Molecular Characterization of Colistin Resistance in Pseudomonas aeruginosa Isolates: Insights from a Study in Bangladesh
Izaz Mia MD1, Nooriya Haque2*, Shamsuzzaman SM3, Tarafder Mohammad Atiquzzaman4, Jannatun Fatema5, Raihan Raihan6
1Lecturer, Department of Microbiology, Manikganj Medical College, Manikganj, Dhaka, Bangladesh
2Assistant Professor (CC), Department of Microbiology, Green Life Medical College, Dhaka, Bangladesh
3Professor, Department of Microbiology, Dhaka Medical College, Dhaka, Bangladesh
4Medical Officer, Department of Paediatric Surgery, Bangladesh Medical University, Dhaka, Bangladesh
5Medical Officer, Baliati Union Health Sub Center, Saturia, Manikganj, Dhaka, Bangladesh
6Medical Officer, Department of ENT & Head-Neck Surgery, Dhaka Medical College & Hospital, Dhaka, Bangladesh
*Corresponding author: Nooriya Haque, Assistant Professor (CC), Department of Microbiology, Green Life Medical College, Dhaka, Bangladesh.
Received: 03 November 2025; Accepted: 11 November 2025; Published: 25 November 2025
Article Information
Citation: Izaz Mia, Nooriya Haque, Shamsuzzaman SM, Tarafder Mohammad Atiquzzaman, Jannatun Fatema, Muhammad Raihan. Molecular Characterization of Colistin Resistance in Pseudomonas aeruginosa Isolates: Insights from a Study in Bangladesh. Archives of Clinical and Biomedical Research. 9 (2025): 496-501.
View / Download Pdf Share at FacebookAbstract
Background: Pseudomonas aeruginosa is a major opportunistic pathogen responsible for severe hospital-acquired infections. Increasing antimicrobial resistance, particularly to colistin, which is the last-resort therapy for multidrug-resistant (MDR) Gram-negative bacteria, poses a critical challenge.
Objective: This study aimed to determine the prevalence, resistance profile, and colistin resistance genes among P. aeruginosa isolates from clinical samples.
Methodology: This cross-sectional study was conducted at the Department of Microbiology and Immunology at Dhaka Medical College Hospital (DMCH), Dhaka, Bangladesh. The research was conducted over one year, from January to December 2022. A total of 330 clinical specimens were subjected to culture and sensitivity testing. PCR screened colistin-resistant isolates for pmrA, pmrB, pmrC, phoP, and phoQ genes.
Results: Culture positivity was observed in 64.24% of samples, with wound swabs and pus showing the highest rate (72.41%), followed by endotracheal aspirates (70.00%). P. aeruginosa constituted 24.05% of isolates, predominantly from wound and urine samples. High resistance rates were noted for ciprofloxacin (88.23%), ceftazidime (84.31%), gentamicin (68.62%), and carbapenems (imipenem 56.86%, meropenem 60.78%). Colistin resistance was detected in 25.49% of isolates. Among these, 56.86% were MDR, 21.56% extensively drug-resistant (XDR), and 13.72% pandrugresistant (PDR). The pmrA gene was most frequent (46.15%), followed by pmrB (38.46%), pmrC and phoP (30.76% each), and phoQ (23.07%).
Conclusions: The predominance of MDR and XDR P. aeruginosa underscores an urgent need for antimicrobial stewardship and surveillance. Detection of multiple regulatory genes in colistin-resistant isolates suggests complex molecular mechanisms requiring continued genomic monitoring.
Keywords
<p>Pseudomonas aeruginosa; Molecular characterization; Bangladesh; Colistin resistance</p>
Article Details
1. Introduction
The increasing prevalence of multidrug-resistant (MDR) bacterial pathogens represents a major global health concern, limiting therapeutic options and leading to high morbidity and mortality rates [1]. Among these pathogens, Pseudomonas aeruginosa is particularly problematic due to its remarkable ability to acquire and express diverse resistance mechanisms, rendering many classes of antibiotics ineffective. This opportunistic pathogen is associated with severe infections in immunocompromised and critically ill patients, including ventilator-associated pneumonia, bloodstream infections, urinary tract infections, and wound infections [2].
With the diminishing efficacy of β-lactams, aminoglycosides, and fluoroquinolones, colistin (polymyxin E) has re-emerged as a last-line therapeutic option against MDR and XDR P. aeruginosa [3]. Colistin acts by disrupting the integrity of the bacterial outer membrane through binding to lipid A of lipopolysaccharides. However, increasing reports of colistin resistance have raised alarm, as resistance to this drug severely compromises treatment outcomes and leaves clinicians with few viable alternatives [4]. These resistance mechanisms have been reported in various Gram-negative microorganisms, including Salmonella enterica, K. pneumoniae, A. baumannii, P. aeruginosa, and E. coli. They are involved in the two-component system genes phoP/phoQ and pmrA/pmrB [5,6], PhoQ and PmrB proteins possess tyrosine kinase activity, which phosphorylates the regulator protein (PhoP or PmrA), activates the pmrHFIJKLM operon, and finally modifies the surface of bacteria by adding L-Ara4N or pEtN to lipid A [7]. PhoP/PhoQ is also regulated by the ColR/ColS and CprR/CprS systems. Mutations in these regulatory systems can lead to overexpression of PhoP/PhoQ in P. aeruginosa [8]. ParR/ParS is also involved in colistin resistance in P. aeruginosa, with upregulation of the LPS modification operon at sub-inhibitory concentrations of polymyxins [9]. The two-component systems found in P. aeruginosa are PhoP/PhoQ and PmrA/PmrB [10].
In this study, we investigated the amino acid substitution of PmrA-PmrB and PhoP-PhoQ in colistin-nonsusceptible P. aeruginosa (CNPA). Understanding the genetic basis of colistin resistance will aid in enhancing surveillance, promoting antimicrobial stewardship, and formulating strategies to curb the dissemination of resistant strains.
2. Materials and Methods
2.1 Study design and setting
This cross-sectional study was conducted in the Department of Microbiology and Immunology at Dhaka Medical College Hospital (DMCH), Dhaka, Bangladesh. The research was carried out over a year, from January to December 2022.
2.2 Isolation and Identification of P. aeruginosa
Clinical specimens, including wound swabs, urine, wound swabs, pus, tracheal aspirates, sputum, blood, and other body fluids, submitted to the Microbiology Laboratory at DMCH were processed for bacterial isolation. A total of 51 consecutive, non-duplicate isolates of P. aeruginosa were obtained from hospitalized patients during the study period. Phenotypic identification of P. aeruginosa was done by observing colony morphology on blood agar (white or cream-coloured, smooth to mucoid colonies, hemolytic), on MacConkey agar (generally form colourless colonies), Gram staining (gram negative bacilli), produces blue or green pigment and biochemical tests like-Oxidase test (positive), catalase tests (positive), TSI agar (butt-red, slant-red, no H2S or gas production), urease production (negative), indole test (negative), motility (motile) und citrate utilization test (positive). Growth at 42°C on agar was also done for confirmation of P. aeruginasa. For quality control, the reference strain P. aeruginosa ATCC 27853 was used during culture, biochemical testing, and phenotypic confirmation of clinical isolates. The study received ethical approval from the institutional review board of Dhaka Medical College, and written informed consent was secured from all participating patients.
2.3 Antimicrobial Susceptibility Testing
The antimicrobial susceptibility profiles of the Klebsiella pneumoniae isolates were determined using the standard Kirby-Bauer disk diffusion method on Mueller-Hinton agar (Oxoid Ltd., UK) [11]. A range of commercially prepared antibiotic discs was employed, including Amikacin (30 µg), Aztreonam (30 µg), Ceftazidime (30 µg), Cefepime (30 µg), Colistin (10 µg), Ciprofloxacin (5 µg), Gentamicin (10 µg), Imipenem (10 µg), Meropenem (10 µg), Fosfomycin (200µg), Piperacillin-tazobactam (100 µg/10 µg). Interpretation of inhibition zones was carried out under guidelines established by the Clinical and Laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST). To identify ESBL-producing strains, the double-disk synergy test was applied. Furthermore, isolates were categorized as multidrug-resistant (MDR), extensively drug-resistant (XDR), or pandrug-resistant (PDR) based on criteria outlined by the Centers for Disease Control and Prevention (CDC) [12,13].
2.4 MDR
Multidrug-resistant (MDR) isolates were defined as those that showed resistance to three or more classes of antipseudomonal agents (carbapenems, fluoroquinolones, penicillins/cephalosporins, and aminoglycosides) [13].
2.5 ESBL
ESBL production in all of the isolates was detected by the double disk synergy test as described by Jarlier [14]. Synergy was determined between a disk of amoxiclav (20µg amoxicillin and 10µg clavulanic acid) and a 30µg disk of each third-generation cephalosporin. The test antibiotic was placed 20 mm apart on a lawn culture of the isolate under test on Mueller-Hinton agar. The test organism was considered to produce ESBL if the zone size around the antibiotic disk increased towards the amoxiclav disk. This criterion also fulfills the CLSI guidelines [15]. This increase occurs because the clavulanic acid present in the amoxiclav disk inactivates the ESBL produced by the test organism [16].
2.6 Detection of colistin-resistant genes by Polymerase Chain Reaction (PCR)
Conventional polymerase chain reaction (PCR) was used to detect Colistin-resistant genes (pmrA, pmrB, PmrC, phoP, phoQ) in P. aeruginosa isolates. Genomic DNA was extracted using the boiling method. Amplified PCR products were visualized by agarose gel electrophoresis to confirm the presence of target genes.
2.7 Data analysis
All collected data were analyzed using IBM SPSS Statistics version 23. Categorical variables were summarized as frequencies and percentages. Graphs and visualizations were generated using the matplotlib library in Python.
3. Result
A total of 330 clinical samples were processed during the study period. Out of these, 212 (64.24%) yielded positive microbial growth. The culture positivity rate varied notably across sample types. The highest positivity was observed in wound swabs and pus samples (72.41%), followed by endotracheal aspirates (70.00%), and urine samples (58.33%). The lowest rates were found in blood (45.00%) and sputum (46.66%) samples (Table 1).
|
Sample Type |
Total Samples (n) |
Culture Positive (n) |
Culture Positive (%) |
|
Wound swab and pus |
145 |
105 |
72.41 |
|
Urine |
120 |
70 |
58.33 |
|
Endotracheal aspirate |
30 |
21 |
70.00 |
|
Blood |
20 |
9 |
45.00 |
|
Sputum |
15 |
7 |
46.66 |
|
Total |
330 |
212 |
64.24 |
Differences between the growth rate of various clinical samples are statistically significant (p<0.0001).
Table 1: Culture positivity from different clinical samples (N = 330).
A Chi-square test for independence was performed to assess whether the culture positivity rate was associated with sample type. The analysis revealed a statistically significant association between sample type and culture positivity (χ² = 13.42, df = 4, p = 0.009).
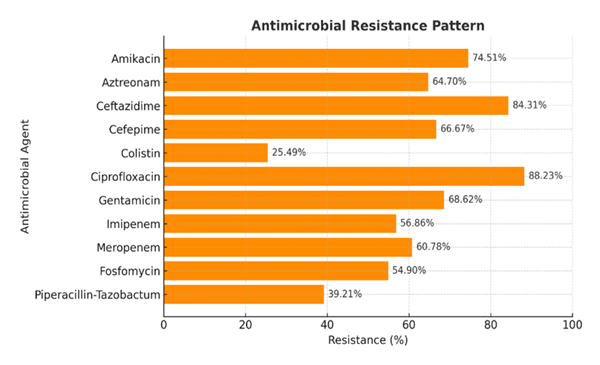
Among the 212 culture-positive samples, 51 (24.05%) were P. aeruginosa (Table 2), of which 45 (88.23%) were resistant to ciprofloxacin, 43 (84.31%) were resistant to ceftazidime, 20 (39.21%) were resistant to piperacillin-Tazobactum, and 13 (25.49%) were resistant to colistin (Figure 1).
|
Organisms |
WS & Pus (105) n (%) |
Urine (70) n (%) |
ETA (21) n (%) |
Blood (09) n (%) |
Sputum (07) n (%) |
Total (212) N (%) |
|
P. aeruginosa |
35 (33.33) |
12 (17.14) |
03 (14.28) |
01 (11.11) |
0 (0.00) |
51 (24.05) |
|
E. coli |
16 (15.23) |
41 (58.57) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
57 (26.88) |
|
Klebsiella spp |
16 (15.23) |
03 (4.28) |
06 (28.57) |
0 (0.00) |
05 (71.42) |
30 (14.15) |
|
Acinetobacter spp |
10 (9.52) |
04 (5.71) |
07 (33.33) |
02 (22.22) |
02 (28.57) |
25 (11.79) |
|
Enterobacter spp |
06 (5.71) |
03 (4.28) |
03 (14.28) |
02 (22.22) |
0 (0.00) |
14 (6.60) |
|
Proteus mirabilis |
05 (4.76) |
02 (2.85) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
07 (3.30) |
|
Proteus vulgaris |
01 (0.95) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
01 (0.47) |
|
Salmonella spp |
0 (0.00) |
0 (0.00) |
0 (0.00) |
03 (33.33) |
0 (0.00) |
03 (1.41) |
|
Staph. aureus |
14 (13.33) |
02 (2.85) |
02 (9.52) |
01 (11.11) |
0 (0.00) |
19 (8.96) |
|
Other Pseudomonas spp. |
02 (1.90) |
01 (1.42) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
03 (1.41) |
|
Coagulase-negative Staphylococcus |
0 (0.00) |
02 (2.85) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
02 (0.94) |
Table 2: Distribution of organisms isolated from different samples by biochemical tests (N = 212).
Among the 51 P. aeruginosa isolates, 29 (56.9%) were multidrug-resistant (MDR), 11 (21.6%) were extensively drug-resistant (XDR), and 7 (13.7%) were pan-drug resistant (PDR). A chi-square goodness-of-fit test demonstrated that the distribution of resistance patterns was significantly different from equal proportions (χ² = 17.53, df = 2, p < 0.001), with MDR being the predominant phenotype (Table 3).
Here, pmrA, pmrB, pmrC, phoP, and phoQ genes from different isolates of colistin-resistant P. aeruginosa were analyzed by PCR. Among 13 colistin-resistant isolates, 6 (46.15%) were positive for pmrA, 5 (38.46%) for pmrB, 4 (30.76%) for pmrC, 4 (30.76%) for phoP, and 3 (23.07%) for phoQ gene (Table 4).
|
Types of resistance |
n (%) |
|
MDR |
29 (56.86) |
|
XDR |
11 (21.56) |
|
PDR |
07 (13.72) |
Table 3: Types of antibiotic resistance patterns among the isolated P. aeruginosa (N = 51).
|
Samples |
pmr A n (%) |
pmr B n (%) |
pmr C n (%) |
pho P n (%) |
pho Q n (%) |
|
Wound swab & Pus (N = 8) |
4 (50.00) |
2 (25.00) |
1 (12.50) |
2 (25.00) |
1 (12.5) |
|
Urine (N = 2) |
0 (0.00) |
1 (50.00) |
2 (100.00) |
1 (50.00) |
0 (0.00) |
|
ETA (N = 2) |
1 (50.00) |
2 (100.00) |
0 (0.00) |
1 (50.00) |
1 (50.00) |
|
Blood (N = 1) |
1 (100.00) |
0 (0.00) |
1 (100.00) |
0 (0.00) |
1 (100.00) |
|
Sputum (N = 0) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
|
Total (N = 13) |
6 (46.15) |
5 (38.46) |
4 (30.76) |
4 (30.76) |
3 (23.07) |
Table 4: Distribution of pmrA, pmrB, pmrC, phoP, and phoQ genes among the colistin-resistant P. aeruginosa by PCR (N = 13).
4. Discussion
330 clinical samples were analyzed in the present study to determine the distribution and antimicrobial resistance profile of Pseudomonas aeruginosa, with particular emphasis on colistin resistance and associated regulatory genes. The findings revealed a concerning positivity rate of 64.24% for bacterial growth across the clinical samples, pointing to a considerable infection burden in the studied population, which is consistent with the findings of Munny et al. [17]. Notably, wound swabs and pus samples exhibited the highest culture positivity at 72.41%, followed closely by endotracheal aspirates, which showed a 70.00% positivity rate which is similar to the study of Siddiqua et al. [18]. This trend aligns with P. aeruginosa's known preference for moist environments and its opportunistic role as a pathogen, particularly among vulnerable patients, including those with burns, chronic wounds, and conditions leading to ventilator-associated pneumonia.
Among the culture-positive isolates, P. aeruginosa accounted for 24.05%, representing a significant proportion of the Gram-negative isolates. This prevalence aligns with previous reports highlighting P. aeruginosa as a major nosocomial pathogen responsible for a wide spectrum of infections, including wound, urinary, respiratory, and bloodstream infections. The relatively higher isolation rate from wound swabs and pus (33.33%) compared to other samples suggests that P. aeruginosa remains a predominant cause of wound and soft-tissue infections due to its intrinsic resistance mechanisms and its ability to form biofilms. About 90% of the P. aeruginosa isolates of this study were obtained from only three important specimens, e.g., wound swab, pus, and urine. Similar results had been obtained in India, reported by Andhale et. al. [19] and Pathi et. al. [20] in different studies.
Antimicrobial susceptibility testing revealed alarmingly high resistance rates among P. aeruginosa isolates. Ciprofloxacin resistance was most frequent (88.23%), followed by ceftazidime (84.31%), gentamicin (68.62%), cefepime (66.67%), and aztreonam (64.70%). Resistance to carbapenems, namely imipenem (56.86%) and meropenem (60.78%), further underscores the limited therapeutic options available for treating these infections. Gill et al. [21] reported that 70% P. aeruginosa were resistant to ceftazidime and 80% were resistant to amikacin, which is in agreement with the present study. Bhatt et al. [22] in India reported 77% resistance to ceftazidime which is also almost similar to the present study.
Although colistin resistance was relatively lower (25.49%), this is still clinically concerning, given that colistin is considered a last-resort antibiotic against multidrug-resistant (MDR) Gram-negative bacteria. Similar trends have been observed in Abd El-Baky et al. in Egypt (21.3%) [23].
More than half (56.86%) of the isolates exhibited a multidrug-resistant (MDR) phenotype, while 21.56% were extensively drug-resistant (XDR) and 13.72% were pandrug-resistant (PDR). Gill et al. [21] reported that 50% isolates were multidrug resistant, which is almost similar to the findings in the present study (21). Owlia et al. [24] in Iran reported that 54.5% of isolates were multidrug resistant and 33% were extensively resistant, which are also similar to the findings of this study. The predominance of MDR strains indicates a significant threat to effective antimicrobial therapy and highlights the urgent need for antimicrobial stewardship and infection control strategies. The emergence of XDR and PDR strains further emphasizes the adaptability of P. aeruginosa in acquiring multiple resistance determinants.
Molecular analysis of the 13 colistin-resistant P. aeruginosa isolates demonstrated the presence of various two-component regulatory system genes associated with colistin resistance, including pmrA, pmrB, pmrC, phoP, and phoQ. Among these, pmrA was the most frequently detected (46.15%), followed by pmrB (38.46%), pmrC and phoP (30.76% each), and phoQ (23.07%). Mostofa et al. found that 61.53% colistin-resistant P. aeruginosa were positive for pmrA, 38.5% for pmrB, 38.46% for phoP and 23.07% for phoQ [25]. Goli et al. [26] in Iran showed that the mutation of pmrB gene and expression change of pmrAB or phoPQ have occurred in colistin-resistant isolates [26]. In P. aeruginosa, mutations occurring in two-component regulatory systems are the primary mechanisms attributed to the development of resistance against colistin, according to Olaitan et al. [27]. These genes are known to mediate lipid A modification of lipopolysaccharides, leading to reduced colistin binding affinity and resistance. The detection of multiple resistance-associated genes within individual isolates suggests a complex regulatory interplay that contributes to colistin resistance. Similar findings have been reported in studies where mutations or overexpression of these genes were correlated with resistance phenotypes in P. aeruginosa.
5. Conclusion
The present study provides a comprehensive overview of the prevalence, antimicrobial resistance patterns, and molecular mechanisms underlying colistin resistance in Pseudomonas aeruginosa isolated from diverse clinical samples. The high culture positivity rate and the predominance of P. aeruginosa among Gram-negative isolates highlight its continued role as a major nosocomial pathogen, particularly in wound and soft-tissue infections. The alarming resistance rates to commonly used antibiotics, including fluoroquinolones, β-lactams, aminoglycosides, and carbapenems, reflect the growing challenge in managing P. aeruginosa infections within clinical settings.
Although the observed rate of colistin resistance was comparatively lower, its presence indicates significant clinical concern, as it is the last-line therapeutic option. The detection of multidrug-resistant, extensively drug-resistant, and pandrug-resistant phenotypes underscores the urgent need for enhanced antimicrobial stewardship and effective infection control measures. Molecular characterization further revealed the involvement of two-component regulatory system genes, particularly pmrA, pmrB, pmrC, phoP, and phoQ, in mediating colistin resistance, which is consistent with global reports implicating these genes in lipid A modification and reduced colistin susceptibility.
Overall, these findings emphasize the adaptive potential of P. aeruginosa and the necessity of continuous surveillance, rational antibiotic use, and molecular monitoring to mitigate the spread of resistant strains and preserve the efficacy of existing antimicrobial agents.
Ethical Clearance:
Ethical approval for this study was granted by the Institutional Review Board.
Financial Support and Sponsorship:
None.
Funding:
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Conflicts of Interest:
The authors declare no conflicts of interest.
References
- Haque N, Shamsuzzaman S, Mohammad Atiquzzaman T, et al. Uncovering Carbapenem Resistance: A Molecular Look at Klebsiella pneumoniae in Clinical Samples. Arch Microbiol Immunology 9 (2025): 1-10.
- Moradali MF, Ghods S, Rehm BHA. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front Cell Infect Microbiol 7 (2017): 1-10.
- Bialvaei AZ, Samadi Kafil H. Colistin, Mechanisms and Prevalence of Resistance. Curr Med Res Opin 31 (2015): 707-721.
- Aghapour Z, Gholizadeh P, Ganbarov K, et al. Molecular Mechanisms Related to Colistin Resistance in Enterobacteriaceae. Infect Drug Resist 12 (2019): 965-975.
- Needham BD, Trent MS. Fortifying the Barrier: The Impact of Lipid A Remodelling on Bacterial Pathogenesis. Nat Rev Microbiol 11 (2013): 467-481.
- Olaitan AO, Morand S, Rolain JM. Mechanisms of Polymyxin Resistance: Acquired and Intrinsic Resistance in Bacteria. Front Microbiol 5 (2014): 1-10.
- Ayoub Moubareck C. Polymyxins and Bacterial Membranes: A Review of Antibacterial Activity and Mechanisms of Resistance. Membranes 10 (2020): 181.
- Gutu AD, Sgambati N, Strasbourger P, et al. Polymyxin Resistance of Pseudomonas aeruginosa phoQ Mutants Is Dependent on Additional Two-Component Regulatory Systems. Antimicrob Agents Chemother 57 (2013): 2204-2215.
- Fernández L, Gooderham WJ, Bains M, et al. Adaptive Resistance to the “Last Hope” Antibiotics Polymyxin B and Colistin in Pseudomonas aeruginosa Is Mediated by the Novel Two-Component Regulatory System ParR-ParS. Antimicrob Agents Chemother 54 (2010): 3372-3382.
- McPhee JB, Lewenza S, Hancock REW. Cationic Antimicrobial Peptides Activate a Two-Component Regulatory System, PmrA-PmrB, That Regulates Resistance to Polymyxin B and Cationic Antimicrobial Peptides in Pseudomonas aeruginosa. Mol Microbiol 50 (2003): 205-217.
- Bauer AW, Kirby WMM, Sheris JC, et al. Antibiotic Susceptibility Testing by a Standardized Single Disk Method. Am J Clin Pathol 45 (1966): 493-496.
- Wayne PA. Clinical and Laboratory Standards Institute: Performance Standards for Antimicrobial Susceptibility Testing: 20th Informational Supplement. CLSI Document M100–S20 (2018).
- Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin Microbiol Infect 18 (2012): 268-281.
- Jarlier V, Nicolas MH, Fourneir G. Extended Spectrum β-Lactamases Conferring Transferable Resistance to Newer β-Lactam Agents in Enterobacteriaceae: Hospital Prevalence and Susceptibility Patterns. Rev Infect Dis 10 (1998): 867-878.
- Clinical Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. Sixteenth International Supplement. CLSI Document M100–S16, Wayne PA (2007).
- Aggarwal R, Chaudhary U, Bala K. Detection of Extended-Spectrum β-Lactamase in Pseudomonas aeruginosa. Indian J Pathol Microbiol 51 (2008): 222-224.
- Munny NN, Shamsuzzaman SM, Hossain T. In Vitro and In Vivo Evaluation of Antibiotic Combination Against Multidrug Resistant Enterobacter Species Isolated from Patients of a Tertiary Care Hospital, Bangladesh. Am J Infect Dis Microbiol 9 (2021): 98-105.
- Siddiqua M, Alam AN, Akter S, et al. Antibiotic Resistance Pattern in Pseudomonas aeruginosa Isolated from a Private Medical College Hospital. KYAMC J 9 (2018): 16-19.
- Andhale JD, Misra RN, Gandham NR, et al. Incidence of Pseudomonas aeruginosa with Special Reference to Drug Resistance and Biofilm Formation from Clinical Samples in Tertiary Care Hospital. J Pharm Biomed Sci 6 (2016): 387-391.
- Pathi B, Mishra SN, Panigrahi K, et al. Prevalence and Antibiogram Pattern of Pseudomonas aeruginosa in a Tertiary Care Hospital from Odisha, India. Transworld Med J 1 (2013): 77-80.
- Gill JS, Arora S, Khanna SP, et al. Prevalence of Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Pseudomonas aeruginosa from a Tertiary Level Intensive Care Unit. J Global Infect Dis 8 (2016): 155-160.
- Bhatt P, Rathi KR, Hazra S, et al. Prevalence of Multidrug Resistant Pseudomonas aeruginosa Infection in Burn Patients at a Tertiary Care Centre. Indian J Burn 236 (2015): 56-59.
- Abd El-Baky RM, Masoud SM, Mohamed DS, et al. Prevalence and Some Possible Mechanisms of Colistin Resistance among Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa. Infect Drug Resist 13 (2020): 323-332.
- Owlia P, Nosrati R, Alaghehbandan R, et al. Antimicrobial Susceptibility Differences among Mucoid and Non-Mucoid Pseudomonas aeruginosa Isolates. GMS Hyg Infect Control 9 (2014): Doc13.
- Mostofa HA, Shamsuzzaman SM, Hasan MM. Colistin Susceptibility Pattern in Gram-Negative Bacilli Isolated from Patients of Dhaka Medical College Hospital with Distribution of Antibiotic Resistance Genes among Them. Microbiology (2020): 1-10.
- Vaez H, Salehi-Abargouei A, Ghalehnoo Z, et al. Multidrug Resistant Pseudomonas aeruginosa in Iran: A Systematic Review and Meta-Analysis. J Global Infect Dis 10 (2018): 212–218.
- Olaitan AO, Morand S, Rolain JM. Mechanisms of Polymyxin Resistance: Acquired and Intrinsic Resistance in Bacteria. Front Microbiol 5 (2014): 1-10.


 Impact Factor: * 5.8
Impact Factor: * 5.8 Acceptance Rate: 71.20%
Acceptance Rate: 71.20%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 