification Kit (Applied Biosystems) (Lot No. 1807261) [17] with the help of PCR thermal cycler GeneAmp 9700 [18] following the protocols recommended by the manufacturer and described in the studies. This kit contains Reaction mixture, Primer set and Taq Gold Polymerase enzyme. Primer Set contains locus-specific 6-FAMTM, VICTM, NEDTM and PETTM dye-labeled and unlabeled primers in the buffer. The primers amplify the STR loci CSF1PO, D2S1338, D3S1358, D5S818, D7S820, D8S1179, D13S317, D16S539, D18S51, D19S433, D21S11, FGA, TH01, TPOX, vWA and gender marker Amelogenin.
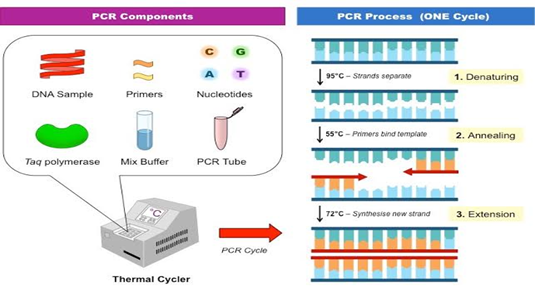
Reaction mixture used for PCR was prepared by adding Reaction mix 10.5 µl, Primer set 5.5 µl and Taq Gold DNA Polymerase 0.55 µl. The extracted DNA sample 10 µl was added to it. The DNA was amplified in 28 cycles using PCR machine selecting 94.0 ºC, 59.0 ºC and 72.0 ºC as temperatures of denaturing, annealing and extension respectively (Table 2) (Figure 2).
The amplified DNA samples were kept at 60.0 ºC for an hour and then at 4.0 ºC till the separation of STRs (Table 2). PCR produces millions of DNA fragments of different sizes. Amplified products were separated and detected using 3500 Genetic Analyzer [19] and analyzed using GeneMapper® ID-X Software V 1.5. The separation of different fragments of DNA molecules on the basis of their sizes was achieved by capillary electrophoresis. Simultaneous amplification of 16 STR Loci was completed and analyzed [20, 21]. DNA profiles obtained were interpreted by comparing with each other.
|
Step |
AmpliTaq Gold Enzyme Activation |
PCR |
PCR Final Step |
PCR product till separation of STRs |
||
|
Hold |
CYCLE (28 cycles) |
Hold |
||||
|
Denaturation |
Anneal |
Extend |
||||
|
Temperature |
95 ?C |
94 ?C |
59 ?C |
72 ?C |
60 ?C |
4 ?C |
|
Time |
11 min |
1 min |
1 min |
1 min |
60 min |
∞ |
Table 2: PCR Protocol used for amplification of DNA
Figure 2: Process of Polymerase Chain Reaction
Results
The DNA extracted from roots of the hairs found in fist of the deceased and reference blood sample of the accused woman was typed for 15 STR Loci and gender specific Amelogenin locus using PCR Amplification technique [22]. Table 3 shows comparative study of the DNA profiles obtained after genotyping of both samples. It shows that the DNA profile obtained from hair roots is of female origin and matched with the DNA profile obtained from reference blood sample of the accused woman.
Discussion
In 1984, Dr. Alec Jeffreys developed DNA fingerprinting technique used to identify the individuals. He found that certain regions of DNA contained repeated DNA sequences. The regions with repeat units that are 2-6 base pair in length are called Short Tandem Repeats. The first STR multiplexes developed was quadraplex created by Forensic Science Services (FSS) that comprises four STR Loci. Now-a-days, there are different multi locus commercial kits available and used by researchers to solve the crimes so as to give justice to the society.
In this case, the police received a call from a villager that an unknown male of about 70 years was found dead under the bridge of a river nearby village of Nandurbar District, Maharashtra. He had sustained head injuries and seemed to be murdered by someone. After observation of crime scene, police registered crime under the section of Indian Penal Code 302. From the mobile found in the pocket of deceased, police called his daughter. She told the police that her father who lived in Gujrat had gone to collect money from a female aged 40 who was residing in the village of Nandurbar district of Maharashtra. They had an illegal relationship. She convinced him to come to her place leaving Gujrat and invest his money in her farm as there was abundant water. He trusted her and gave about 2 lakhs for farming and also supported her relatives by giving loans to fulfill their needs. But later on, when he didn’t gain any profit in farming he asked her to return back his money. Instead of returning the money, she killed him on the bridge of river by hitting a pestle on his head and threw his body under the bridge with the help of her relative. During interrogation, the police asked her about the victim however she misguided them by saying that the victim had gone back to Gujrat. While performing autopsy, the doctors found a bunch of 6 to 7 long hairs in the victim’s fist. Morphologically the hair appeared of the female origin. DNA profiling of the two hair bulbs detected along with reference blood of the accused woman gave the DNA Genotyping Profiles which upon comparison concluded that both the DNA profiles are similar. Use of AmpFlstr® Identifiler Kit helped to get the nuclear DNA profile from trace sample. Thus the DNA profiling proved to be a strong evidence against the accused to get justice for the deceased and his relatives.
Conclusions
When the accused strongly denies the crime and only trace evidence is obtained to solve the mystery, it is skill of the analyst to get the DNA profile from the available biological sample without wasting it. Further, if the sample is received to the laboratory in proper condition, it becomes somewhat easy to perform the analysis. This is one of the most helpful techniques to solve the complicated heinous crimes.
|
STR LOCUS |
GENOTYPE |
|
|
Hair roots (from the fist of deceased) |
Reference Blood (Accused woman) |
|
|
D8S1179 |
10,14 |
10,14 |
|
D21S11 |
30,33.2 |
30,33.2 |
|
D7S820 |
8,11 |
8,11 |
|
CSF1PO |
10,10 |
10,10 |
|
D3S1358 |
15,18 |
15,18 |
|
THO1 |
6,9 |
6,9 |
|
D13S317 |
12,12 |
12,12 |
|
D16S539 |
9,11 |
9,11 |
|
D2S1338 |
19,19 |
19,19 |
|
D19S433 |
12,12 |
12,12 |
|
vWA |
18,19 |
18,19 |
|
TPOX |
8,8 |
8,8 |
|
D18S51 |
12,15 |
12,15 |
|
AMELOGENIN |
X,X |
X,X |
|
D5S818 |
10,11 |
10,11 |
|
FGA |
21,23 |
21,23 |
Table 3: The results of DNA typing
Acknowledgement
We are thankful to the Director General, Legal and Technical, Mantralaya, Mumbai, Home Department, Government of Maharashtra, India for his guidance and encouragement all the time extended to us.
References
- Lanning KA, Michaud AL, Bisbing RE, Springer FA, and Tridico SR. Scientific Working Group on Materials Analysis Position on Hair Evidence. Journal of Forensic Sciences 54 (2009): 1198–1202.
- Fischer H, Szabo S, Scherz J, Jaeger K, Rossiter H, et al. Essential Role of the Keratinocyte-Specific Endonuclease DNase1L2 in the Removal of Nuclear DNA from Hair and Nails. Journal Of Investigative Dermatology 131 (2011): 1208-1215.
- Wilson, D Polanskey J, Butler, JA DiZinno, J. Replogle, et al. Extraction, PCR amplification and sequencing of mitochondria DNA from human hair shafts, BioTechniques 18 (1995): 662-669.
- Szabo S, Jaeger K, Fischer H, Tschachler E, Parson W, et al. In situ labeling of DNA reveals interindividual variation in nuclear DNA breakdown in hair and may be useful to predict success of forensic genotyping of hair. International Journal of Legal Medicine 126 (2012): 63-70.
- Moretti TR, Baumstark AI, Defenbaugh DA, Keys KM, Budowle B, et al. Validation of STR typing by capillary electrophoresis. J Forensic Sci 46 (2001): 661-676.
- Moretti TR, Baumstark AI, Defenbaugh DA, Keys KM, Budowle B, et al. Validation of (STRs) for forensic usage: performance testing of fluorescent multiplex STR systems and analysis of authentic and simulated forensic samples. J Forensic Sci 46 (2001): 647-660.
- Krenke BE, Tereba A, Anderson SJ, Buel E, Culhane S, et al. Validation of a 16-locus fluorescent multiplex system. J Forensic Sci 47 (2002): 773-785.
- Collins PJ, Hennessy LK, Leibelt CS, Roby RK, Reder DJ. Et al. Developmental validation of a single-tube amplification of the 13 CODIS STR loci, D2S1338, D19S433, and amelogenin: the AmpFlSTR Identifiler PCR amplification kit. J. Forensic Sci 49 (2004): 1265-1277.
- Wang D, Chang C, Oldroyd N, Hennessy I. Direct amplification of STRs from blood or buccal cell samples. Forensic Sci Int 2 (2009): 113-114.
- Wang D, Chang C, Lagace R, Calandrol I, Hennessy I. Developmental validation of the AmpFlSTR Identifiler Plus PCR Amplification Kit: an established multiplex assay with improved performance. J Forensic Sci 57 (2012): 453-465.
- Brevnov MG, Pawar HS, Mundt J, Calandro LM, Furtado MR, Shewale JG. Developmental validation of the PrepFiler Forensic DNA Extraction Kit for extraction of genomic DNA from biological samples. J Forensic Sci 54 (2009): 599-607.
- Jason Y. Liu, Chang Zhong, Allison Holt, Robert Lagace, Michael Harrold, et al. AutoMate ExpressTM Forensic DNA Extraction System for the Extraction of Genomic DNA from Biological Samples. J Forensic Sci 57 (2012): 1556-4029.
- Kochl S, Niederstatter H, Parson W. DNA extraction and quantitation of forensic samples using the phenol-chloroform method and real-time PCR. Methods Mol Biol 297 (2005): 13-30.
- Brevnov MG, Mundt J, |Benfield J, Treat-Clemons Labelled, Kalusche G, Meredith J, et al. Automated Forensic DNA Extraction Kit. JALA 14 (2009): 294-302.
- Applied Biosystems Quantifiler user’s manual. Foster City, CA. (2005).
- Barbisin M, Fang R, O’Shea CE, Calandro LM, Furtado MR, et al. Developmental validation of the Quantifiler Duo DNA Quantification kit for simultaneous quantification of total human and human male DNA and detection of PCR inhibitors in biological samples. J Forensic Sci 54 (2009): 305-319.
- Applied Biosystems. AmpFlSTR® Identifiler® PCR amplification kit user’s manual, part # 4323291 Rev. B. Foster City, CA: Applied Biosystems, 2001.
- Mullis K, Faloona F, Scharf S, Saiki R, Horn G, et.al. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold spring Harb Syma Quant Biol 51 (1986): 263-273.
- Applied Biosystems 3500/3500XL Genetic Analyzer User Guide Applied Biosystems, Faster City CA.
- Budowle B, Allen RC. Analysis of amplified fragment length polymorphism (VNTR/STR Loci) for human identity testing. Methods mol Biol 98 (1998): 155-171.
- Gill P, Kimpton CP, Urquhart A, Oldrod N, Millican ES, et.al. Automated short tandem repeat (STR) Analysis in forensic casework-a strategy for the future. Electrophoresis 16 (1995): 1543-1552.
- Hall D, Roy R. An evaluation of direct PCR amplification Croat Med J 55 (2014): 655-661.


 Impact Factor: * 5.8
Impact Factor: * 5.8 Acceptance Rate: 71.20%
Acceptance Rate: 71.20%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 