The LOX1 Gene Polymorphisms and Coronary Artery Disease in North Indian Population
Naindeep Kaur1, Jagtar Singh2, Sreenivas Reddy3*
1Department of Biotechnology, Panjab University, Chandigarh, India
2Department of Biotechnology, Panjab University, Chandigarh, India
3Department of Cardiology, Postgraduate Institute of Medical Education and Research, Chandigarh, India
*Corresponding Author: Sreenivas Reddy, Department of Cardiology, PGIMER 160012, Chandigarh, India
Received: 11 March 2019; Accepted: 12 April 2019; Published: 16 April 2019
Article Information
Citation: Naindeep Kaur, Jagtar Singh, Sreenivas Reddy. The LOX1 Gene Polymorphisms and Coronary Artery Disease in North Indian Population. Archives of Clinical and Biomedical Research 3 (2019): 021-035.
View / Download Pdf Share at FacebookAbstract
Background: The lectin-like oxidized-LDL receptor 1 (LOX1) is considered as a key receptor for internalizing oxidized LDL (oxLDL) and hence influencing manifestation of coronary artery disease (CAD).
Aim: The study aimed to explore the association of LOX1 rs11053646 G/C and rs1050283 C/T polymorphisms and the risk of CAD in North Indian population.
Methods: Angiographically confirmed 500 CAD patients and 500 healthy controls were recruited and genotyped by PCRRFLP.
Results: Multiple logistic regression revealed mutant genotype of both the polymorphisms viz. CC of rs11053646 and TT of rs1050283 to confer CAD risk after adjustment for confounders (p<0.001, OR=3.072, 95% CI (1.765-5.347) and p<0.001, OR=3.487, 95% CI (1.945-6.254) respectively). Risk association was reported in recessive model for both the polymorphisms indicating that two copies of the allele are required for disease manifestation. Stratified analysis on the basis of gender showed risk association in males for both the polymorphisms with the mutant genotype exhibiting highly significant p values. However, only the LOX1 rs11053646 G/C showed the risk association in females in the mutant genotype with OR=3.179, 95% CI (1.071-9.438) and p=0.037. Stratified analysis for age reported that the mutant genotypes for both the polymorphisms significantly contributed to CAD risk in subjects above 40 years of age with OR=3.702, 95% CI (1.962-6.983) and a highly significant p<0.001 for rs11053646 G/C and OR=4.308, 95% CI (2.198-8.445) and p<0.001 for rs1050283 C/T.
Conclusion: The study reveals that LOX1 rs11053646 G/C and rs1050283 C/T polymorphisms increase CAD risk in the
Keywords
<p>Genetic polymorphism; Coronary artery disease; North Indian Population; ARMS-PCR; epidemiology study; LDL metabolism</p>
Article Details
1. Introduction
Coronary artery disease (CAD) has become an epidemic worldwide and a major barrier to sustainable human development. According to “The 2017 Heart Disease and Stroke Statistics update” of the American Heart Association, around 16.5 million people who are above 20 years of age in United States of America suffer from CAD. Not only that, the prevalence increases in both the genders with a gradual increase in the age [1]. The incidence in the developing countries like India is also alarming. There are a number of established key modifiable and non-modifiable factors like age, gender, genetics, smoking, dyslipidemia, hypertension, diabetes, obesity, high-fat diet, physical inactivity, drug abuse, alcohol consumption and mental stress attributing significant risk towards the disease. An individual’s risk of harboring CAD is inflected by the interplay between genetic and lifestyle factors established by the multifactorial nature of CAD. Atherosclerosis is a key event driving the pathogenesis of CAD. The lectin-like oxidized-LDL receptor 1 (LOX1) is a 50 kDa transmembrane receptor expressed on monocytes, macrophages, vascular smooth muscle cells, platelets and fibroblasts [2-4] and considered prime receptor for oxidized LDL (oxLDL). LOX1 was first cloned in 1997 by Sawamura et al. [5]. LOX is a member of C-type lectin family and has four domains: short cytoplasmic N-terminal domain, transmembrane domain, connecting neck domain and long extracellular C-type lectin like domain [2, 6, 7, 8, 9]. The latter is also called the carbohydrate recognition domain, is the functional domain for binding of oxLDL [10] and found to be highly conserved among species [2, 8, 11]. Lectin-like domain is made up of positively charged amino acids which recognize the negative charges on oxLDL (the modifications on the apo B moiety) [9, 10].
LOX1 binds with a greater affinity to oxLDL as compared to unoxidized LDL and is a key molecule accountable for the binding, internalization and degradation of oxLDL in the endothelial cells [2]. This binding triggers a cascade of events leading to formation of foam cells, vascular smooth muscle cells proliferation, platelet activation, collagen degradation and reactive oxygen species generation [12]. Therefore LOX1 plays eminent role in growth of atherosclerosis. The soluble LOX1 has been shown to be increased in vascular carotid plaque in patients with ischaemic stroke [13]. Seven genetic polymorphisms have been reported in the LOX1 gene. One of these polymorphisms viz. the G to C change in exon 4 at position 501 of LOX1 gene results in (Lys/Asn) in codon 167 [14]. The amino acid at 167 resides in C-terminal lectin-like domain and mediates oxLDL binding. Presence of basic residues in this domain strengthens the ligand binding and if these residues get substituted, this will lead to a reduced binding and thus, internalization of oxLDL [12]. Multiple studies report this polymorphic change to be associated with CAD [15-18]. One more polymorphism is reported in 3′ untranslated region (UTR), 188 bp downstream of stop codon in LOX1 resulting into a C to T change [14, 15, 19]. This polymorphic change influences the binding of nuclear proteins (NF-κB) and the polymorphic allele is known to be linked with heightened risk of CAD [15, 16, 19, 20]. Therefore, this study has attempted to explore the possible role of LOX1 rs11053646 and rs1050283 SNPs in CAD in North Indian population and its correlation with other selected parameters.
2. Material and Methods
2.1 Study population
A total of 1000 subjects aged 25-70 years of both the sexes were enrolled to evaluate the role of LOX1 rs11053646 G/C and rs1050283 C/T polymorphisms in CAD. Five hundred patients belonging to the North Indian states (Jammu and Kashmir, Haryana, Chandigarh, Punjab, Himachal Pradesh, New Delhi, Uttaranchal, Uttar Pradesh, Uttarakhand and Rajasthan) visiting the Department of Cardiology at Postgraduate Institute of Medical Education and Research, Chandigarh and angiographically documented CAD with more than 50% stenosis in at least one epicardial coronary artery) were registered as cases. Patients with acute or chronic infection, hepatic dysfunction, renal dysfunction, severe heart failure, hypo or hyperthyroidism, pregnancy and malignancy were excluded. Five hundred healthy individuals satisfying the inclusion criteria (with absence of any cardiac disorder, chronic diseases such as diabetes, hypertension, hypo- or hyperthyroidism, tuberculosis, hepatitis, AIDS, malignancy and pregnancy) were enrolled as controls. Subjects with history of smoking, alcohol consumption and tobacco chewing were also excluded. Majority of the controls were donors at the blood donation camps. Informed consent was obtained from all individual participants included in the study.
2.2 Biometric and biochemical measurements
Anthropometric parameters like height, weight, waist to hip ratio, BMI and blood pressure were noted. Risk factors for CAD like diabetes, hypertension, dyslipidemia, family history, smoking and drinking habits were recorded. Lipid profile, fasting blood glucose, hsCRP, uric acid Apolipoprotein A1 and Apolipoprotein B and were determined by standard biochemical methods.
2.3 DNA isolation and SNP selection and genotyping
Five ml of venous blood sample was collected in EDTA-coated vials, stored at -80°C until genomic DNA isolation using the sodium saline citrate buffer method [21]. Genotyping was done by RFLP-PCR by using primer sequences given in Table 1.
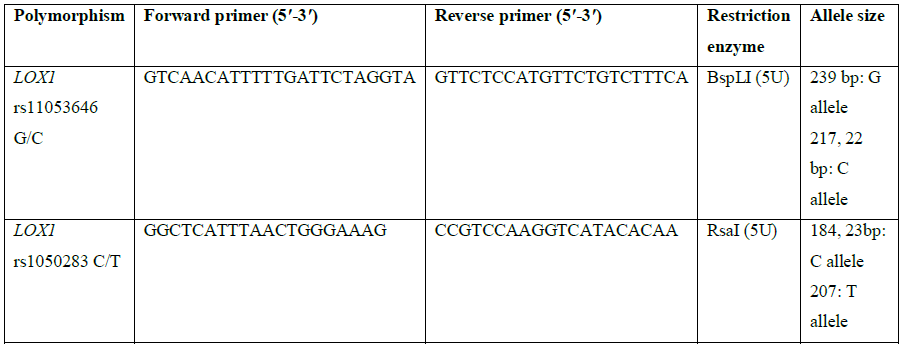
Table 1: Primer sequences and restriction enzymes used for genotyping of LOX1 polymorphisms.
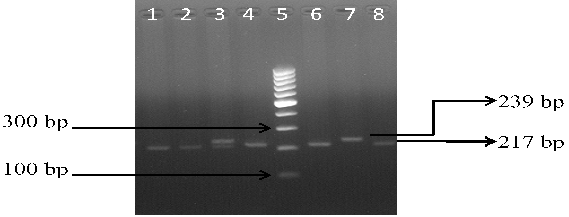
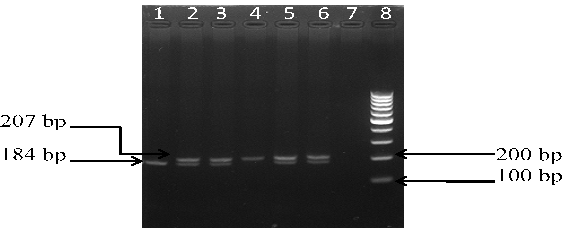
The PCR was carried out in a thermal cycler with a total volume of 25 µl containing: 10 × PCR Buffer, 2.5 mM MgCl2, 1 mg/mL BSA, 50 pmol of each primers, 10 mM of each dNTP, 0.125 U Taq polymerase and 2 µl genomic DNA. The PCR cycles comprised of initial denaturation at 94°C for 5 min, followed by 35 cycles at 94°C for 40 s, 58°C for 35 sec (for rs11053646 G/C) and 60°C for 40 sec (for rs1050283 C/T), 72°C for 40 s, and final extension at 72°C for 10 min. Ten μl of PCR product for both polymorphisms underwent overnight restriction digestion at 37°C with the respective enzymes mentioned in Table 1 and the results were visualized by UV transillumination after running the samples on 3% agarose gel electrophoresis. For rs11053646 G/C polymorphism, the wild-type allele remained uncut as a 239 bp band after digestion. The heterozygote (GC) had bands at 239 bp, 217 bp and 22 bp while the mutant (CC) genotype identified as a 217 bp and 22 bp band (Figure 1). For rs1050283 C/T polymorphism, the wild (CC) genotype was seen as two bands of 184 bp and 23 bp. The heterozygote (CT) was visualised as 207 bp, 184 bp and 23 bp bands while the mutant (TT) genotype was observed as a 207 bp band only (Figure 2). Random retyping of samples was done to check for homology of results.
Lane 1, 2, 4, 6, 8: homozygous mutant CC genotype (217 bp); Lane 3: heterozygous GC genotype (239 and 217 bp); Lane 7: homozygous wild GG genotype (239 bp); Lane 5: 100 bp ladder
Lane 1: homozygous wild CC genotype (184 bp); Lane 2, 3, 5, 6: heterozygous CT genotype (207 and 184 bp); Lane 4: homozygous mutant TT genotype (207 bp); Lane 8: 100 bp ladder
2.4 Statistical analysis
Continuous variables were expressed as the mean ± standard deviation. Chi-square test was used to calculate the difference between baseline characteristics. Multivariate logistic regression was applied to investigate the association of SNP and the susceptibility to CAD, adjusted for age and gender. Furthermore, recessive and dominant models were analyzed. Stratified analysis for gender and age was also done. Odds ratio (OR) and 95% confidence interval (CI) were used for the assessment of association. A p<0.05 was considered as statistically significant for all the analysis. All the statistical analysis was performed with SPSS version 20.0 (SPSS, Inc., Chicago, IL) and Epi Info version 3.4.7 (CDC, Atlanta, GA).
3. Results
The baseline parameters of patients and controls are listed in Table 2. The results revealed a statistically significant variation between the two groups with respect to age, gender, smoking, drinking, waist to hip ratio, lifestyle, family history, dyslipidemia, diabetes, diet, hypertension, occupation, exercise, fasting blood sugar, uric acid, TC, VLDL, LDL, Apo A1, Apo B, but not with total lipids, triglycerides, BMI, HDL and hsCRP.
|
Phenotypic traits |
Controls n (%) |
Cases n (%) |
p |
|
Age (Mean ± SD; years) |
50.95 ± 10.18 |
56.08 ± 9.55 |
<0.001* |
|
Waist to hip ratio |
0.91 ± 0.08 |
0.97 ± 0.16 |
0.001* |
|
Blood Pressure |
|||
|
SBP (mmHg) |
121.13 ± 9.92 |
133.67 ± 15.25 |
0.001* |
|
DBP (mmHg) |
80.35 ± 7.07 |
90.81 ± 13.45 |
0.046* |
|
Gender |
|||
|
Males |
370 (74) |
397 (79.4) |
0.043* |
|
Females |
130 (26) |
103 (20.6) |
|
|
BMI (kg/m2) |
0.069 |
||
|
Underweight ≤18.5 |
5 (1) |
13 (2.6) |
|
|
Normal weight = 18.5-24.9 |
265 (53) |
283 (56.6) |
|
|
Overweight = 25-29.9 |
168 (33.6) |
159 (31.8) |
|
|
Obese ≥ 30 |
62 (12.4) |
45 (9) |
|
|
Smoking status |
|||
|
Non-smoker |
Nil |
327 (65.4) |
<0.001* |
|
Smoker |
173 (34.6) |
||
|
Drinking status |
|||
|
Nondrinker |
Nil |
348 (69.6) |
<0.001* |
|
Drinker |
152 (30.4) |
||
|
Address |
|||
|
Rural |
233 (46.6) |
248 (49.6) |
0.342 |
|
Urban |
267 (53.4) |
252 (50.4) |
|
|
Family history |
|||
|
Nil |
447 (89.4) |
304 (60.8) |
<0.001* |
|
+ve |
53 (10.6) |
196 (39.2) |
|
|
Lifestyle |
|||
|
Active |
431 (86.2) |
352 (70.4) |
<0.001* |
|
Sedentary |
69 (13.8) |
148 (29.6) |
|
|
Occupation |
|||
|
Home sitter/retired |
5 (1) |
111 (22.2) |
<0.001* |
|
Student |
35 (7) |
4 (0.8) |
|
|
Working |
354 (70.8) |
231 (46.2) |
|
|
Housewife |
79 (15.8) |
86 (17.2) |
|
|
Agriculturist |
18 (3.6) |
50 (10) |
|
|
Laborer |
9 (1.8) |
18 (3.6) |
|
|
Diabetes |
|||
|
Negative |
Nil |
353 (70.6) |
<0.001* |
|
Positive |
147 (29.4) |
||
|
Hypercholesterolemia |
|||
|
Negative |
Nil |
500 (100) |
<0.001* |
|
Positive |
|||
|
Hypertension |
|||
|
Negative |
Nil |
500 (100) |
<0.001* |
|
Positive |
|||
|
Exercise |
|||
|
none |
334 (66.8) |
189 (37.8) |
<0.001* |
|
half hour once |
37 (7.4) |
110 (22) |
|
|
half hour twice |
85 (17) |
78 (15.6) |
|
|
one hour once |
0 (0) |
28 (5.6) |
|
|
one hour twice |
44 (8.8) |
95 (19) |
|
|
Diet |
|||
|
Veg |
425 (85) |
385 (77) |
0.001* |
|
Non veg |
75 (15) |
115 (23) |
|
|
CPK MB |
54.46 ± 45.81 |
34.36 ± 41.81 |
<0.001* |
|
CPKNAC |
135.76 ± 99.80 |
102.76 ± 79.10 |
<0.001* |
|
APO A1 |
144.56 ± 26.41 |
121.09 ± 42.36 |
<0.001* |
|
APO B |
98.67 ± 36.54 |
65.94 ± 23.77 |
<0.001* |
|
hsCRP |
4.56 ± 0.20 |
2.35 ± 0.22 |
<0.001* |
|
HDL-C |
52.35 ± 3.12 |
88.06 ± 9.11 |
0.673 |
|
LDL-C |
135.65 ± 22.33 |
76.54 ± 32.50 |
0.005* |
|
VLDL |
48.57 ± 21.08 |
34.83 ± 20.74 |
0.043* |
|
FBG |
82.73 ± 19.30 |
111.56 ± 39.59 |
0.016* |
|
URIC ACID |
9.80 ± 3.45 |
6.26 ± 5.10 |
0.006* |
|
TC |
275.76 ± 53.49 |
144.95 ± 37.22 |
0.045* |
|
TRIGLYCERIDES |
198.23 ± 82.45 |
145.37 ± 65.62 |
0.468 |
|
TL |
512.18 ± 116.05 |
436.05 ± 112.36 |
0.353 |
BMI-body mass index; TC-total cholesterol; TG-triglycerides; HDL-C-high-density lipoprotein cholesterol; LDL-C-low density lipoprotein cholesterol; FBG-fasting blood glucose; SBP-systolic blood pressure; DBP-diastolic blood pressure; *=significant
Table 2: Demographic characteristics of the studied population.
Distribution of allele frequencies of the selected polymorphism followed the Hardy-Weinberg Equilibrium. Comparing the allelic frequencies of LOX1 rs11053646 G/C and rs1050283 C/T polymorphisms, wild allele (G) in rs11053646 G/C was found to be more prevalent in cases (64.7%) as compared to controls (62.2%) and wild allele (C) in rs1050283 C/T (63.1%) was found to be marginally more prevalent in cases (63.1%) than in the control patients (62.1%) (Table 3). Likewise, the mutant allele (C) in rs11053646 G/C and (T) in rs1050283 C/T had higher frequency among the controls with OR=1.111, 95% CI (0.92-1.34) and non-significant p=0.245 in rs11053646 G/C while a remarkable protective association in rs1050283 C/T with OR=0.68, 95% CI (0.56-0.82) and p<0.001. In LOX1 rs11053646 G/C, homozygous (GG) genotype was found to have greater frequency in controls (46.2%) than patients (40.2%). The heterozygous (GC) showed higher prevalence in cases (49%) than controls (32%) with OR=1.447, 95% CI (0.848-2.466) and non-significant p=0.175. The homozygous mutant (CC) genotype also showed higher prevalence in controls (21.8%) than the patients (10.8%) with OR=3.072, 95% CI (1.765-5.347) and high risk association p<0.001. Significant protective association was seen in dominant model with OR=0.643, 95% CI (0.445-0.923) and p=0.018 whereas risk was seen in recessive model with OR=2.052, 95% CI (1.243-3.388) and p=0.005 respectively.
The genotypic frequencies in LOX1 rs1050283 C/T polymorphism showed the homozygous (CC) genotype to have increased trend among the controls (45.4%) in comparison to the patients (35.2%). The heterozygous (CT) genotype had higher occurrence among the cases (55.8%) than controls (33.4%) with OR=1.248, 95% CI (0.704-2.213) and non-significant p=0.449. The homozygous mutant (TT) genotype showed higher prevalence in controls (21.2%) than CAD patients (9.0%) with OR=3.487, 95% CI (1.945-6.254) and p<0.001. The analysis of the dominant model revealed a protective association with OR=0.477, 95% CI (0.328-0.693) and p<0.001 whereas OR=2.093, 95% CI (1.224-3.579) and p=0.007 was observed in recessive model (Table 3).
|
Genotypes |
Controls |
Cases |
Multiple logistic regression analysis |
|
|
500 (%) |
500 (%) |
p |
OR (95% CI) |
|
|
LOX1 rs11053646 G/C |
||||
|
GG |
231 (46.2) |
201 (40.2) |
(ref.) |
- |
|
GC |
160 (32) |
245 (49) |
0.175 |
1.447 (0.848-2.466) |
|
CC |
109 (21.8) |
54 (10.8) |
<0.001* |
3.072 (1.765-5.347) |
|
Alleles G |
622 (62.2) |
647 (64.7) |
(ref.) |
- |
|
C |
378 (37.8) |
353 (35.3) |
0.245 |
1.111 (0.92-1.34) |
|
Dominant Model |
||||
|
GG |
231 (46.2) |
201 (40.2) |
(ref.) |
- |
|
GC+CC |
269 (53.8) |
299 (59.8) |
0.018* |
0.643 (0.445-0.923) |
|
Recessive Model |
||||
|
GC+GG |
391 (78.2) |
446 (89.2) |
(ref.) |
- |
|
CC |
109 (21.8) |
54 (10.8) |
0.005* |
2.052 (1.243-3.388) |
|
LOX1 rs1050283 C/T |
||||
|
CC |
227 (45.4) |
176 (35.2) |
(ref.) |
- |
|
CT |
167 (33.4) |
279 (55.8) |
0.449 |
1.248 (0.704-2.213) |
|
TT |
106 (21.2) |
45 (9) |
<0.001* |
3.487 (1.945-6.254) |
|
Alleles C |
621 (62.1) |
631 (63.1) |
(ref.) |
|
|
T |
379 (37.9) |
369 (36.9) |
<0.001* |
2.150 (1.620-2.860) |
|
Dominant Model |
||||
|
CC |
227 (45.4) |
176 (35.2) |
(ref.) |
- |
|
CT+TT |
273 (54.6) |
324 (64.8) |
<0.001* |
0.477 (0.328-0.693) |
|
Recessive Model |
||||
|
CT+CC |
394 (78.8) |
455 (91) |
(ref.) |
|
|
TT |
106 (21.2) |
45 (9) |
0.007* |
2.093 (1.224-3.579) |
Ref. =reference; OR=odds ratio; CI=confidence interval; %=frequency; *=significant
Table 3: Genotypic and allelic frequencies of LOX1 rs11053646 G/C and rs1050283 C/T polymorphism in controls and cases.
Further, the data was stratified on the basis of age. The mutant genotypes considerably contributed to CAD risk in subjects above 40 years of age with OR=3.702, 95% CI (1.962-6.983) and a highly significant p<0.001 for rs11053646 G/C and OR=4.308, 95% CI (2.198-8.445) and p<0.001 for rs1050283 C/T (Table 4). Similarly, data was stratified on the basis of gender to figure out any gender specific associations. The LOX1 rs11053646 G/C and rs1050283 C/T polymorphism both revealed risk association in males with the mutant genotype exhibiting highly significant p values with OR=3.020, 95% CI (1.584-5.758) and OR=3.712, 95% CI (1.903-7.239) respectively (Table 4). However, only the LOX1 rs11053646 G/C showed the risk association in females in the mutant genotype with OR=3.179, 95% CI (1.071-9.438) and p=0.037.
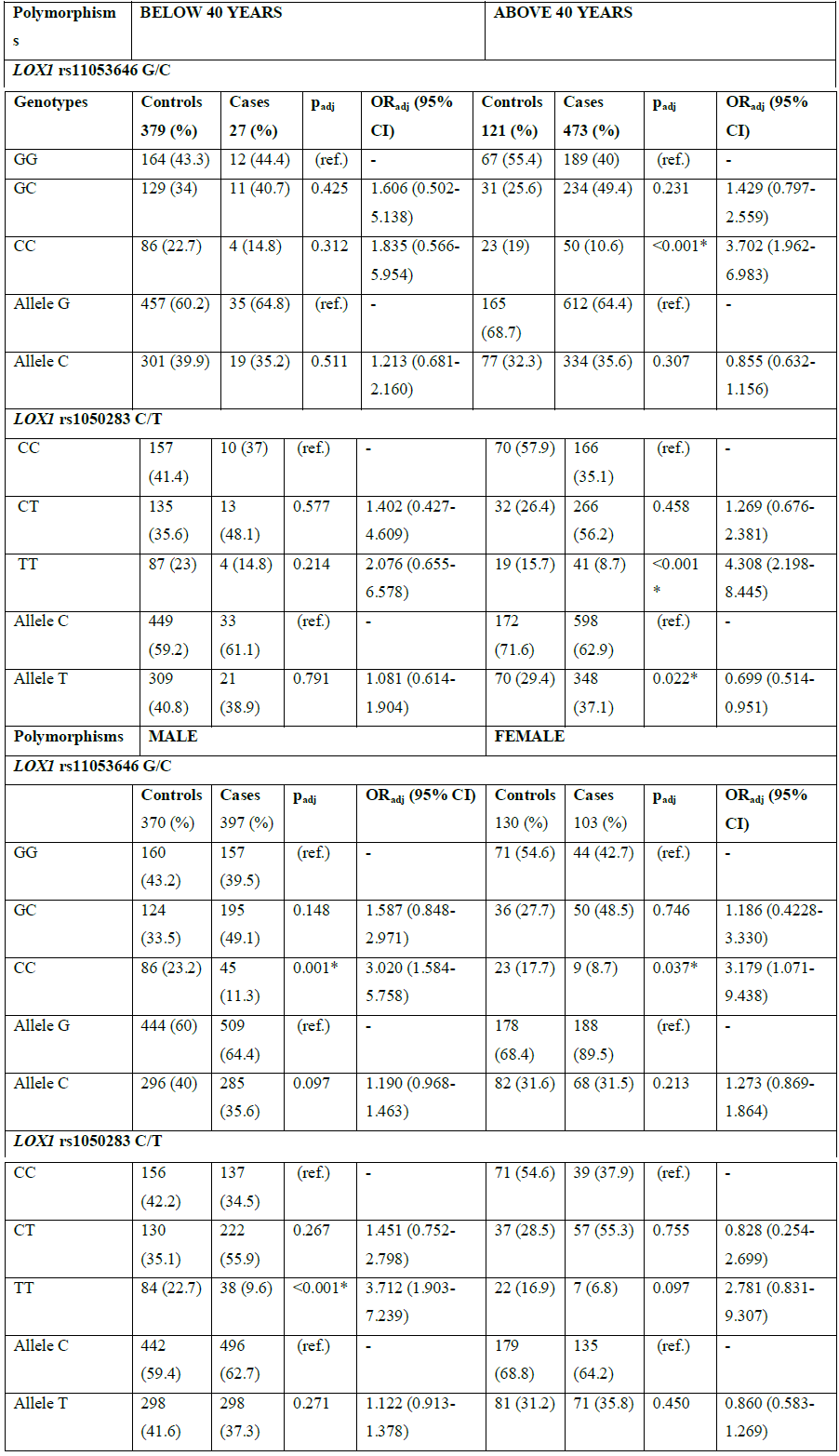
ref.=reference; OR=odds ratio; CI=confidence interval; %=frequency; *=significant
Table 4: Genotype and allele distributions for the age and gender stratified analysis for LOX1 polymorphisms.
Also, statistical analysis was performed to see the role of smoking as risk factor for CAD. Data revealed significant risk association with rs11053646 G/C in both the heterozygous and mutant genotypes with OR=3.982, 95% CI (1.644-5.496) and p=0.032 and OR=5.471, 95% CI (1.739-7.929) and p=0.027 respectively. Likewise, the risk association with heterozygous and mutant genotype was reported for rs1050283 C/T (OR=2.973, 95% CI (0.635-3.493) and p=0.002 and OR=4.930, 95% CI (1.060-6.520) and p=0.022 respectively) (Table 5).
|
Polymorphisms |
Non-Smoker 327 (%) |
Smoker 173 (%) |
P |
OR (95% CI) |
|
LOX1 rs11053646 G/C |
||||
|
GG |
131 (40.1) |
70 (40.5) |
(ref.) |
- |
|
GC |
158 (48.3) |
87 (50.3) |
0.032* |
3.982 (1.644-5.496) |
|
CC |
38 (11.6) |
16 (9.2) |
0.027* |
5.471 (1.739-7.929) |
|
Allele G |
440 (67.3) |
227 (65.6) |
(ref.) |
- |
|
Allele C |
234 (32.7) |
119 (34.4) |
0.646 |
1.805 (0.349-9.326) |
|
LOX1 rs1050283 C/T |
||||
|
CC |
116 (35.5) |
60 (34.7) |
(ref.) |
- |
|
CT |
181 (55.4) |
98 (56.6) |
0.002* |
2.973 (0.635-3.493) |
|
TT |
30 (9.2) |
15 (8.7) |
0.022* |
4.930 (1.060-6.520) |
|
Allele C |
413 (63.1) |
218 (63) |
(ref.) |
- |
|
Allele T |
241 (36.9) |
128 (37) |
0.993 |
1.281 (0.224-7.317) |
ref.=reference; OR=odds ratio; CI=confidence interval; %=frequency; *=significant
Table 5: Association of LOX1 polymorphisms with smoking.
4. Discussion
In this study, the LOX1 rs11053646 G/C and rs1050283 C/T polymorphisms were investigated and risk association towards CAD in a North Indian population was observed. Genotyping was performed by PCR-RFLP and results revealed the mutant genotype in both the polymorphisms conferred risk towards CAD in the studied population with statistical significance and risk was also observed under the recessive model (Table 2). During initiation of atherosclerosis, endothelial cells majorly express LOX1 but as the atherosclerotic lesions build up, the expression could also be seen on vascular smooth muscle cells and macrophages after being induced by oxLDL, pro-oxidative and biomechanical stimuli and pro-inflammatory cytokines [4, 22-25], LOX1 expression is upregulated in various disease conditions such as atherosclerosis [4, 26, 27], hypertension [28] and myocardial infarction [4]. LOX1 further promotes the secretion of various pro-inflammatory cytokines like IL-6, MCP-1, IL-8 or TNF-α by NF-κB pathway hence, promoting the plaque formation [29-30].
A G>C change in the coding region of the LOX1 at 501 position (rs11053646 G/C) has been identified that substitutes lysine at 167 position in the C terminal domain to asparagine. Functional analysis studies report that this substitution leads to a reduced oxLDL binding and uptake in vitro. Also, an altered oxLDL-induced LOX1 expression is also observed [22, 31]. Mango examined the rs11053646 G/C polymorphism and reported minor allele frequency lower in the MI group than controls (9% vs. 18%) and pointed towards the protective role of C allele [15]. In a study on Turkish population [32], the frequency of GG genotype and the G allele was found to be higher in patients than controls (p<0.05), while frequency of the CC genotype was high in control group but our results showed the frequency of GG was high in controls but the G allele was more prevalent in the control group. The results of the present piece of work also reveal lesser frequency of C allele in the patient group but no significant association could be observed. In a study conducted on Indian population, a significant predisposition towards CAD for this SNP was reported (p<0.001, OR=1.99 and 95% CI (1.42-2.78)) [33]. A Japanese study on 102 MI patients and 102 controls revealed significant risk association of rs11053646 with MI with OR=2.89, 95% CI (1.51-5.53) and p=0.002 in their study [17] which is comparable to our findings (OR=3.072, 95% CI (1.765-5.347) and p<0.001). In contrast to the above reported studies, [34] failed to report any association between rs11053646 polymorphism and acute MI in Italian population of 350 patients and 327 controls, [14] also reported no association between this polymorphism in a Japanese population comprising of 235 patients with ischaemic cerebrovascular disease in 274 age and gender matched healthy controls. In the current work, a significant correlation with the rs11053646 G/C polymorphism and the lipid profile was observed but [32] have failed to find any relationship with the lipid levels.
The LOX1 rs1050283 C/T resides 188 bp downstream to stop codon in LOX1 gene in the UTR and can affect transcription or modulate exon splicing or influence the binding affinity to the putative regulatory element [14, 19]. Thus, population studies have been conducted to uncover whether LOX1 rs1050283 C/T has a role in CAD but associations remains controversial. A study done by [35] exhibited a significant risk association between LOX1 rs1050283 T allele and reduced plasma LOX1 levels in a Caucasian and Afro-Caribbean population. The contribution of the polymorphism to CAD risk has been supported by two Italian studies. One study by Mango et al. [15] comprised of 150 acute MI patients and 103 controls reported significant risk association of rs1050283 C/T with OR of 3.74 and the findings were supported by Novelli et al. [20] in their study on 496 subjects revealing the minor allele T is related with increased risk towards the disease. The data was also in concordance to the study reported by Chen et al. [19] on the American population. In the present study, risk association has been observed with mutant genotype and under recessive model indicating that two copies of the polymorphic allele are required for disease manifestation. Examining the allelic frequencies has also revealed a significant risk association towards CAD in the studied population (Table 3). However, in a study comprising of 329 CAD patients and 331 controls from Indian population, no association was observed for this polymorphism with CAD (OR=1.13, 95% CI (0.89-1.43) and p=0.30) [33]. Nevertheless, this association with the disease risk and any of the parameters has not been supported by different studies on the Italian population [34, 36]. But significant association of smoking with CAD in Turkish population has been reported which is in consensus to the results given by our study [37]. They enrolled 83 cases and 99 controls and showed significant association with the three genotypes and SBP, HDL, VLDL and TG which is similar to the results obtained in the present piece of work.
5. Strengths and Limitations
The present study was performed in a well characterized set of individuals. Only those patients that were angiographically confirmed to have CAD were enrolled as cases. Even the control subjects underwent a close examination by the physician keeping in mind the inclusion and the exclusion criteria. Strict caution was observed that all the subjects belonged to the North Indian descent. This study documents the allelic and genotypic frequencies for the LOX1 rs11053646 G/C and rs1050283 C/T polymorphisms and risk association with CAD. Moreover, age and gender specific risk associations are also reported. However genotyping only two polymorphisms of the LOX1 is a major drawback of this study. Genotyping multiple SNPs along with the linkage analysis would have led to a better understanding to elucidate the role of LOX1 polymorphisms in CAD. Studies at transcriptional and translational level will add more to the existing knowledge. The risk associations with respect to age and gender demand more research to quantify the sex hormones at different intervals of time so as to find the missing links and hence have a robust CAD risk assessment in the North Indian population.
6. Conclusion
The current study revealed significant risk association of LOX1 rs11053646 G/C and rs1050283 C/T polymorphisms with CAD in the North Indian population and moreover the risk associations were age and gender dependant also. There is an utmost requirement to have a proper SNP data regarding the genetic polymorphisms throughout the world that can be implemented for genetic and molecular biomarker identification and in the field of personalized medicine. The inter- and intra-genic interactions also need to be studied to better understand the disease initiation and progression and hence develop better strategies to combat the disease.
Acknowledgments
All the authors are thankful to the participants who consented to be a part of the study.
Conflict of Interest
None
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
References
- Sanchis-gomar F, Perez-quilis C, Leischik R, Lucia A. Epidemiology of coronary heart disease and acute coronary syndrome. Annals of translational medicine 4 (2016).
- Vecchione L, Gargiul E, Borgiani P, Predazzi I, Mango R, et al. Genotyping OLR1 gene: a genomic biomarker for cardiovascular diseases. Recent patents on cardiovascular drug discovery 2 (2007): 147-151.
- Mingyi C, Narumiya S, Masaki T, Sawamura T. Conserved C-terminal residues within the lectin-like domain of LOX-1 are essential for oxidized low-density-lipoprotein binding. Biochemical Journal 355 (2001): 289-296.
- Kataoka H, Kume N, Miyamoto S, Minami M, Morimoto M, et al. Oxidized LDL modulates Bax/Bcl-2 through the lectinlike Ox-LDL receptor-1 in vascular smooth muscle cells. Arteriosclerosis, thrombosis, and vascular biology 21 (2001): 955-960.
- Sawamura T, Kume N, Aoyama T, Moriwaki H, Hoshikawa H, et al. An endothelial receptor for oxidized low-density lipoprotein. Nature 386 (1997): 73.
- Chen M, Masaki T, Sawamura T. LOX-1, the receptor for oxidized low-density lipoprotein identified from endothelial cells: implications in endothelial dysfunction and atherosclerosis. Pharmacology and therapeutics 95 (2002): 89-100.
- Chen M, Sawamura T. Essential role of cytoplasmic sequences for cell-surface sorting of the lectin-like oxidized LDL receptor-1 (LOX-1). Journal of molecular and cellular cardiology 39 (2005): 553-561.
- Chen XP, Du GH. Lectin-like oxidized low-density lipoprotein receptor-1: protein, ligands, expression and pathophysiological significance. Chinese medical journal 120 (2007): 421-426.
- Tate SI. Oxidized low-density lipoprotein receptor, LOX-1, on the endothelial cell-the receptor structure and functions of LOX-1 in atherogenesis. J Biol Macromol 7 (2007): 11-22.
- Mehta JL, Chen J, Hermonat PL, Romeo F, Novelli G. Lectin-like, oxidized low-density lipoprotein receptor-1 (LOX-1): a critical player in the development of atherosclerosis and related disorders. Cardiovascular research 69 (2006): 36-45.
- Murphy JE, Tedbury PR, Homer-vanniasinkam S, Walker JH, Ponnambalam S. Biochemistry and cell biology of mammalian scavenger receptors. Atherosclerosis 182 (2005): 1-15.
- Ellis TJ. LOX-1 genotype, dietary fat intake, and aerobic exercise training: influence on endothelial function, oxidative stress, lipoprotein-lipids, and soluble LOX-1 (2006).
- Skarpengland T, Skjelland M, Kong XY, Skagen K, et al. Increased Levels of Lectin?Like Oxidized Low?Density Lipoprotein Receptor?1 in Ischemic Stroke and Transient Ischemic Attack. Journal of the American Heart Association 7 (2018): 006479.
- Hattori H, Sonoda A, Sato H, Ito D, Tanahashi N, et al. G501C polymorphism of oxidized LDL receptor gene (OLR1) and ischemic stroke. Brain research 1121 (2006): 246-249.
- Mango R, Clementi F, Borgiani P, Forleo G, Federici M, et al. Association of single nucleotide polymorphisms in the oxidised LDL receptor 1 (OLR1) gene in patients with acute myocardial infarction. Journal of Medical Genetics 40 (2003): 933-936.
- Ohmori R, Momiyama Y, Nagano M, Taniguchi H, Egashira T, et al. An oxidized low?density lipoprotein receptor gene variant is inversely associated with the severity of coronary artery disease. Clinical cardiology 27 (2004): 641-644.
- Tatsuguchi M, Furutani M, Hinagata JI, Tanaka T, Furutani Y, et al. Oxidized LDL receptor gene (OLR1) is associated with the risk of myocardial infarction. Biochemical and biophysical research communications 303 (2003): 247-250.
- Wang L, Yanuck D, Beecham A, Gardener H, Slifer S, et al. A candidate gene study revealed sex-specific association between the OLR1 gene and carotid plaque. Stroke 42 (2011): 588-592.
- Chen Q, Reis SE, Kammerer C, Craig WY, Lapierre SE, et al. Genetic variation in lectin-like oxidized low-density lipoprotein receptor 1 (LOX1) gene and the risk of coronary artery disease. Circulation 107 (2003): 3146-3151.
- Novelli G, Borgiani P, Mango R, Lauro R, Romeo F. Further evidence that polymorphisms of the OLR1 gene are associated with susceptibility to coronary artery disease and myocardial infarction. Nutrition, Metabolism and Cardiovascular Diseases 17 (2007): 7-8.
- Roe BA, Crabtree JS, Khan AS. DNA isolation and sequencing, Wiley-Blackwell (1996).
- Hofmann A, Brunssen C, Morawietz H. Contribution of lectin-like oxidized low-density lipoprotein receptor-1 and LOX-1 modulating compounds to vascular diseases. Vascular pharmacology S1537-1891 (2017): 30171-30174.
- Morawietz H. LOX-1 and atherosclerosis: proof of concept in LOX-1–knockout mice. Am Heart Assoc 100 (2007): 1534-1536.
- Catar R, Muller G, Heidler J, Schmitz G, Bornstein S, et al. Low-density lipoproteins induce the renin-angiotensin system and their receptors in human endothelial cells. Hormone and Metabolic Research 39 (2007): 801-805.
- Ogura S, Kakino A, Sato Y, Fujita Y, Iwamoto S, et al. LOX-1. Circulation Journal 73 (2009): 1993-1999.
- Kume N, Kita T. Roles of lectin-like oxidized LDL receptor-1 and its soluble forms in atherogenesis. Current opinion in lipidology 12 (2001): 419-423.
- Mitra S, Goyal T, Mehta JL. Oxidized LDL, LOX-1 and atherosclerosis. Cardiovascular Drugs and Therapy 25 (2011): 419.
- Nagase M, Ando K, Nagase T, Kaname S, Sawamura T, FUJITA T. Redox-sensitive regulation of lox-1 gene expression in vascular endothelium. Biochemical and biophysical research communications 281 (2001); 720-725.
- Xu S, Ogura S, Chen J, Little PJ, Moss J, et al. LOX-1 in atherosclerosis: biological functions and pharmacological modifiers. Cellular and Molecular Life Sciences 70 (2013): 2859-2872.
- Pirillo A, Norata GD, Catapano AL. LOX-1, OxLDL, and atherosclerosis. Mediators of inflammation 2013 (2013): 152786.
- Biocca S, Falconi M, Filesi I, Baldini F, Vecchione L, et al. Functional analysis and molecular dynamics simulation of LOX-1 K167N polymorphism reveal alteration of receptor activity. PLoS One 4 (2009): e4648.
- Kurnaz O, Aydo?an HY, Isbir CS, Tekeli A, Isbir T. Is LOX-1 K167N polymorphism protective for coronary artery disease? in vivo 23 (2009): 969-973.
- Tripathi R, Tewari S, Ramesh V, Agarwal S. Oxidized LDL receptor 1 (OLR1) SNPs and CAD: a case-control association study in a North Indian population. Journal of Biological Research-Thessaloniki 18 (2012): 328-331.
- Trabetti E, Biscuola M, Cavallari U, Malerba G, Girelli D, et al. On the association of the oxidised LDL receptor 1 (OLR1) gene in patients with acute myocardial infarction or coronary artery disease. European journal of human genetics 14 (2006): 127-130.
- Brinkley TE, Kume N, Mitsuoka H, Brown MD, Phares DA, et al. Variation in the human lectin?like oxidized low?density lipoprotein receptor 1 (LOX?1) gene is associated with plasma soluble LOX?1 levels. Experimental physiology 93 (2008): 1085-1090.
- Sentinelli F, Filippi E, Fallarino M, Romeo S, Fanelli M, et al. The 3′-UTR C> T polymorphism of the oxidized LDL-receptor 1 (OLR1) gene does not associate with coronary artery disease in Italian CAD patients or with the severity of coronary disease. Nutrition, metabolism and cardiovascular diseases 16 (2006): 345-352.
- Kurnaz O, Akadam-teker AB, Yilmaz-aydogan H, Tekeli A, Isbir T. The LOX-1 3′ UTR188CT polymorphism and coronary artery disease in Turkish patients. Molecular biology reports 39 (2012): 4351-4358.
Article Views: 2128
Journal Statistics




Discover More: Recent Articles