History, Structure, Epidemiology and Molecular Typing of Staphylococcal Cassette Chromosomes Mec (Sccmec) involved in Multiple Resistances to Beta-Lactams in the Genus Staphylococcus: an Overview
Ouédraogo Ganamé Abasse1*, Kaboré Boukaré1, Cissé Hama1, Zongo Oumarou1, Ouédraogo Henri Sidabéwindin1, Bassolé Imael Henri Nestor3, Traoré Yves1, Tchoumbougnang François2, Savadogo Aly1
1Laboratory of Applied Biochemistry and Immunology, University Joseph Ki-ZERBO, 03 BP 7021 Ouagadougou 03, Burkina Faso
2Institute of fisheries and aquatic Sciences at Yabassi, University de Douala, Cameroon
3Laboratory of Epidemiology and Surveillance of Foodborne Diseases, University Joseph Ki-ZERBO, 03 BP 7021 Ouagadougou 03, Burkina Faso
*Corresponding author: Ouédraogo Ganamé Abasse, Laboratory of Applied Biochemistry and Immunology, University Joseph Ki-ZERBO, 03 BP 7021 Ouagadougou 03, Burkina Faso.
Received: 16 August 2022; Accepted: 26 August 2022; Published: 14 September 2022
Article Information
Citation: Ouédraogo Ganamé Abasse, Kaboré Boukaré, Cissé Hama, Zongo Oumarou, Ouédraogo Sidabéwindin Henri, Bassolé Imael Henri Nestor, Traoré Yves, Tchoumbougnang François, Savadogo Aly. History, Structure, Epidemiology and Molecular Typing of Staphylococcal Cassette Chromosomes Mec (Sccmec) involved in Multiple Resistances to Beta-Lactams in the Genus Staphylococcus: an Overview. Archives of Clinical and Biomedical Research 6 (2022): 809-827.
View / Download Pdf Share at FacebookAbstract
Staphylococcal cassette chromosomes mec (SCCmec) are chromosomal mobile genetic determinants that confer to the Staphylococcus strain hosting a multi-resistance to antibiotics and metallic trace elements. The objective of this study was to review the literature on the history, structure, epidemiology and characterization techniques of SCCmec. Data were collected from articles in scientific journals through Google scholar and PubMed engines. In 1999, was characterized the first SCCmec type on genome of S. aureus N315. A type of SCCmec is characterized by the complex type ccr and the class complex mec. The first classification took place in 2001. Today, there are 15 recognized SCCmec types. SCCmec type IV is the most disseminated and predominates on the continents. The application of PCR and Illimina or MinION sequencing have revolutionized SCCmec typing, which is one of the main methods of epidemiological surveillance of methicillin resistance Staphylococcus aureus. MRSA are the most threatening bacteria in the hospital environment and cause enormous problems in community settings and in animal health. SCCmec typing is a very effective means of characterizing the genetic determinants of resistance in MRSA. Popularization of SCCmec typing through their applications in routines
Keywords
<p>Antibiotics; Methicillin Resistance; Staphylococcus; SCCmec; Typing</p>
Article Details
1. Introduction
Staphylococci are ubiquitous bacteria that represent one of the major threats in bacterial infections in both human and animal health [1-3]. The species Staphylococcus aureus (S. aureus) is the second cause of bacterial human pathologies after Escherichia coli [4,5]. In addition to their high prevalence across investigations around the world, Staphylococcus aureus has been closely linked to the history of control and epidemiology of bacterial resistance to antibiotics. In this effect, the first antibiotic (Penicillin G) was discovered in the 1940s, the deductions of which began following the accidental contamination by Penicillium notatum on a culture of S. aureus carried out by Sir Alexander Fleming Scottish biologist on September 3, 1928 [5]. But the first bacterial resistance to Penicillin G was officially reported in 1942 in a strain of S. aureus which showed sensitivities to penicillinase produced by Penicillium notatum [6]. Thus a molecule of semi-synthesized antibiotic <<methicillin>> insensitive to the action of penicillinase was set up in 1959 to remedy this phenomenon of resistance. But, immediately after a year and a few months (in 1961) in England, the first bacterial resistance to methicillin was reported in a strain of S. aureus [7]. Methicillin-resistant (MRSA) strains of S. aureus are also resistant to almost all beta-lactams [8]. Indeed, resistance to methicillin in Staphylococcus aureus is caused by the modification of penicillin-binding proteins (PLP) possessing an enzymatic activity (trans-peptidases, carboxypeptidases or glycosyltransferases) involved in the synthesis of the bacterial wall and possessing an affinity for beta-lactams [10,11]. These modified PLPs bear the name of PLP2a, which has very little affinity with beta-lactams. The work performed allowed the complete sequencing of the genetic determinant of PLP2a (the mec gene). Today, there are 4 types (A, B, C, and D) of known guy genes. Among these 4 types of mec gene, 3 types of the 4 (A, B and C) have been characterized in S. aureus and type D has been reported in Macrococcus caseolyticus [11]. The mec genes in S. aureus are mostly chromosomal and integrate into the bacterial genome to express themselves through the coordinated action of a mobile genetic complex called staphylococcal cassette chromosome mec (SCCmec) with a molecular weight ranging from 21 to 67 kb [12]. A staphylococcal cassette chromosome would be the genetic unit for the transfer and expression of virulence and/or resistance genes in the genus Staphylococcus [14,15]. SCCmec types confer resistance to Staphylococcus strains against beta-lactam residues [15]. It was in 1999 that the first SCCmec was described in the strain of S. aureus N315 but it was in 2001 that their classification was initiated [10]. After the description of the first SCCmecs in patients in hospital settings, subsequent investigations reported that MRSA carriers of SCCmecs were equipped with genes on their SCCmecs that allowed them to adapt and proliferate also in community settings and in animals. Epidemiologically, a diversity of SCCmec has been reported in strains of S. aureus in Europe, America, Africa and Asia (Table 2) [17,18]. The proliferation of SCCmecs and their roles in multi-antibiotic resistance in MRSA have become public health issues being addressed by top global health officials. Thus, an international structure in charge of the follow-up in the world of SCCmec called <<the International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements>> (IWG-SCC) was created. Since 2009, the IWG-SCC has shown the guidelines to follow for the nomenclature of types and subtypes of SCCmec [18]. Today, the IWG-SCC has registered 15 types of SCCmec, of which the last SCCmec was type XV (7A) and was characterized in strain NV_1 by [19]. This study aims to conduct an overview through retrospective data from official publications on Staphylococcal Cassette Chromosome mec implicated in multi-resistance to beta-lactams in Staphylococcus. Specifically to: (i) provide information on the history and structuring of SCCmec types (ii) detail on the structural functioning and epidemiology of SCCmec (iii) list the application of technology for the characterization of molecular weight of SCCmec.
2. Methodology
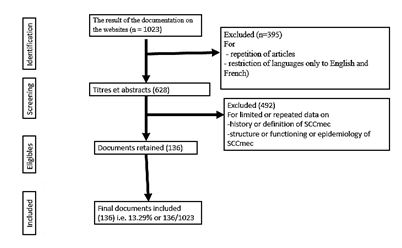
Relevant documents on the subject were searched on websites through Google scholar and Pub Med. The documentation lasted between September 2021 to February 2022. The expressions and keywords that were used during this documentary research were only related to the specific objectives. These words and expressions concerned ''the work of Sir Alexander Fleming'' for the discovery of the first antibiotic, ''definition and history and resistance to methicillin'', ''expression of the mecA gene and the resistance of MRSA to the metallic traces elements and other families of antibiotics other than beta-lactams'' for the operation of the SCCmec cassette, ''definition and types of Staphylococcal cassette chromosome mec'' for a general view of SCCmec and ''the prevalence of MRSA and SCCmec in continents and by country'' of which for 87 countries were surveyed and more than half were excluded with reasons listed in Figure 1.
3. Results and Discussion
History of Drug Resistance and Early Descriptions of Sccmec Types
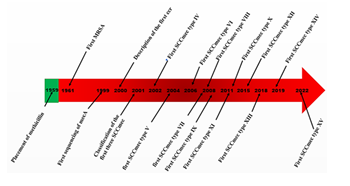
Methicillin was the first semi-synthesized antibiotic developed in 1959 for the treatment of infections with penicillinase-producing strains of S. aureus. Its routine use in hospitals had very short success, because in 1961 the first methicillin-resistant strain was reported in England [7]. After this hard blow on the fight against bacterial resistance to antibiotics, epidemiological investigations on the surveillance of MRSA and the determination of the genetic supports responsible for this resistance. But it was in 1980 that the first chromosomal fragment (DNA) responsible for resistance to methicillin was reported in the Japanese MRSA strain N315. This genetic determinant with a molecular weight of approximately 30 kb was named mecDNA. Following this discovery, the mecA gene was detected on mecDNA and was sequenced in a clone of the same Japanese strain. In the same decade 1980-1990 reasoning led to believe that S. aureus susceptible to methicillin (SASM) and MRSA formed two different bacterial species. But it has been demonstrated in a study based on 23s-RNA ribotyping of community MRSA strains (collected in Tokyo in 1959), MRSA and SASM strains from 19 hospitals in Japan (collected 1982 and 1983 entries), and MRSA strains from the Malaya hospital center (collected between 1987 and 1989) that MRSA and SASM form the same bacterial species [20]. Indeed, the SASM chromosome would simply be a support for the insertion of the mecA gene for possible acquisition of resistance to methicillin [20]. During 1990-1995, it was reported that the functioning of the mecA gene depended on the state of expression of another gene placed upstream of the mecA gene; it is the regulatory gene of the mecA gene named mecI. This brings the genetic determinant of methicillin resistance back to a mecI-mecR1-mecA complex. However, it has been reported that on the mecDNA of some SARMs, the mecI gene is absent and it is substituted by an IS1272 insertion sequence. Thus, the classification of mecDNAs in MRSA was based solely on the differences of the genes that make up the mec complex. Elsewhere, the complete sequencing of the genome of the Japanese strain N315 (isolated in 1982) by Kuroda et al., (2001) and compared with the chromosomal fragment (at position 1274-1933) of the MRSA strain NCTC 8325, made it possible to bring more information on the mecDNA determinant [21]. This study reported that beyond mecDNA, there are other genes on the mec chromosome that may contribute to the determinism of methicillin resistance. Thus, all of this large DNA fragment weighing approximately 52 kb where a mec complex carrying the mecA gene is located is called Staphylococcal cassette chromosome mec (SCCmec). A year after the description of the first SCCmec, Katayama et al., (2000) reported that on the SCCmec fragment of the S. aureus strain N315, there is a set of genes forming a complex <<the chromosomal recombinase (ccrA/B) cassette>> other than the nearby mec complex, which is responsible for the recombination of the various genes on SCCmec [22]. The diversity of the nucleotide sequences of this new complex made it possible to set up the first bases of the classification of SCCmec. This classification takes into account, on the one hand, the type of the ccr complex and, on the other hand, the class of the mec complex. In this effect, Ito et al. (2001) reported in strains NCTC 10442, N315 and 85/2082 three types of SCCmec respectively classified as SCCmec type I, SCCmec type II and SCCmec type III. Thus, SCCmec type I harbors the mec class B complex whose gene sequence is “IS1272-ΔmecR1-mecA-IS431” and the type 1 ccr complex (with the ccrA1 and ccrB1 genes) (table 1). SCCmec type II and type III were all positive for the mec class A complex and showed homology differences on the nucleotide sequences of their ccr complex. The ccr type 2 (ccrA2/B2) and ccr type 3 (ccrA3/B3) complexes have been identified respectively in SCCmec type II and SCCmec type III [10]. As for SCCmec type IV, it was immediately described in the work of [23]. It was following hybridization techniques with the nucleotide sequences of the first three types of SCCmec that Ma's team found perfect homology with the sequences of the mec class B complex and rates of 96.2 and 98.2 % homology to the ccrA2 and ccrB2 genes respectively [23]. In another investigation, Ito et al., (2004) described a new class of the mec complex in a WIS-encoded community-acquired MRSA strain (WBG8318). This class of the mec complex is characterized by the appearance on the mec complex sequence of a new copy of the IS431 insertion gene to the left of the mecA gene; this was called the mec class C2 complex, the gene structure which is IS431-mecA-ΔmecR1-IS431 [24]. The nucleotide sequence of the ccr complex of this SCCmec presented a great difference of homology with the other types of the ccr complex already known. This new complex thus sequenced was noted ccr type 5 complex carrying the ccrC1 gene [24]. The new SCCmec cassette carrying the mec class C2 complex and the ccr type 5 complex has been recognized by IWG-SCC and classified as SCCmec type V. The determination of the structure of the SCCmec type VI has gone through many twists and turns before being finalized in 2006 with the publication of the work of Chongtrakool et al., (2006) on a new classification of SCCmec [25]. Indeed, Oliveira et al. (2001) had characterized the ccr type 4 complex (with the ccrA4 and ccrB4 genes) and the mec class B complex in the strain S. aureus HDE288 [26]. The structure of this SCCmec (ccrA/B4 and mec class B complex) was considered until 2005 as a new variant of SCCmec type IV before being accepted by IWG-SCC as a different SCCmec [25]. In 2008, other levels of knowledge on new genetic sequences of SCCmec in MRSA were reached. Indeed, the mec class C1 complex which had been described in Staphylococcus haemolyticus (SH621) by Katayama et al. (2001) was also characterized on the SCCmec of the MRSA strain JCSC6082 in 2008 in Sweden [27]. One of the important differences between the class C1 and class C2 mecs complexes would be the orientation of the IS431 insertion gene located downstream of the mecA gene. On the gene sequences of the two complexes, the mecA genes are oriented in opposite directions. In the same survey Berglund et al. (2008) salso reported the ccr type 5 complex on the same SCCmec that had been described in SCCmec type V. This ccr type 5 complex was almost identical (99.9%) to the ccr complex of the SCCmercure strain 85/2082 [27-29]. Finally the structure of the SCCmec of MRSA JCSC6082 described by Berglund et al. (2008), is recognized as a new type of SCCmec and classified as SCCmec type VII. In the same year, the work of Zhang et al. (2009) reported in a Canadian strain C10682 a new type of SCCmec structured by a combination of the mec class A complex and the ccr type 4 complex. For this purpose, the two complexes had been reported separately on other SCCmecs, but this was the first time both were worn by the same SCCmec [29]. This characterized SCCmec was therefore recognized as SCCmec type VIII. The genes of the mec class A complex of SCCmec type VIII showed 100% genetic homology to those of the mec class A complex of SCCmec type II in strain N315. As for its ccrA4 and ccrB4 genes of its ccr complex, they showed homologies of 89.6% and 94.5% respectively to those of SCCmec type VI of the strain HDE288. Furthermore Li et al. (2011) carried out screening surveys for SCCmec in Staphylococcus strains isolated from participants of the 19th International Swine Veterinary Conference in Denmark in 2006. During this study, Li et al. (2011) characterized two other types of SCCmec in two strains of MRSA encoded JCSC6943 and JCSC6945 respectively isolated from a Thai participant and a Canadian participant [30]. The first SARM JCSC6943 carries in its genetic heritage a SCCmec harboring on the one hand a ccr type 1 complex which carries the ccrA1 and ccrB1 genes and on the other hand a mec class C2 complex. This new combination of the ccr type 1 complex and the mec class C2 complex on the same SCCmec is identified by IWG-SCC as SCCmec type IX. However, the second SARM JCSC6945 had a SCCmec that carried a ccr type 7 complex having ccrA1 and ccrB6 genes and its mec complex was identified as class C1.2. This mec complex is distinct from those of the other class C1 mecs complexes, by its weight (6422 bp) unlike that of SCCmec type VII (7212 bp) and the direction of orientation of its genes opposite to those of SCCmec type VII and SCCmec type I. The SCCmec of MRSA JCSC945 has been recognized in this sense as SCCmec type X [30]. The same year, as part of the epidemiological surveillance of MRSA strains in cattle and humans in Denmark, García-Álvarez et al. (2011) reported a mecA gene (in a MRSA strain IGA251 called mec AIGA251) having a homology of 70% of the mecA genes previously described [31]. In addition, the team reported a class (E) of the mec complex described for the first time in a strain of Staphylococcus, but it should be noted that this complex had been described on a plasmid pMCCL2 of a strain of Macrococcus caseolyticus JSCS5402 by [32]. This mec complex is characterized by the gene sequence “mecI-mecR1-mecA-blaZ” and is designated by the class E mec complex[13,31]. The ccrA1 and ccrB3 genes were characterized on the ccr complex carried by the SCCmec of the same MRSA IGA251 strain. This ccr complex is thus called type 8. The SARM IGA251 strain however harbors a SCCmec different from the previous SCCmecs described and was identified by IWG-SCC as being SCCmec type XI. In 2015, another variant of the ccrC complex was reported by Wu et al., (2015) in a strain of MRSA (MRSA-BAO1611) isolated from cow's milk in the Chinese North-West zone. This complex carried a particular ccrC2 gene having nucleotide homologies of 62.6% to 69.4% with those of the ccr1 genes already reported in SCCmec types V and VII [33]. Thus, the ccr complex carrying this ccrC2 gene was classified as a ccr type 9 complex. This strain harbored a SCCmec which, in addition to ccr type 9, also possesses the mec class C2 complex. This new combination of the ccr type 9 and mec class C2 complexes on the same SCCmec has been recognized as a new SCCmec and has been classified as SCCmec type XII by IWG-SCC. In the same dynamism of epidemiological studies of MRSA, Baig et al. (2018) in Denmark reported in a clone of MRSA-ST152, a SCCmec harboring the ccr type 9 complex and the mec class A complex. This was also new to IWG-SCC and was identified as SCCmec type XIII. The ccrC2 gene of this type 9 ccr complex reported by Baig et al. (2018) had nucleotide homologies of 68.5% to 70.2% of the sequences of the ccC1 genes identified on SCCmec V and VII [34]. The mecA complex hosted by this SCCmec presented a gene sequence IS431-mecI-mecR1-mecA-IS43. A new type of SCCmec (type XIV) was reported by Urushibara et al. (2020) in one of their publications in two strains of MRSA (SC640 and SC792) in Hokkaido in Japan [35]. This SCCmec presented a combination of mec class A complex and ccr type 5 complex. If these complexes had already been described in other types of SCCmec, such as mec class A complex in SCCmec types II, III and VIII or the complex ccr type 5 in SCCmec types V and VII, this DNA fragment harbors a new combination of these complexes, described for the first time. In addition, this DNA also carried an SCC-like harboring two gene clusters including Arginine Catabolic Mobile Element (ACME-arc) and Sespermine/spermine-N1-acetyltransferase (speG). The ACME and speG genes give strains resistance to certain hostile environmental factors such as acidity (from ACME-arc), polyamine toxicity (from speG) [35,36]. Sabat et al. (2021) reported a new class of the mec complex on a variant of SCCmec type IV hosted by an S. epidermidis strain IVUMCG335 in the Netherlands. Indeed, on this new mec class B4 complex, a plasmid pUB110 is inserted into the structure of the mec class B complex of SCCmec type IV. Thus, the new mec complex has the following structure IS431-ΔmecR1-mecA-IS431-pUB110-IS431-ΨIS1272 contrary to the previously described structure IS1272-△mecR1-mecA-IS431 [37]. However, it is not a new type of SCCmec according to the IWG-SCC, but a subtype of SCCmec type IV. Until 2020, the IWG-SCC has registered 14 types of SCCmec whose classification is based on the combination of 5 classes of mec complex (A, B, C1, C2 and E) and 8 types of ccr complex. In January 2022, Wang et al. (2022) in China in their publication reported a new type of SCCmec. This is SCCmec XV with a class A mec complex (mecI-mecR1-mecA-IS431) and a type 7 ccr complex (A1B6) [19]. Figure 2 illustrates a few years marking the history of meticillin resistance in S. aureus and the first characterizations of SCCmec types.
Table 1: Types of Staphylococcal cassette chromosome mec (SCCmec).
Structure and Nomenclature of Staphylococcal Cassette Chromosome mec (SCCmec)
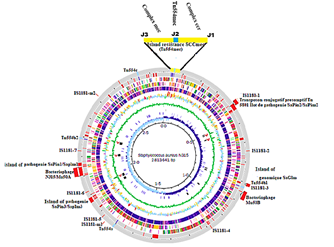
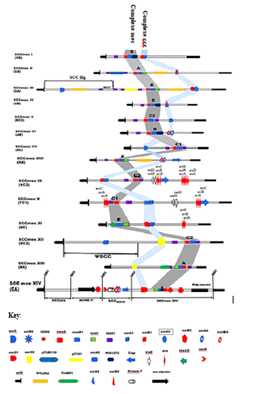
The management of Staphylococcus is ubiquitous and its different species manage to adapt to the different, often very difficult conditions of the environments they colonize [9]. This adaptability of these strains has found first in-depth explanations with the complete sequencing of the genome of the strain S. aureus N315 (Figure 3) by [21]. Indeed, the Staphylococcus genome in general is made up of two distinct functional domains. Most of their genome contains the genes that ensure the bacteria's vital functions. The second part of the genome is made up of accessory and mobile genetic elements such as plasmids, transposons, prophages or islands of pathogenesis carrying most of the genes associated with virulence factors and antibiotic resistance [17,18,38]. In this part, this study will focus on the description of the SCCmec and these constituents. SCCmec is part of the mobile genetic elements that confer resistance to Staphylococcus against beta-lactams following the pressure of beta-lactams. Each SCCmec is characterized by two complexes, the mec complex and the ccr complex. A SCCmec has three joining sequences (J1, J2 and J3) complementary to their structures, which delimit the regions occupied by the two complexes (Figure 3).
The mec Complex
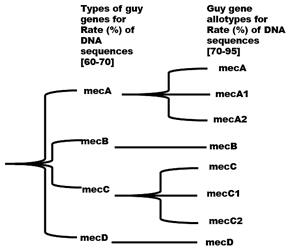
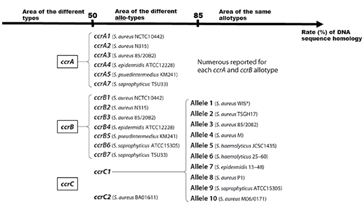
The mec complex includes the mec gene (A or C) of approximately 2.1 kb, regulatory genes (mecI encodes a transcriptional repressor of mecA and mecR1 encodes the MecR1 protein) and insertion sequences (IS431 or IS1272) [28]. The MecR1 protein detects the presence of beta-lactams by to its extracellular domain. Once the antibiotic binds, there is activation of the intracellular domain which acquires protease activity and degrades the repressor MecI, thus promoting the expression of mecA. These regulatory genes can be intact or truncated, mutations occurring in these regulatory genes can affect the level of methicillin resistance. In bacteria, there are 4 types of mec gene including mecA, mecB, mecC and mecD. These 4 mec genes share between 60 and 70% nucleotide sequence homology and can also be subdivided into allotypes which share between 70 and 95% sequence homology between them (Figure 4). Indeed, a mec gene is classified as a new type or a new allotype, if it has respectively nucleotide similarity rates between 60 and 70% or between 70% and 95% to the others. Thus, the identity of the mec gene combined with those of the regulatory genes and insertion sequences carried by the DNA of the mec complex, make it possible to determine the class of the mec complex. The IWG-SCC currently recognizes five mec complex classes (from A to E) in staphylococci.
The Recombinase Gene Complex (ccr)
Recombinases are responsible for the recombinaison and mobility of the genes on cassette. The recombinases gene complex consists of either a pair of genes, ccrA and ccrB combined, or a single ccrC gene [39]. Five types of SCCmec including types V, VII, XII, XIII and XIV which harbor the ccrC gene in their ccr complex. The other ten carry the ccrA and ccrB gene pair (Table 1). Figure 5 illustrates the classification of ccr complexes recognized by IWG-SCC. The classification of ccr complexes is based on the homology rates of their nucleotide sequences. Two ccr complexes would be classified in two types of ccr complex, if the rate of their nucleotide homologies is less than or equal to 50%. However, if this rate is strictly between 50-85%, the two complexes are said to be of different allotype. And the ccr complexes are classified in the same allotype if the homology rate of their sequences is greater than or equal to 85%. As for the function of the ccr complex, the genes of the ccr complex code for a recombinase of integration or excision of the gene sequences of the SCCmec [40]. The integrating role of recombinases is similar to that of bacteriophage integrases. In addition, the ccr complex also has the ability to cleave DNA to allow the exchange of gene fragments and recombinations between the two attachment sites [41]. There are other gene fragments also carried by the SCCmecs, which accompany the ccr complex in the different functions. This is the case of inverted repeated sequences (or inverted repeats) which have a role of excising but do not allow integration.
3.5 Junction Regions
Each SCCmec has in its structure three zones which delimit the positions of the two complexes. These areas are today called junction regions (J) numbered J1, J2 and J3. On the front panel these regions bore the designations LC (for J1) located between the extreme right and the ccr complex, CM (for J2) positioned between the ccr complex and the mec complex and IR (for J3) between the mec complex and the far left of the SCCmec structure [18]. The junctions harbor certain accessory elements such as plasmids (pUB110) or transposons (Tn554) or other genes which confer resistance to other antibiotics. In the classification of SCCmec, the components of junctions serve to distinguish allotypes of a type of SCCmec [41,42].
3.6 SCCmec Nomenclature
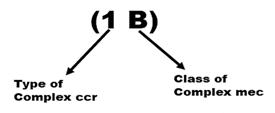
SCCmecs are identified into different types of SCCmecs based on the class of the mec complex and the type of ccr complex they harbor. Indeed, a type of SCCmec is named by two radicals in brackets, the first of which is designated by an Arabic numeral corresponding to the type of the ccr complex and the second is in uppercase alphabet letter which corresponds to the complex. Figure 6 illustrates a representation of SCCmec type I. The IWG-SCC has recognized and classified between 2000 and 2020, 14 types of SCCmec. It was not until January 2022 that the fifteenth type of SCCmec was identified with the work of [19]. This sets the total at fifteen types of SCCmec.
3.6.1 SCCmec Type I or (1B): This SCCmec was first described in 2021 in the strain of S. aureus NCTC10442 (Genbank number AB033763) isolated since 1961 in England. It is one of the genetic carriers conferring resistance to beta-lactams, predominant in commensal MRSA (HA-MRSA) [43]. SCCmec type I harbors the class B mec complex with the ccr type 1 complex carrying the ccrA1 and ccrB1 genes (Figure 7). Its IA allotype relates to its J3 junction region, the plasmid pUB110. In SARM-NCTC10442, the SCCmec is 34359 nucleotides long. In general, a variety of accessory elements are incorporated into the structure of the SCCme type I, which contribute to its operation. These include 41 coded DNA sequences (CDS) with 36 ORFs (Open Reading Frames), 2 mobile sequences and repeated regions. In one of their investigations, Lakhundi and Zhang (2018) oriented these last departures and others on the structure of the SCCmec type I; upstream of the ccr complex (one repeated region and 17 CDS), at the level of the ccr complex (the two ccrA1 and ccrB1 genes and 7 CDS), on the mec complex (the mecR1 and mecA genes, 2 mobile elements, 2 repeated regions and 10 CDS), downstream of the mec complex (a repeated region and 3 CDS) and the ORFs sequences in the ends [41]. The functions of most of these ORFs remain unknown, however, exceptionally, CEO10 is the ORF which codes for the polypeptide plasmid-sequence surface protein (PLs) and two other ORFs on PLs, code for the glycerophosphoryl diester phosphodiesterase and the putative transposase.
3.6.2 SCCmec Type II or (2A): It has a large size of approximately 53017 nucleotides and is frequent in commensal strains. SCCmec II is characterized by this combination of the class A mec complex and the type 2 ccr complex on its structure (Figure 7). It also hosts several other fragments including 4 repeated regions, 3 mobile elements and 51 CDS. Thus, upstream of the ccr complex (a repeated region and 15 CDS), at the level of the ccr complex (the ccrA2 and ccrB2 genes and 16 CDS), between the two complexes (a mobile element and 12 CDS), at the level of the mec complex (the mecA, mecR1, mecI and IS431 genes and 5 CDS) and at the downstream end of the mec complex (one mobile element, 2 repeat regions and 8 CDS). This cassette also has other remarkable elements such as at the J3 junction, the copy of the Staphylococcal plasmid pUB110 and an ORF (pre) coding for the recombination enzyme for pUB110. In addition, on the J1 junction there is a regulatory gene kdp or between the complexes there is a transposon Tn554 involved in resistance to streptomycin and erythromycin. Investigations have reported a diversity of SCCmec type II subtypes, including SCCmec types IIA, IIB, IIC, IID or IIE. However, the first SCCmec II was described in 1999 in Japan in MRSA strain N315 (with Genbank accession number D86934) [44].
3.6.3 SCCmec Type III or (3A): it is a very large cassette with 66891 nucleotides and it is among the most characterized SCCmec in commensal MRSA. SCCmec III harbors the class A mec complex and the type 3 ccr complex (Figure 7). SCCmec III carries 10 repeat regions, 6 elements and 97 CDS (with 22 CDS of unknown function). These structures are located upstream of the ccr complex (a repeated region and 2 CDS), on the ccr complex (the ccrA3 and ccrB3 genes and 11 CDS), between the complexes (a mobile element and 18 CDS), on the mec complex (the mecA, mecR1, mecI and IS431 genes, a mobile element, 2 repeat regions and 9 CDS) and downstream of the mec complex (4 mobile elements, 7 repeat regions and 52 CDS) [10]. On the J3 junction there is a transposon Tn554 which carries genes encoding resistance to erythromycin and a copy of the plasmid pUB181 harboring genes encoding resistance to tetracycline and mercury. The J2 junction carries a transposon ΨTn554 coding for cadmium resistance determinants. The first SCCmec III was identified in 2001 in the MRSA strain 85/2082 which had been isolated in 1985. Today several subtypes of SCCmec have been identified, including SCCmec subtypes IIIA, IIIB, IIIC, IIID and IIIE [45].
3.6.4 SCCmec Type IV or (2B): It was described for the first time in 2002 following the work of Ma et al. (2002) in the United States of America. During this investigation, two subtypes of SCCmec IV (SCCmec IVa and SCCmec IVb) were characterized respectively in MRSA JCSC1986 (Genbank accession number AB063173) and MRSA JCSC1978 (Genbank accession number ABO63172). SCCmec IV are characterized by the combination of the class B mec complex and the type 2 ccr complex (Figure 7). Subtype IVa harbors two mobile elements, 4 repeat regions and 22 CDS (of which 17 CDS have unknown functions). These structures are distributed upstream of the ccr complex (one repeated region and 4 CDS), on the ccr complex (the ccrA2 and ccrB2 genes and 6 CDS), on the mec complex (the mecA, IS1272 and mecR1 genes, 2 mobile elements, 2 repeat regions and 5 CDS) and downstream of the mec complex (one repeat region and 3 CDS). This subtype is approximately 24244 nucleotides in size. As for subtype IVb, it has a small size of 20916. SCCmec IV is the type of SCCmec that has more variants including subtypes IVa, IVb, IVc, IVd, IVe, IVf, IVg, IVh, IVi, IVj, IVk, IVl, IVm, IVn and Ivo.
3.6.5 SCCmec Type V or (5C): It has a size of 27638 nucleotides and its high prevalence have been reported in community-acquired MRSA. SCCmec V is characterized by the combination of the mec class C complex and the ccr type 5 complex (with the ccrC1 gene) [24]. It relates to its structure 6 repeated regions, 2 mobile elements and 23 CDS (of which 15 CDS have unclarified functions). These different elements are located upstream of the mec complex (a repeated region and a CDS), at the level of the mec complex (the mecA, mecRI and IS431 genes, 2 mobile elements and 4 repeated regions), between the complexes (2 CDS), on the ccr complex (the ccrC1 genes and 6 CDS) and downstream of the ccr complex (one repeat region and 7 CDS). SCCmec V was first identified in 2004 in MRSA strain JCSC3624 and has no antibiotic resistance gene other than the mecA gene. SCCmec type V has very few subtypes including Va and Vb variants [47].
3.6.6 SCCmec Type VI or (4B): This type of cassette is characterized by a combination of the class B mec complex (IS1272-△mecR1-mecA-IS431) and the ccr type 4 complex (Figure 7). The structure of its ccr complex has a composition almost identical to that of the ccr complex type 3. The region downstream of its ccr complex has a 99% similarity with that of SCCmec type I. SCCmec VI has been described for the first in 2001 following the work of Oliveira et al. (2001) in the MRSA strain HDE288 (Genbank accession number AF411935). SCCmec VI had been identified as a variant of SCCmec IV and subsequently redefined in 2006 as SCCmec VI by the IWG-SCC [26].
3.6.7 SCCmec Type VII or (5C1): it has a size of approximately 26753 nucleotides and is more characterized in community-acquired MRSA strains. SCCmec type VII is characterized by the combination of the class C1 mec complex and the type 5 ccr complex (Figure 7). Its structure also contains 2 repeat regions and 29 CDS (with 16 CDS having unclear functions). Along its structure present upstream of the ccr complex (a repeated region and 3 CDS), on the complex (the ccrC1 gene and 6 CDS), between the complexes (8 CDS), on the mec complex (the IS431 gene sequences -mecA-△mecR1-IS431 and 4 CDS) and downstream of the mec complex (one repeat region and 5 CDS). It also carries three Tnp genes, two of which code for the IS431 transposase enzyme and the other codes for the IS12960D transposase B protein. SCCmec was first described in 2008 in MRSA strain JCSC6082 (Genbank accession number AB373032). Very few subtypes of SCCmec VII have been characterized. However, SCCmec VIIa and VIIb subtypes have been demonstrated in several infestations [35,48].
3.6.8 SCCmec Type VIII or (4A): It is a medium-sized cassette of about 32184 nucleotides and its highest prevalence is in MRSA acquired from animals and food (LA-acquired) [49,50]. SCCmec VIII is characterized by a combination of the class A mec complex and the type 4 ccr complex (Figure 7). Its structure also carries 6 repeated regions, a mobile element and 36 CDS (with 14 CDS with unknown functions). These last elements are distributed upstream of the mec complex (2 repeated regions and 2 CDS), on the mec complex (the sequence mecI-mecR1-mecA-IS431, a mobile element, 2 repeated regions and 6 CDS), between the complexes (19 CDS), on the ccr complex (the ccrA4 and ccrB4 genes) and downstream of the ccr complex (2 repeat regions and 5 CDS). Its J1 junction carries 5 ORFs including a code for a putative membrane protein and a truncated ccr gene. The J2 junction hosts 7 ORFs including ermA which codes for rRNA adenine N-6-methyltransferase, aad9 which codes for streptomycin 3'-adenyltransferase, tmpA which codes for transposase A, tmpB expressing for transposase B, tmpC for transposase C the other two ORFs are mobile elements of transposon Tn554. SCCmec VIII was first described in 2008 in the MRSA C10682 strain isolated in 2003 in Canada. Reported SCCmec VIII subtypes are VIIIa, VIIIb, and VIIIc [50].
3.6.9 SCCmec Type IX or (1C1): it is 43675 nucleotides long and is characterized by a combination of class C2 mec complex and the type 1 ccr complex (Figure 7). Other most remarkable elements reported on its structure, 6 repeated regions, 2 mobile elements and 42 CDS (with 22 ORFs with unknown functions). The gene sequences on the SCCmec IX are distributed upstream of the ccr complex (a repeated region and 3 CDS), on the ccr complex (the ccrA1 and ccrB1 genes and 6 CDS), between the complexes (5 CDS), on the mec complex (the sequence IS431-mecA-△mecR1-IS431, 2 mobile elements, 4 repeat regions and 5 CDS) and downstream of the mec complex (one repeat region and 20 CDS). Its J1 junction presents several resistance genes to metallic trace elements. This is the case of the CadD and CadX genes which encode the cadmium-binding protein and the cadmium variant-resistant protein respectively. The copB gene on J1 codes for copper-transporting ATPase which mediates copper detoxification. The J1 junction also harbors genes for arsenic resistance including ArsR, ArsA, ArsB, ArsC and ArsD which code respectively for the enzymes, arsenical resistance operon repressor, arsenical pump-driving ATPase, arsenical pump-membrane protein, arsenical reductase and arsenical resistance operon trans-acting. The SCCmec IX cassette was first identified in 2011 in Denmark in MRSA strain JCSC6943 (Genbank accession number AB505628).
3.6.10 SCCmec Type X or (7C1): This is a large cassette of approximately 50803 nucleotides, identified by the combination of the class C1 mec complex and the type 7 ccr complex (Figure 7). It also harbors 6 repeat regions, 2 mobile elements, 54 CDS (with 33 CDS having unclear functions) and several metal trace element resistance genes [42]. The SCCmec X is structured; upstream of its mec complex (one repeat region and 6 CDS), on the mec complex (the sequence IS431-mecA-△mecR1-IS431, 2 mobile elements, 4 repeat regions and 5 CDS), between the complexes (4 CDS), on the ccr complex (the ccrA1 and ccrB6 genes and 5 CDS) and downstream of the ccr complex (one repeat region and 30 CDS). On the J1 junction, it harbors the insert sequence ISSha1, an arsRBC operon for the expression of the arsB, arsC and arsR genes encoding arsenic detoxification. The same Junction carries the copB and cadD/X genes encoding resistance to copper and cadmium respectively. Its J3 junction also carries the arsB, arsC and arsR genes for the same arsenic detoxification functions. It was first characterized in 2011 in Denmark in MRSA strain JCSC945 (Genbank accession number AB505630) (Table 1).
3.6.11 SCCmec Type XI or (8E): it has a weight of 29.4 kb and is identified by the combination of the mec E complex and the ccr type 8 complex (Figure 7). SCCmec XI has a gene sequence mecA gene (blaZ-mecA-mecR1-mecI) with 69% nucleotide homology to other mecA genes. Its J1 and J2 junctions host ORF sequences that code for membrane export proteins and lipases [41]. The J1 junction also carries arsenic resistance genes. The SCCmec XI cassette was first characterized in 2011 in the MRSA clone CC130 (LGA251). This MRSA was isolated in 2007 in Denmark. SCCmec XI is a particularly common cassette in MRSA circulating in Europe and its incidence is more of animal origin [51].
3.6.12 SCCmec Type XII or (9C2): This is a cassette weighing approximately 25 kb located between DR2 (repeated sequence 2 or direct repeats 2) and DR3 of the mec DNA fragment. In total, the SCCmec XII cassette carries 31 ORFs (from the 31st ORF to the 62nd of the mec DNA fragment) [33]. It is characterized by the combination of the class C2 mec complex and the type 9 ccr complex (Figure 7). The mec complex (located between 44th ORF and 50th ORF) carries the ΔmvaS, ugpQ, maoC, mecA and ΔmecR1 genes and at the ends, insert sequences ΔIS431-1 and ΔIS431-2. The mec DNA carrying the SCCmec XII cassette is characterized on its structure by another cassette located between DR1 and DR2 called pseudo-SCC. This pseudo-SCC has a weight of 24.3 kb with 30 ORFs along its length and carries the ccrA1 genes which would appear to be a semi-complex mec type 1. Studies have reported on the structure of SCCmec XII several other antibiotic resistance genes. Than mecA including, blaZ (coding for beta-lactamases), aadA, aadE and aacA-aphD (resistance to Aminoglycosides), lnuB (resistance to macrolide/lincosamide/streptogramin), tetL (resistance to tetracyclines), dha1 and fexA (resistance to phenicols) [52]. MRSA harboring SCCmec XII are generally of animal origin. The first characterization of the SCCmec XII cassette was in 2015 in the MRSA strain BAO1611 (Genbank number KR187111) isolated from cow's milk [33].
3.6.13 SCCmec Type XIII or (9A): It has a weight of 32.3 kb and is characterized by a combination of the class A mec complex and the type 9 ccr complex (Figure 7). The SCCmec XIII cassette harbors on its J2 junction a transposon Tn4001 which carries the aac(6')-aph(2'') resistance genes to gentamycin. It also carries the plasmid pSaa619 which harbors the blaZ gene coding for resistance to beta-lactams. The SCCmec cassette was first described in 2018 in the MRSA strain ST152 isolated in Denmark [34].
3.6.14 SCCmec Type XIV or (5A): This is a cassette weighing approximately 41 kb and characterized by the class A mec complex and the type 5 ccr complex (Figure 7). It is potted between DR4 and DR5 of a chromosomal fragment having three pseudo cassettes [35]. The first pseudo cassette located between DR1 and DR2 has a weight of 12kb and carries a pseudo-SCC lacking with genes that code for the plasmin-sensitive protein (ΨSCCpls). The second pseudo-cassette has a weight of 14 kb, located between DR2 and DR3 and hosts an arc cluster (ACMEII') which codes for the resistance to the acidity of the medium. The third pseudo cassette has a weight of 14 kb, located between DR3 and DR4, it carries the speG gene which codes for resistance to the effects of polyamines, the copA gene which codes for the copper-transforming ATPase protein (for resistance to copper ), the protein F gene (teichoic acid bio-synthesis protein) and the ccr type 4 complex genes (ccrA4-/B4). The SCCmec XIV cassette was characterized for the first time in 2020 in the MRSA strain SC792 in Japan.
3.7 Some Epidemiological Data on SCCmec
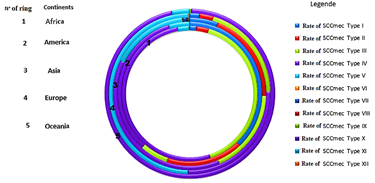
Staphylococcal Cassettes Chromosomes mec are highly diverse mobile genetic carriers (Figure 7) that confer multi-resistance staphylococcal beta-lactams [53,54]. These bacterial gene pools are public health concerns with very little new data on global distribution. Nevertheless, some representative investigations allow insights into the distribution and prevalence of SCCmec types on the continents [55]. In this part, the review is limited to recent data reported in publications found in official journals. In Africa, certain types of SCCmec (table 2) have been identified in the Staphylococcus genus predominated by Staphylococcus aureus species isolated from humans as well as from animals or environmental samples [56-58]. The distribution of SCCmec types is not uniform, however we can see the presence of SCCmec type III and type IV with high prevalence in the different areas of the African continent (Figure 8). Sekyere and Mensah (2019) made the same findings during their investigation of the literature on the molecular epidemiology and mechanism of antibiotic resistance in Enterococcus spp., Staphylococcus spp. and Streptococcus spp. in Africa. They reported a predominance of strains harboring SCCmec IV (747/4437) followed by SCCmec III (305/4437), SCCmec II (163/4437), SCCmec V (135/4437) and SCCmec I (79/4437). The proliferation and dissemination of SCCmec would be favored by the easy acquisition mechanisms (transformation, transition and conjugation) of these cassettes between strains of the genus Staphylococcus on the one hand and on the other hand by the ability of these strains to resist environmental factors [16,53,59]. Indeed, the Staphylococcal Cassettes Chromosome (SCC) can carry other genes such as the mer gene, the ACME gene or the speG gene in addition to the mecA gene, whose expressions would allow Staphylococcus carriers to resist high levels of mercury (mer gene) acidity (ACME gene), polyamine toxicity (speG gene) [40,59,60]. Others investigations in Africa have reported Staphylococcus strains harboring both SCCmec type III or type IV with SCCmer [61] or with the ACME gene [62]. In Europe, Staphylococcus harboring SCCmec IV have the greatest distribution in the different countries (Table 2) and SCCmec IVa and IVc subtypes have been the most reported [63-65]. This predominance was confirmed during an investigation of Bartels et al. (2020) in Northern Europe on 466 MRSA from Denmark (354), France (10), Norway (24), Sweden (27) and England (51)[66]. They reported that 94% of the 466 MRSA harbored SCCmec and 100% of these SCCmec were type IVa. As in Africa or Europe, in the rest of the world Staphylococcus carrying SCCmec IV predominates (Figure 8) in human or animal infections and food contamination or healthcare activity surfaces. On the other continents, several authors have come to the conclusion of the predominance of Staphylococcus harboring various subtypes of SCCmec IV and by the high frequency of cohabitation of SCCmec IV and the ACME, speG and mer genes on their mecDNA [41,67 -70].
Table 2: MR-CoNs (coagulase-negative methicillin-resistant Staphylococcus), MRSA (methicillin-resistant Staphylococcus aureus).
Note: Investigations in Table 2 reported MRSAs and MR-CoNs that harbored mec and ccr complexes not recognized by the IWG-SCC and MRSAs or MR-CoNs whose SCCmec cassettes were not determined, which explain the discrepancies between the numbers of MRSA or MR-CoNs and the rates of characterized SCCmec.
3.8 Application of Technology for the Characterization of SCCmec
Methicillin-resistant Staphylococcus, especially MRSA, are the strains that undoubtedly represent the greatest threats in the fight against bacterial multi-resistance to antibiotics in hospitals [71]. This threat also affects the community and veterinary sectors. To this end, their management is generally very complex due to confusion in practice about the type of MRSA that causes the infection or contamination. However, molecular typing techniques for MRSA have been proposed and over time, rapid and effective screening models have been put in place, all of which contribute to knowledge of the molecular epidemiology and evolution of MRSA [72,73]. Among these techniques, MLST (multi locus sequence type), spa typing (protein A sequence) and SCCmec typing are the most used for determining the types and subtypes of MRSA [8,74]. For typing efficiency, very often techniques are combined. In this part, the study will stop on the SCCmec typing. SCCmec typing is a technique that consists of determining types and subtypes of the SCCmec genetic determinants, which carry the mecA gene and other genes for resistance to antibiotics and metallic trace elements (Figure 8), carried by MRSA. The first SCCmec typing techniques were methods of hybridization of mecA gene and Tn554 transposon probes based on restriction enzyme digestion with ClaI genomic DNA. This technique has been used since 1988, for the identification of repeated DNA elements (characterized by insert elements) of the genome of the Bordetella pertussis strain, well before the first nomenclatures of SCCmec cassettes [75]. It was first used to characterize mecDNA fragments with the work of [76]. Today, improved methods with digestive restriction enzymes combined with PCR (polymerase chain reaction) are applied to SCCmec typings [77-79]. This is the case of the SCCmec typing method based on PCR amplification of the ccrB genes (of the ccr complex) with RFLP (restriction fragment length polymorphism) or the case of the ME-AFLP method. After the 2000s, methods solely based on multiplex PCR were applied to SCCmec typings. This technology uses a set of multiple primer pairs to simultaneously amplify multiple DNA targets in a single PCR reaction, rather than a single primer pair to amplify a single DNA target. Thus, in 2002 the multiplex PCR technique was applied for the first time to characterize types I to IV of SCCmec [80]. Subsequently, it was used for the characterization of SCCmec types and subtypes. Thus, in 2007 several methodologies of the multiplex PCR technique were used by Kondo et al. (2007) for the characterization of SCCmec type I to V with their variants. In recent years, it is the technique that seems to predominate in research centers, for the characterization of types and subtypes of SCCmec [15,77,81-83]. In addition to the two previous SCCmec typing methods, real-time PCR was the third method applied since 2004 with the work performed by Francois et al., (2004) in Switzerland [84]. Real-time PCR is based on the detection and quantification of a fluorescent contribution whose emission is directly proportional to the quantity of amplicons generated during the PCR reaction. Initially, this technique had very little success in SCCmec typing, because the first tests were not considered innovative to the knowledge already acquired [78]. Later, with the use of padlock probes, it gave great success to the application by real time PCR in SCCmec typing thanks to its diagnostic efficiency of trace elements on regions of the mecA, ccrB and ccrC genes [85]. This technology has been used to characterize the types and subtypes of SCCmec in several investigations [61,86,87]. In the last decade, new sequencing technologies (Illimina or MinION) are used in the determination of SCCmec types and subtypes. The principle of Illimina sequencing is based first on the amplification of DNA fragments of 100 to 500 bp coupled to regions complementary to the oligonucleotides of the plate, and then sequenced using a fluorescent emitter with lengths specific to the types of nucleotides. Baig et al. (2018) have combined these two technologies to describe for the first time the SCCmec type XIII.
4. Conclusions
The staphylococcal cassette chromosome mec are mobile genetic carriers harboring genes for resistance to antibiotics and metallic trace elements and virulence genes. They are present in Staphylococcus and are characterized by the mec and ccr complex genes. Until January 2022, fifteen types of SCCmec numbered from I to XV, have been described with their subtypes. Epidemiological studies have reported the existence of multi-resistant Staphylococcus carrying SCCmec on all continents. The global spread of SCCmec is predominated by SCCmec type IV. During the last three decades, technological methods have made the characterization of SCCmec types rapid and increasingly precise, presenting SCCmec typing as one of the best options for monitoring the epidemiology and managing Staphylococcus Methicillin-resistant.
Author Contributions
Conceptualization, Ouédraogo G. Abasse, Savadogo Aly and Tchoumbougnang François; methodology, Ouédraogo G. Abasse; software, Ouédraogo G. Abasse; validation, Ouédraogo S. Henri and Kaboré Boukaré; formal analysis, Ouédraogo G. Abasse and Kaboré Boukaré; resources, Ouédraogo G. Abasse; writing review and editing, Ouédraogo G. Abasse, Cissé Hama, Zongo Oumarou; visualization, Cissé Hama, Bassolé I. H. Nestor; supervision, Bassolé I. H. Nestor, Tchoumbougnang François, Savadogo Aly; project administration, University Joseph KI-ZERBO and University of Douala; funding acquisition, AFRIDI.
Funding
This overview was funded by AFRIDI. The internet connection costs of the activities were financed by the AFRIDI structure.
Acknowledgments
Joseph KI-ZERBO University and the University of Douala have made available to us the premises and internet research tools.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Anas MB, Almousawi EA, Alhatami ON, et al. Characterization and molecular evaluation of Staphylococcus aureus isolated from poultry and dairy cattle milk in Iraq, 3rd Int. Sci. Conf. Alkafeel Univ. (ISCKU 2021) (2022):
- Strauß L, Stegger M, Eberechi P, et al. Origin , evolution , and global transmission of community-acquired Staphylococcus aureus PNAS (2017): E10596-E10604.
- Nguyen DB, Fernanda CL, Ruth B, et al. Invasive Methicillin-Resistant Staphylococcus aureus Infections Among Patients on Chronic Dialysis in the United States , 2005 – 2011. Infect. Dis 57 (2013): 1393-400.
- Klevens RM, Morrison MA, Nadle J, et al. Methicillin-Resistant Staphylococcus aureus Infections in the United States. JAMA 298 (2007): 1763-1771.
- Bréchot C. La lettre de l’Institut Pasteur Antibiotiques?: quand les bactéries, Le Dossier (2014): 12.
- Touaitia R. Staphylococcus aureus résistant à la méthicilline?: Emergence et mécanismes de résistance. Univrsite Badji Mokhtar – Annaba (2016).
- Jevons MP. Celbenin’ -resistant Staphylococci. Med. J 1 (1961): 124-125.
- Enright MC, Robinson DA, Randle G, et al. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). PNAS 99 (2002).
- Pekana A, Green E. Antimicrobial Resistance Profiles of Staphylococcus aureus Isolated from Meat Carcasses and Bovine Milk in Abattoirs and Dairy Farms of the Eastern Cape, South Africa. J. Environ. Res. Public Heal 15 (2018).
- Ito T, Katayama Y, Asada K, et al. Structural Comparison of Three Types of Staphylococcal Cassette Chromosome mec Integrated in the Chromosome in Methicillin-Resistant Staphylococcus aureus. Agents Chemother 45 (2001): 1323-1336.
- Tchamba CN. Les staphylocoques d’origine animale?: génétique de la résistance à la méticilline et évaluation thérapeutique des bactériophages dans le cadre des mammites bovines Staphylococci. Université de Liège (2021).
- Zhang K, Mcclure J, Elsayed S, et al. Novel Multiplex PCR Assay for Characterization and Concomitant Subtyping of Staphylococcal Cassette Chromosome mec Types I to V in Methicillin-Resistant Staphylococcus aureus Novel Multiplex PCR Assay for Characterization and Concomitant Subtyping of Stap. Clin. Microbiol 43 (2005): 5026-5033.
- Shore AC, Deasy EC, Slickers P, et al. Detection of Staphylococcal Cassette Chromosome mec Type XI Carrying Highly Divergent mecA , mecI , mecR1 , blaZ , and ccr Genes in Human Clinical Isolates of Clonal Complex 130 Methicillin-Resistant Staphylococcus aureus. Agents Chemother 55 (2011): 3765-3773.
- Kondo Y, Ito T, Ma XX, et al. Combination of Multiplex PCRs for Staphylococcal Cassette Chromosome mec Type Assignment?: Rapid Identification System for mec , ccr , and Major Differences in Junkyard Regions. Agents Chemother 51 (2007): 264-274.
- Bhowmik D, Chetri S, Das BJ, et al. Distribution of virulence genes and SCCmec types among methicillin-resistant Staphylococcus aureus of clinical and environmental origin: a study from community of Assam, India. BMC Res. Notes 14 (2021): 1-7.
- Shittu AO, Kenneth O, Solayide A, et al. Antibiotic resistance and molecular epidemiology of Staphylococcus aureus in Nigeria. BMC Microbiol 11 (2011).
- Ghebremedhin B, Olugbosi MO, Raji AM, et al. Emergence of a Community-Associated Methicillin-Resistant Staphylococcus aureus Strain with a Unique Resistance Profile in Southwest Nigeria. Clin. Microbiol 47 (2009): 2975-2980.
- IWG-SCC. Classification of Staphylococcal Cassette Chromosome mec (SCC mec): Guidelines for Reporting Novel SCC mec Elements. Agents Chemother 53 (2009): 4961-4967.
- Wang W, Yue H, Michelle B, et al. Novel SCC mec type XV ( 7A ) and two pseudo-SCC mec variants in foodborne MRSA in China. Antimicrob. Chemother 7 (2022): 7-8.
- Hiramatsu K, Kondo N, Ito T. Genetic basis for molecular epidemiology of MRSA, Infect. Chemother vol. 2 (1996): 117-129.
- Kuroda M, Toshiko O, Uchiyama I, et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357 (2001): 1225-1240.
- Katayama Y, Ito T, Hiramatsu K. A New Class of Genetic Element, Staphylococcus Cassette Chromosome mec , Encodes Methicillin Resistance in Staphylococcus aureus. Agents Chemother 44 (2000): 1549-1555.
- Ma XX, Ito T, Chuntima T, et al. Novel Type of Staphylococcal Cassette Chromosome mec Identified in Community-Acquired Methicillin-Resistant Staphylococcus aureus. Strains 46 (2002): 1147-1152.
- Ito T, Ma XX, Takeuchi F, et al. Novel Type V Staphylococcal Cassette Chromosome mec Driven by a Novel Cassette Chromosome Recombinase , ccrC. Agents Chemother 48 (2004): 2637-2651.
- Chongtrakool P, Ito T, Ma XX, et al. Staphylococcal Cassette Chromosome mec ( SCC mec ) Typing of Methicillin-Resistant Staphylococcus aureus Strains Isolated in 11 Asian Countries?: a Proposal for a New Nomenclature for SCC mec Elements. Agents Chemother 50 (2006): 1001-1012.
- Oliveira DC, Milheiric C. Redefining a Structural Variant of Staphylococcal Cassette Chromosome mec, SCCmec Type VI. Agents Chemother 50 (2006): 3457-3459.
- Berglund C, Ito T, Ikeda M, et al. Novel type of staphylococcal cassette chromosome mec in a methicillin-resistant Staphylococcus aureus strain isolated in Sweden. Agents Chemother 52 (2008): 3512-3516.
- Katayama Y, Ito T, Hiramatsu K. Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: Role of IS431-mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin-resistant Staphylococcus haemolyticus. Agents Chemothervol 45 (2001): 1955-1963.
- Zhang K, Mcclure J, Elsayed S, et al. Novel Staphylococcal Cassette Chromosome mec Type , Tentatively Designated Type VIII , Harboring Class A mec and Type 4 ccr Gene Complexes in a Canadian Epidemic Strain of Methicillin-Resistant Staphylococcus aureus. Agents Chemother 53 (2009): 531-540.
- Li S, Skov RL, Han X, et al. Novel Types of Staphylococcal Cassette Chromosome mec Elements Identified in Clonal Complex 398 Methicillin-Resistant Staphylococcus aureus Antimicrob. Agents Chemother vol. 55 (2011): 3046-3050.
- García-Álvarez L, Holden ATG, Lindsay H, et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: A descriptive study. Lancet Infect. Dis 11 (2011): 595-603.
- Baba T, Kuwahara-Arai K, Uchiyama I, et al. Complete genome sequence of Macrococcus caseolyticus strain JSCS5402, reflecting the ancestral genome of the human-pathogenic staphylococci. Bacteriol 191 (2009): 1180-1190.
- Wu Z, Li F, Liu D, et al. Novel type XII staphylococcal cassette chromosome mec harboring a new cassette chromosome recombinase, ccrC2, Agents Chemother 59 (2015): 7597-7601.
- Baig S, Johannesen TB, Overballe-petersen S, et al. Infection , Genetics and Evolution Novel SCC mec type XIII ( 9A ) identi fi ed in an ST152 methicillin-resistant Staphylococcus aureus. Genet. Evol 61 (2018): 74-76.
- Urushibara N, Aung MS, Kawaguchiya M, et al. Novel staphylococcal cassette chromosome mec (SCCmec) type XIV (5A) and a truncated SCCmec element in SCC composite islands carrying speG in ST5 MRSA in Japan, Antimicrob. Chemother 75 (2020): 46-50.
- Srishtee H, Xiqi A, Li A, et al. crossm Staphylococcus epidermidis MSCRAMM SesJ Is Encoded in. MBio 11 (2020): e02911-19.
- Saba AJ, Bathoorn E, Becker K, et al. Staphylococcal cassette chromosome mec containing a novel mec gene complex , B4. J Antimicrob Chemother (2021): 1-5.
- Wysocka M, Monteiro T, de Pina C, et al. Whole-genome analysis uncovers loss of blaZ associated to carriage isolates belonging to MRSA clone ST5-VI in Cape Verde Magdalena. Glob. Antimicrob. Resist 21 (2021): S2213-7165.
- Founou LL, Founou RC, Allam M, et al. Genome analysis of methicillin ? resistant Staphylococcus aureus isolated from pigs?: Detection of the clonal lineage ST398 in Cameroon and South Africa. Wiley 398 (2019): 512-525.
- Eibach D, Michael N, Hogan B, et al. Nasal Carriage of Staphylococcus aureus among Children in the Ashanti Region of Ghana. PLoS One 12 (2017): e0170320.
- Lakhundi S, Zhang K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Microbiol. Rev 31 (2018): e00020-18.
- Liu J, Chen D, Peters BM, et al. Microbial Pathogenesis Staphylococcal chromosomal cassettes mec ( SCC mec ): A mobile genetic element in methicillin-resistant Staphylococcus aureus. Pathog 101 (2016): 56-67.
- Alghizzi M, Shami A, The prevalence of Staphylococcus aureus and methicillin resistant Staphylococcus aureus in milk and dairy products in Riyadh, Saudi Arabia. Saudi J. Biol. Sci (2021).
- Ito T, Katayama Y, Hiramatsu K. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus Antimicrob. Agents Chemother 43 (1999): 1449-1458.
- Oosthuysen WF. Molecular characterisation of methicillin- resistant Staphylococcus aureus (MRSA) from South Africa. University of the Witwatersrand, Johannesburg (2007).
- Ma XX, Ito T, Tiensasitorn C, et al. Novel Type of Staphylococcal Cassette Chromosome mec Identified in Community-Acquired Methicillin-Resistant Staphylococcus aureus Antimicrob. Agents Chemother 46 (2004): 1147-1152.
- Jin Y, Zhou W, Yin Z, et al. The genetic feature and virulence determinant of highly virulent community-associated MRSA ST338-SCCmec Vb in China. Microbes Infect 10 (2021): 1052-1064.
- Aung MS, Urushibara N, Kawaguchiya M, et al. Clonal diversity of methicillin-resistant Staphylococcus aureus (MRSA) from bloodstream infections in northern Japan: Identification of spermidine N-acetyltransferase gene (speG) in staphylococcal cassette chromosomes (SCCs) associated with type II and IV. Glob. Antimicrob. Resist 24 (2021): 207-214.
- Tao C, Chen J, Hsu B, et al. Molecular Evaluation of Traditional Chicken Farm-Associated Bioaerosols for Methicillin-Resistant Staphylococcus aureus Antibiotics 10 (2021): 1-13.
- Marty E, Bodenmann C, Buchs J, et al. Prevalence of antibiotic resistance in coagulase-negative staphylococci from spontaneously fermented meat products and safety assessment for new starters. J. Food Microbiol 159 (2012): 74-83.
- Sahin-tóth J, Albert E, Juhász A, et al. Science of the Total Environment Prevalence of Staphylococcus aureus in wild hedgehogs ( Erinaceus europaeus ) and first report of mecC -MRSA in Hungary. Total Environ 815 (2022): 152858.
- Chen CJ, Tsai-Ling YL, Lu CT, et al. Clinical and molecular features of MDR livestock-associated MRSA ST9 with Staphylococcal Cassette Chromosome mec XII in humans. Antimicrob. Chemother 73 (2018): 33-40.
- Mcclure J, Conly JM, Zhang K. Characterizing a Novel Staphylococcal Cassette Chromosome mec with a Composite Structure from a Clinical Strain of. Agents Chemother 65 (2021): e00777-21.
- Bolte J, Zhang Y, Wente N, et al. Comparison of phenotypic and genotypic antimicrobial resistance patterns associated with Staphylococcus aureus mastitis in German and Danish dairy cows. Dairy Sci 103 (2020): 3554-3564.
- Rahi A, Kazemeini H, Jafariaskari S, et al. Genotypic and Phenotypic-Based Assessment of Antibiotic Resistance and Profile of Staphylococcal Cassette Chromosome mec in the Methicillin-Resistant Staphylococcus aureus Recovered from Raw Milk. Drug Resist 13 (2020): 273-283.
- Egyir B, Guardabassi L, Nielsen SS, et al. Prevalence of nasal carriage and diversity of Staphylococcus aureus among inpatients and hospital staff at Korle Bu Teaching Hospital, Ghana. Med. Res 1 (2013): 189-193.
- Mahomed TG, Kock MM, Masekela R, et al. Genetic relatedness of Staphylococcus aureus isolates obtained from cystic fibrosis patients at a tertiary academic hospital in Pretoria, South Africa. Rep 8 (2018): 1-6.
- Mekhloufi OA, Chieffi D, Hammoudi A, et al. Resistance of Staphylococcus aureus and Methicillin-Resistant Staphylococcus aureus ( MRSA ) Isolated from Algerian Ready to Eat Foods. Toxins (Basel) 13 (2021): 1-17.
- Budri PE, Shore AC, Coleman DC, et al. Observational cross-sectional study of nasal staphylococcal species of medical students of diverse geographical origin , prior to healthcare exposure?: prevalence of SCCmec , fusC , fusB and the arginine catabolite mobile element ( ACME ) in the absence. BMJ Open 8 (2018): e020391.
- Braulke WWC, Strommenger CCB, Heuck GWD, et al. Emergence of methicillin-resistant Staphylococcus aureus with Panton – Valentine leukocidin genes in central Europe. Eur J Clin Microbiol Infect Dis 24 (2005): 1-5.
- Aouati H, Hadjadj L, Aouati F, et al. Emergence of Methicillin-Resistant Staphylococcus aureus ST239 / 241 SCCmec -III Mercury in Eastern Algeria. Pathogens 10 (2021): 1-14.
- Egyir B, Guardabassi L, Monecke S, et al. Journal of Global Antimicrobial Resistance Methicillin-resistant Staphylococcus aureus strains from Ghana include. Med. Res 3 (2015): 26-30.
- Gostev V, Ivanova K, Kruglov A, et al. Comparative genome analysis of global and Russian strains of community-acquired methicillin-resistant Staphylococcus aureus ST22, a ‘Gaza clone’. J. Antimicrob. Agents 57 (2021): 106-264.
- Klein S, Boutin S, Heeg K, et al. Genomic structure of ST8-t008 USA300 and USA300-LV MRSA in the Rhine-Neckar Region, Germany, 2012–2018. J. Antimicrob. Agents 57 (2021): 106312.
- Abreu R, Rodríguez-Álvarez C, Lecuona M, et al. Increased antimicrobial resistance of MRSA strains isolated from pigs in Spain between 2009 and 2018. Sci 6 (2019).
- Bartels MD, Worning P, Andersen LP, et al. Repeated introduction and spread of the MRSA clone t304 / ST6 in northern Europe. Microbiol. Infect (2020): 4-8.
- Di Gregorio S, Haim S, Vallenilla V, et al. Genomic Epidemiology of CC30 Methicillin-Resistant Staphylococcus aureus Strains from Argentina Reveals Four Major Clades with Distinctive Genetic Features. mSphere 6 (2021): e01297-20.
- Cuiabano R, Leme P, José P, et al. Staphylococcus aureus skin and soft tissue infections in Latin America?: a systematic review. Brazilian J. Iinfectious Dis 5 (2021): 1-9.
- Mitevska E, Wong B, Surewaard BGJ, et al. The Prevalence, Risk, and Management of Methicillin-Resistant Staphylococcus aureus Infection in Diverse Populations across Canada?: A Systematic Review. Pathogens 10 (2021).
- Khan S, Marasa BS, Sung K. Genotypic Characterization of Clinical Isolates of Staphylococcus aureus from Pakistan. Pathogens 10 (2021).
- Achek R, Hotzel H, Cantekin Z, et al. Emerging of antimicrobial resistance in staphylococci isolated from clinical and food samples in Algeria. BMC Res. Notes, vol. 11 (2018): 1-7.
- Kaya H, Hasman H, Larsen J, et al. SCCmec Finder, a Web-Based Tool for Typing of Staphylococcal Cassette Chromosome mec in Staphylococcus aureus Using Whole-Genome Sequence Data. mSphere, vol. 3 (2018): 1-9.
- Monecke S, Baier V, Coombs GW, et al. Genome sequencing and molecular characterisation of Staphylococcus aureus ST772-MRSA-V, ‘bengal Bay Clone. BMC Res. Notes 6 (2013): 0-6.
- Monecke S, Slickers P, Gawlik D, et al. Molecular Typing of ST239-MRSA-III From Diverse Geographic Locations and the Evolution of the SCCmec III Element During Its Intercontinental Spread. Microbiol 9 (2018).
- Martha LH, Mclafferty AD, Erik RH. Nucleotide Sequence and Characterization of a Repetitive DNA Element from the Genome of Bordetella pertussis with Characteristics of an Insertion Sequence. Gen. Microbiol 134 (1988): 2297-2306.
- Leski T, Oliveira D, Trzcinski K, et al. Clonal Distribution of Methicillin-Resistant Staphylococcus aureus in Poland. Clin. Microbiol 36 (1998): 3532-3539.
- Havaei SA, Halaji M, Vidovic S, et al. Prevalence and Genotyping of Methicillin-Resistant and - Susceptible Staphylococcus aureus Strains Isolated from Patients in a University Hospital , Isfahan , Iran. Jundishapur J Microbiol 10 (2017): e13571.
- Turlej A, Hryniewicz W, Empel J. Staphylococcal Cassette Chromosome mec (SCC mec) Classification and Typing Methods?: an Overview. Polish J. Microbiol 60 (2011): 95-103.
- Gilbert M, MacDonald J, Gregson D, et al. Outbreak in Alberta of community-acquired (USA300) methicillin-resistant Staphylococcus aureus in people with a history of drug use, homelessness or incarceration Mark. CMAJ 175 (2006): 149-154.
- Oliveira DC, De Lencastre H. Multiplex PCR Strategy for Rapid Identification of Structural Types and Variants of the mec Element in Methicillin-Resistant Staphylococcus aureus. Agents Chemother 46 (2002): 2155-2161.
- Kondo S, Phokhaphan P, Tongsima S, et al. Molecular characterization of methicillin-resistant Staphylococcus aureus genotype ST764 - SCC mec type II in Thailand. Rep (2022): 1-8.
- Chanayat Y, Akatvipat A, Bender Jeff B, et al. The SCCmec Types and Antimicrobial Resistance among Methicillin-Resistant Staphylococcus Species Isolated from Dogs with Superficial Pyoderma. Sci 8 (2021): 1-10.
- Funaki T, Yasuhara T, Kugawa S, et al. SCC mec typing of PVL- positive community-acquired Staphylococcus aureus ( CA- MRSA ) at a Japanese hospital. Heliyon (2019): e01415.
- Francois P, Renzi G, Pittet D, et al. A Novel Multiplex Real-Time PCR Assay for Rapid Typing of Major Staphylococcal Cassette Chromosome mec J. Clin. Microbiol 42 (2004): 3309-3312.
- Kurt K, Alderborn A, Nilsson M, et al. Multiplexed genotyping of methicillin-resistant Staphylococcus aureus isolates by use of padlock probes and tag microarrays. Clin. Microbiol 47 (2009): 577-585.
- Galia L, Ligozzi M, Bertoncelli A, et al. Real-time PCR assay for detection of Staphylococcus aureus, Panton- Valentine Leucocidin and Methicillin Resistance directly from clinical samples. Microbiology 5 (2019): 138-146.
- Sadeghi Y, Salami SA, Kananizadeh RP, et al. Real-time PCR followed by high-resolution melting analysis – a new robust approach to evaluate SCC mec typing of methicillin-resistant Staphylococcus aureus. Microbiol 14 (2019): 155-164.
- Singh-moodley A, Lowe M, Mogokotleng R, et al. Diversity of SCC mec elements and spa types in South African Staphylococcus aureus mecA-positive blood culture isolates. BMC Infect. Dis 20 (2020): 1-12.
- Singh-moodley A, Strasheim W, Mogokotleng R, et al. Unconventional SCC mec types and low prevalence of the Panton-Valentine Leukocidin exotoxin in South African blood culture Staphylococcus aureus surveillance isolates, 2013-2016. PLoS One 14 (2019): e0225726.
- Alseqely M, Foot MN, Khalil A, et al. Association between fluoroquinolone resistance and MRSA genotype in Alexandria, Egypt. Rep 11 (2021): 1-9.
- Masaisa F, Kayigi E, Seni J, et al. Antibiotic Resistance Patterns and Molecular Characterization of Methicillin- Resistant Staphylococcus aureus in Clinical Settings in Rwanda. Am J Trop Med Hyg 99 (2018): 1239-1245.
- Shittu AO, Taiwo FF, Froböse NJ, et al. Genomic analysis of Staphylococcus aureus from the West African Dwarf ( WAD ) goat in Nigeria. Resist. Infect. Control 10 (2021): 1-12.
- Vitali LA, Petrelli D, Lamikanra A, et al. Diversity of antibiotic resistance genes and Staphylococcal Cassette Chromosome mec elements in faecal isolates of coagulase-negative staphylococci from Nigeria. BMC Microbiol 14 (2014): 1-8.
- McClure A, Jo-Ann JM, Osahon C, et al. A Novel Assay for Detection of Methicillin-Resistant Staphylococcus aureus Directly from Clinical Samples. Microbiol 11 (2020): 1-16.
- Dhaouadi S, Soufia L, Campanile F, et al. Prevalence of meticillin-resistant and -susceptible coagulase-negative staphylococci with the first detection of the mecC gene among cows, humans and manure in Tunisia. J. Antimicrob. Agents 55 (2020): 105826.
- Paterson GK. Low prevalence of livestock-associated methicillin-resistant Staphylococcus aureus clonal complex 398 and mecC MRSA among human isolates in North-West England. Appl. Microbiol 128 (2020): 1785-1792.
- Fisher EA, Paterson GK. Journal of Global Antimicrobial Resistance Prevalence and characterisation of methicillin-resistant staphylococci from bovine bulk tank milk in England and Wales. Med. Res 22 (2020): 139-144.
- Xu Z, Shah HN, Misra R, et al. The prevalence, antibiotic resistance and mecA characterization of coagulase negative staphylococci recovered from non-healthcare settings in London, UK. Resist. Infect. Control 7 (2018): 1-10.
- Luisa M, Dotto G, Mondin A, et al. Comparative Immunology, Microbiology and Infectious Diseases Prevalence and characterization of methicillin-resistant Staphylococcus pseudintermedius from symptomatic companion animals in Northern Italy?: Clonal diversity and novel sequence types. Immunol. Microbiol. Infect. Dis 66 (2019): 101331.
- Pirolo M, Gioffrè A, Visaggio D, et al. Prevalence , molecular epidemiology , and antimicrobial resistance of methicillin- resistant Staphylococcus aureus from swine in southern Italy. BMC Microbiol (2019): 1-12.
- Silva V, Ribeiro J, Rocha J, et al. High Frequency of the EMRSA-15 Clone ( ST22-MRSA-IV) in Hospital Wastewater. Microorganisms 147 (2022): 1-10.
- Román F, Mendez-Echevarria A, Rosa TD, et al. Characterization of methicillin-resistant Staphylococcus aureus strains colonizing the nostrils of Spanish children. Wile Microbiol. Open 10 (2021): 1-11.
- Krapf M, Müller E, Reissig A, et al. Molecular characterisation of methicillin-resistant Staphylococcus pseudintermedius from dogs and the description of their SCC mec elements. Microbiol 233 (2019): 196-203.
- Demirci M, Yigin A, Ekici S. In silico MLST, SCCmec AND SPA typing of strains and determination of resistance genes. J Basic Clin Heal. Sc 3 (2021): 171-178.
- Wu S, Huang J, Zhang F, et al. Prevalence and characterization of food-related methicillin-resistant Staphylococcus aureus (MRSA) in China Shi. Microbiol (2019): 1664-302x.
- Li X, Huang T, Xu K, et al. Molecular characteristics and virulence gene profiles of Staphylococcus aureus isolates in Hainan, China. BMC Infect. Dis 19 (2019): 1-12.
- Liang B, Tu C, Tan C, et al. Prevalence and characterization of Staphylococcus aureus isolated from women and children in Guangzhou, China. Microbiol 9 (2018): 1-9.
- Goudarzi M, Navidinia M, Dadashi M, et al. First report of methicillin-resistant Staphylococcus aureus carrying the mecC gene in human samples from Iran?: prevalence and molecular characteristics. New Microbes New Infect 39 (2021): 100832.
- Ahmadishoar S, Kazemi N, Sadeghi J, et al. Gene Reports Molecular epidemiology of clinical isolates of methicillin-resistant Staphylococcus aureus by multilocus sequence typing in northwestern ( Tabriz ) and southern ( Kerman ) of Iran?: The emergence of MRSA ST4848-SCC mec III. Gene Reports 24 (2021): 101212.
- Dehbashi S, Tahmasebi H, Zeyni B, et al. Regulation of virulence and β-lactamase gene expression in Staphylococcus aureus isolates: cooperation of two-component systems in bloodstream superbugs. BMC Microbiol 21 (2021): 1-12.
- Bakthavatchalam V, Karthick Y, Shoma V, et al. Genomic Portrait of Community-Associated Methicillin-Resistant Staphylococcus Aureus ST772-SCCmec V Lineage From India. Sq (2021): 1-19.
- Sivaraman GK, Muneeb KH, Sudha S, et al. Prevalence of virulent and biofilm forming ST88-IV-t2526 methicillin-resistant Staphylococcus aureus clones circulating in local retail fish markets in Assam, India. Food Control 127 (2021): 108098.
- Watanabe A, Watanabe T, Kokeguchi S, et al. Environmental survey of Methicillin-Resistant Staphylococci in a Hospital in Japan. Biocontrol Sci 26 (2021): 137-145.
- Mitsuboshi S, Yamaguchi T, Seino H, et al. Regional outbreak of methicillin-resistant Staphylococcus aureus ST2725-t1784 in rural Japan. Control Hosp. Epidemiol 42 (2021): 1294-1296.
- Albarrag A, Shami A, Almutairi A, et al. Prevalence and Molecular Genetics of Methicillin-Resistant Staphylococcus aureus Colonization in Nursing Homes in Saudi Arabia. J. Infect. Dis. Med. Microbiol 2020 (2020): 6.
- Goes ICRDS, Romero LC, Turra AJ, et al. Prevalence of nasal carriers of methicillin-resistant Staphylococcus aureus in primary health care units in Brazil. Rev Inst Med Trop São Paulo 63 (2021): e14.
- Teixeira NB, Magno C, Branco C, et al. Molecular characterization of methicillin - resistant Staphylococcus aureus among insulin-dependent diabetic individuals in Brazil. Clin. Microbiol. Antimicrob 20 (2021): 1-12.
- Oliveira CMC, De Yasmin MA, Alisson MFSA, et al. Description and Interrelationship Analysis of the Phenotypic and Genotypic Characteristics of MSSA and MRSA Strains Isolated from Healthcare Workers in North-eastern Brazil. Sqaure (2021): 1-17.
- Negrete-González C, Turrubiartes-Martínez E, Galicia-Cruz OG, et al. High prevalence of t895 and t9364 spa types of methicillin-resistant Staphylococcus aureus in a tertiary-care hospital in Mexico?: different lineages of clonal complex 5. BMC Microbiol 20 (2020): 1-11.
- Yosainix PCG, Patricia CGD, Lozano-Zarain C, et al. Genotyping of Antimicrobial Resistance and Virulence in Staphylococcus Isolated from Food of Animal Origin in Mexico. Indian J Microbiol (2018): 2-5.
- Ortíz-gil MÁ, Velazquez-meza ME, Echániz-aviles G, et al. Tracking methicillin-resistant Staphylococcus aureus clones in a hospital in Southern Mexico. Salud Publica Mex 62 (2020): 186-191.
- Bernier-Lachance MAJ, Arsenault J, Usongo V, et al. Prevalence and characteristics of Livestock- Associated Methicillin-Resistant Staphylococcus aureus (LA-MRSA) isolated from chicken meat in the province of Quebec. PLoS One 15 (2020): e0227183.
- Bastidas B, Méndez MV, Vásquez Y, et al. Typification of the Staphylococcal Chromosome Cassette of methicillin- resistant Staphylococcus aureus in the State of Aragua, Venezuela. Rev Peru Med Exp Salud Publica 37 (2020): 239-245.
- Lucía IS, Camila CH, Jaycia VK, et al. Brief report frequency of community-acquired methicilin-resistant Staphylococcus aureus in Peru. Rev Peru Med Exp Salud Publica 38 (2021): 313-317.
- Vanegas JM, Salazar-ospina L, Gallego MA, et al. A longitudinal study shows intermittent colonization by Staphylococcus aureus with a high genetic diversity in hemodialysis patients. J. Med. Microbiol 311 (2021): 151471.
- Beukers AG, Newton P, Hudson B, et al. A multicentre outbreak of ST45 MRSA containing deletions in the spa gene in New South Wales, Australia. Antimicrob Chemother (2020): 1-5.
- Ma GC, Worthing KA, Gottlieb T, et al. Molecular characterization of community-associated methicillin-resistant Staphylococcus aureus from pet dogs. Wiley 00 (2019): 1-9.
- Van Hal SJ, Steinig EJ, Andersson P, et al. Global Scale Dissemination of ST93?: A Divergent Staphylococcus aureus Epidemic Lineage That Has Recently Emerged from Remote Northern Australia. Microbiol 9 (2018): 1-11.


 Impact Factor: * 5.8
Impact Factor: * 5.8 Acceptance Rate: 71.20%
Acceptance Rate: 71.20%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 