A Sub-Hour Point-of-Care Testing of SARS-CoV-2 Variants Identified by XNA-Based RT-qPCR
Jonathan Li#, 1, Hong Huang#, 2, Shuo Shen#, 1, Andrew Y. Fu*, 1, Ang Cao2, Qing Sun1, Daniel Cataldi1, Sabrina Li2, Chuanyi M. Lu3, Aiguo Zhang1, Michael Y. Sha*, 1
1DiaCarta Inc, 4385 Hopyard Road, Suite 100, Pleasanton, CA 94588, USA
2Coyote Bioscience, Alameda, CA 94501, USA
3University of California San Francisco and VA Health Care System, San Francisco, CA 94121, USA
#These authors contributed equally
*Corresponding author: Michael Y. Sha, Andrew Y. Fu. DiaCarta Inc, 4385 Hopyard Road, Suite 100, Pleasanton, CA 94588, USA.
Received: 04 December 2023 Accepted: 11 December 2023 Published: 20 December 2023
Article Information
Citation: Jonathan Li, Hong Huang, Shuo Shen, Andrew Y. Fu, Ang Cao, Qing Sun, Daniel Cataldi, Sabrina Li, Chuanyi M. Lu, Aiguo Zhang, Michael Y. Sha. A Sub-Hour Point-of-Care Testing of SARSCoV- 2 Variants Identified by XNA-Based RT-qPCR. Archives of Microbiology and Immunology. 7 (2023): 487-497.
View / Download Pdf Share at FacebookAbstract
Point-of-care testing (POCT) is a medical diagnostic testing at the time and place of patient care, or called bedside or near-the-patient testing. POCT has been fast-growing and showed tremendous potential in the era of precision diagnostics and personal medicine. This is exemplified by the numerous and widely-used fast antigen POCT tests (15-min only) during the COVID-19 pandemic globally. During the COVID-19 pandemic peak period while the major variants appeared, developing a fast POCT detection and identification method of SARS-CoV-2 variants attracted our interest at DiaCarta. We combined the POCT PCR system from Coyote Bioscience and our established XNA-based RT-qPCR method to detect and identify the major early virus variants as a Proof-of-Concept feasibility study. Our multiplex molecular assay uses RT-qPCR with the aid of xenonucleic acids (XNA) as molecular-clampers which are artificial genetic polymers with Watson-Crick base-pairing capability and robust DNA/RNA-binding ability, high stability, and particularly the appealing specificity --- even a single-base mismatch in XNA/DNA duplex can result in a drop of 10 – 18 °C in the melting temperature. This allows for specific and reliable detection of the SARS-CoV-2 variants, based on the unique combination of characteristic mutations on the viral spike protein domain. Our data showed that XNA-based RT-qPCR by Flash20 instrument successfully detected SARS-CoV-2 Delta and Delta Plus variants from infected or break-through patients. Furthermore, Flash20 POCT instrumentation allows for extraction-free PCR procedure. The turn-around time for Delta Variant kit on Flash20 is less than one hour --- only about 50 minutes of POCT versus regularly 4 hours for the non-POCT assay. As this pilot feasibility study, SARS-CoV-2 variants were rapidly detected with great sensitivity and specificity (PPV 100% and NPV 100%) in a randomly chosen set of clinical samples.
Keywords
<p style="text-align:justify">Point-of-Care testing (POCT), SARS-CoV-2 variant, reverse-transcription real-time PCR (RT-qPCR), molecular-clamping, xeno nucleic acid (XNA)</p>
Article Details
1. Background
In the era of molecular precision diagnostics, point-of-care testing (POCT) has been growing significantly and rapidly. There is an estimate that the global point of care diagnostics market in terms of revenue was approximately $45 billion in 2022 and may reach $75 billion by 2027, growing ~10% annually [1], in the conventional range of point-of-care diagnostic products (blood glucose test, pregnancy test etc). But ever since COVID19 pandemic swept quickly worldwide 3+ years ago, this growing process is even further accelerated. In the field of SARS-CoV-2 virus testing, there are mainly three areas: 1) immunoassay tests for the virus protein (antigen) detection, 2) molecular PCR tests for SARS-CoV-2 viral nucleic acids (RNA) detection, and 3) sequencing mainly NGS of viral genome especially for monitoring new emerging strains or variants of SARS-CoV-2. Because of high simplicity in use, very low cost and quickly available result (within 15 minutes) using the lateral-flow microfluidic devices, the “antigen test” became very popular and has been substantially used worldwide as a new POCT product, with the estimated market of from $28 billion to $14 billion in 2021-2028 period when the pandemic has gradually phased out over time. [2, 3] Unlike the antigen POCT assays, usually the other two areas are more limited toward POCT applications due to more sophisticated instrumentation and operation, and higher expense. Especially for the virus variant detection and monitoring, NGS is the primary method even if it is expensive, complicated, and time-consuming; actually, it is mainly used by public health authorities and large research institutes worldwide to conduct the pandemic surveyance to watch for the new variants of concern (VOC) as well as variants of interest (VOI).
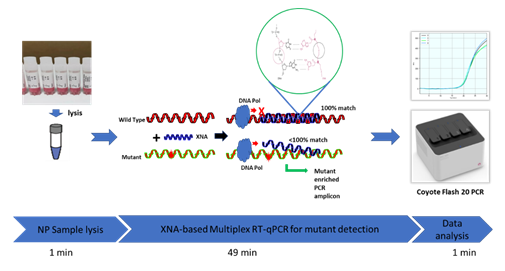
However, thinking on the other side -- for the conventional PCR tests, usually a centralized lab facility, costly instruments, skilled lab staff, and purposeful sample pretreatment are required. Additionally, clinical sample transportation and nucleic acid extraction procedure require longer turnaround time; furthermore, PCR reagents usually need cold chain logistics which increase the cost and risk further. Proper POCT assays can avoid or circumvent all these limitations or issues. Nevertheless, WHO has highlighted the need for the development of point-of-care diagnostics for fast SARS-CoV-2 detection. [4] Encouragingly, nearly a hundred of POCT tests for detection of SARS-CoV-2 have been spotted and summarized by The Foundation for Innovative Diagnostics, CoviSwiftTM COVID-19 S Plus Rapid PCR Kit was one of the early reported POCT PCR assays. [4]. Coyote Bioscience’s portable PCR instrument and custom-made PCR reagent kit also reflect and represent this new trend in the field. Interestingly, a new report on a retrospective analysis of data from 356 hospitals in England showed that cross infections and intra-hospital infections during Jun-2020 and Feb-2021 accounted for the majority of COVID infections [5]. The POCT assays can help reduce the time and frequency of the patient visits to hospitals. Although the NGS-based assays facilitate variant detection and confirmation, they are expensive, time-consuming and not widely available, considering there is an essentially blank area in POCT PCR assay of SARS-CoV-2 variant identification, in this study we explored the feasibility and potential of applying our recently-developed XNA-based multiplex PCR kit for variant detection to the POCT setting -- tested a superfast method to detect SARS-CoV-2 variants by integrating our molecular-clamping technology and a portable PCR instrument without viral RNA extraction. The principle of this assay design is shown in Figure 1.
2. Materials and Methods
2.1 Study Design and Ethics
De-identified patient nasopharyngeal swab (NPS) and saliva samples (from the leftover of COVID-positive samples stored at -80 °C) were used in the study. All patient specimens were collected in January, February and March in 2021 and tested at the UCSF-affiliated San Francisco VA Health Care clinical laboratory and DiaCarta CLIA-certified clinical laboratory for clinical diagnostic or screening purposes. Besides qualitative RT-PCR results (positive or negative), only PCR cycle threshold (Ct) values were included in study analysis and no patient clinical chart reviews were performed. This study was approved by the institutional review board (IRB) at UCSF (UCSF IRB #11-05207) as a no-subject contact study with waiver of consent and as exempt under category 4.
2.2 Sample Preparation
Option 1: with viral RNA extraction
The viral RNA extraction was conducted according to our published protocol [6]. Briefly, for each patient sample, 200 μL was used in the extraction procedure on automatic nucleic acid extraction instrument MGI SP-960 which can automatically complete the high-throughput extraction within 1.5 hours. Of note, an extraction control (EC) for each batch was included, by spiking 20 μL of EC from the QuantiVirus™ SARS-CoV-2 kit into 180 μL sterile RNase-free water, then underwent the same extraction procedure as above. The RNA output amount for each sample is 30 - 50 μL (eluted out by RNase-free water), of which 15 μL is used for RT-PCR reaction in one test. Extracted RNA samples were cautiously handled throughout the procedure (e.g. under 0-5 °C temperature) to avoid degradation and stored at -80°C if not immediately used.
Option 2: without viral RNA extraction
Pipette 15 μL of the Sample Dilution Buffer (Coyote Bioscience, namely the lysis buffer) into a new sterilized tube or PCR tube. Mix it with 15 μL (1:1 volume ratio) of the Nasopharyngeal/Oropharyngeal swab sample (swab in Storage Buffer). The positive control and negative control were treated identically as the swab specimen. Note: Avoid pipetting the viscous part of specimens. Mix them completely by vortexing and then short-spining (5 -10 seconds) to give the ready-to-use samples for subsequent PCR step.
2.3 PCR Instrument with Point-of-Care Capability
Flash20 (Coyote Bioscience) is a compactly-sized benchtop PCR designed for POCT applications, which is featured by a small size (about 30 cm × 30 cm × 20 cm , L x W x H), a portable weight (4.7 kg), a popular four-dye channels: FAM (emission 470 nm), HEX (emission 525 nm), ROX (emission 580 nm) and CY5 (emission 635 nm). Its typical RT-qPCR test takes only 49 minutes to complete, with 45 amplification cycles of the PCR thermocycling profile recommended from the QuantiVirus™ SARS-CoV-2 Variants Detection kit (Table 1).
Table 1: The RT-qPCR thermocycling profile of the Quanti Virus™ SARS-CoV-2 Variants Detection kit, pre-installed on the Flash20 instrument as “Dia Carta SARS-CoV-2 V1”.
|
Thermocycling Parameters |
Temperature (°C) |
Time (s) |
Cycles |
Data Collection |
|
Reverse Transcription |
53 |
600 |
1 |
OFF |
|
Polymerase Activation |
95 |
120 |
1 |
OFF |
|
Denaturation |
95 |
5 |
45 |
OFF |
|
Annealing and Extension |
60 |
30 |
FAM, HEX, ROX, CY5 |
2.4 Multiplex Primer and Probe Design
The conserved ORF1ab gene [6] and those regions adjacent to N501Y and D614G mutations in the SARS-CoV-2 genome were used to design primers and probes for detecting all SARS-CoV-2 variants of concern. Similarly, the primers and probes for the human RNase P gene (RNP gene) as extraction control was designed for this multiplex RT-PCR. Gene sequences were retrieved from GenBank and GISAID databases for primer and probe design to ensure coverage of all SARS-CoV-2 variant strains. Multiple alignments of the collected sequences were performed using Qiagen CLC Main Workbench 20.0.4., and conserved regions in each target gene were identified using BioEditor 7.2.5. prior to primer and probe designs. Primers and probes were designed to target the most conserved regions of each of the target genes of the viral genome using Primer3plus software and following general rules of real-time PCR design. All primers were designed with a Tm of approximately 60 °C and the probes were designed with a Tm of about 65 °C. The amplicon sizes were designed within the range of 70 - 150 bp for each primer pair to achieve optimal amplification efficiency and detection sensitivity. All designed primers and probes were ordered from Integrated DNA Technologies, Inc. (IDT, Coralville, IA, USA) and LGC Biosearch Technologies (Novato, CA, USA), respectively.
2.5 XNA sequence design and chemical synthesis
Sequence design and chemical synthesis of XNAs are based on our previously described method [7]. The XNAs for SARS-CoV-2 Spike-protein gene mutations T478K and K417N were designed to perfectly match the wild type (WT) sequences to ensure specific blocking of WT amplification. For each target region (T478 and K417 codon respectively in the viral S-gene), a single best XNA was selected based on the wildtype-clamping PCR test and the physicochemical characteristics (sequence length, GC content, purine contentment, theoretical melting temperature of XNA-DNA duplex). All chemicals and solvents were of ACS grade or higher, purchased from Sigma, Fisher Scientific and other commercial sources. XNAs were synthesized via the classic solid-phase peptide synthesis (SPPS) method on an CEM/INTAVIS MultiPep RSi automatic synthesizer. After the solid-phase synthesis procedure, the crude product was obtained following a cleavage/deprotection step in trifluoroacetic acid, then underwent a fast purification procedure by size-exclusion chromatography. The purified XNAs were analyzed and characterized by reverse-phase HPLC (Agilent 1100 HPLC system) and matrix-assisted-laser-desorption-ionization mass spectrometry (Shimadzu Axima MALDI-TOF). [7]
2.6 PCR Assay Procedure
2.6.1 Amplification Reagent Preparation
From the QuantiVirus™ SARS-CoV-2 PCR kit (DiaCarta Inc), take out contents including RT-qPCR Master Mix, Primer/Probe Mix, XNA Mix, Positive Control, Negative Control, Extraction Control and No-Template Control, together with the Sample Dilution Buffer from Coyote Bioscience. Dissolve the reagents completely, mix all contents completely by vortexing, then short-spinning for 5-10 seconds. Prepare the RT-qPCR Mixes as follows: add contents into a new sterile tube to make a bulk solution so that for each PCR reaction, there should be 5.6 μL of RT-qPCR Master Mix, 2.2 μL of Primer/Probe Mix, 2.2 μL of the XNA Mix, totaling 10 μL of this Amplification Reagent for each PCR reaction. Mix well by vortexing for 10 seconds and short-spinning for 10 seconds. Totally 10 μL of the above-prepared Amplification Reagent was dispensed into each PCR tube.
2.6.2 Real-time quantitative RT-PCR (RT-qPCR) on Flash20 Instrument
Add 15 uL of each raw sample (half amount of each sample prepared above) into each prepared PCR tube. Close the caps tightly then vortex and short-spin (10 seconds respectively). Insert each of the PCR tubes into the PCR instrument Flash20 (Coyote Bioscience, 4 PCR slots in Flash20). Record the sample order and position. Use the Flash20’s pre-defined thermocycling profile at a laptop computer to acquire the RT-PCR result in 35 minutes. The PCR instrument setup is as follows, demonstrating the simplicity of its use:
- Enter the interface of Flash20 software, then click the thermocycling profile “DiaCarta SARS-CoV-2 V1” (RT-qPCR parameters listed in Table 1).
- Type the sample information (sample name and number) following the sample slot order.
- Select FAM channel for ORF1ab detection, HEX/VIC channel for K417N mutation detection, ROX channel for Internal Control (RNP gene), CY5 channel for T478K mutation detection.
- Click “Start” to initiate the RT-PCR amplification and real-time detection.
- The amplification curves and Ct values are shown on the interface of software when the reaction is completed.
- Determine and report the result for each sample according to the criteria for Interpretation of Results of the kit.
3. Results
3.1 Concept of Point-of-Care Testing (POCT) for Variant Detection
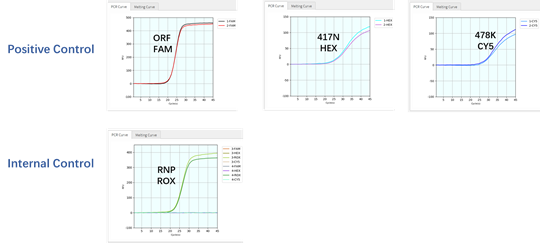
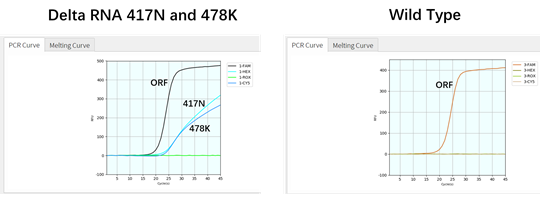
Coyote Bioscience’s Flash20 PCR instrument is designed for POCT application (Figure 1). It could be conveniently used as urgent on-site detection in hospitals, emergency clinics, customs and airports. To apply such POCT platform to rapidly identify SARS-CoV-2 variants, firstly we had two gBlock oligos synthesized, corresponding to two of the main mutations of viral S-gene: T478K and K417N respectively, then tested them with the Variant Detection Kit by RT-qPCR. As expected, the result indicated that the ORF1ab gene as viral detection, RNase P gene as internal control, K471N mutant, and T478K mutant can all be detected by this multiplex assay, in as short as 49 minutes (Figures 1 and 2). After this initial proof, we further applied Twist Bio SARS-CoV-2 RNA References (wildtype, Delta, Delta plus) to RT-qPCR for the variant detection. The results also showed that all three targets (K417N, T478K and ORF1ab) were detectable for Delta-plus variant reference by this assay; only two (T478K and ORF1ab) were detectable for Delta variant reference, according to the unique combination of the characteristic S-gene mutations (Table 2). In contrast, among these three viral targets, only ORF1ab target was detected from the SARS-CoV-2 wildtype reference. This demonstrates that genomic RNAs of Delta variant, Delta-plus variant, and wildtype SARS-CoV-2 virus can be detected and identified by our Variant Detection kit.
Table 2: List of all main mutations detected by QuantiVirus™ SARS-CoV-2 Variants Detection Kit. The Delta Variant Detection Kit includes a subset of mutations only (T478K and K417N) as a single-tube multiplex assay for POCT use.
Figure 2: The DNA oligos as gBlocks (containing two mutants of viral S-gene, one viral ORF1ab wildtype target, and one human reference gene RNP as internal extraction control) were detected and identified by the variant detection kit on Flash20 PCR. Each standard was tested in duplicate at XNA-based RT-qPCR on a Flash20 instrument.
3.2 Verification of SARS-CoV-2 Variant Detection by Coyote’s Flash20 with viral RNA Extraction
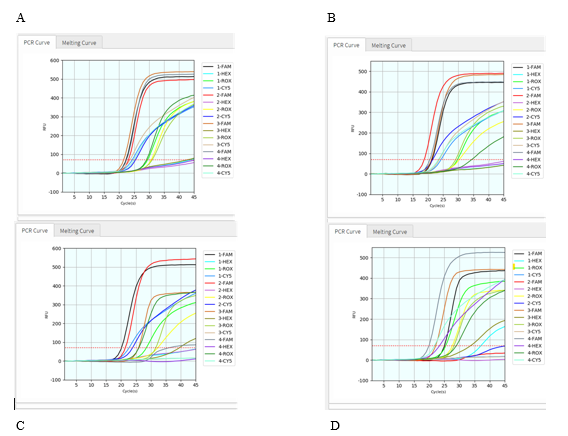
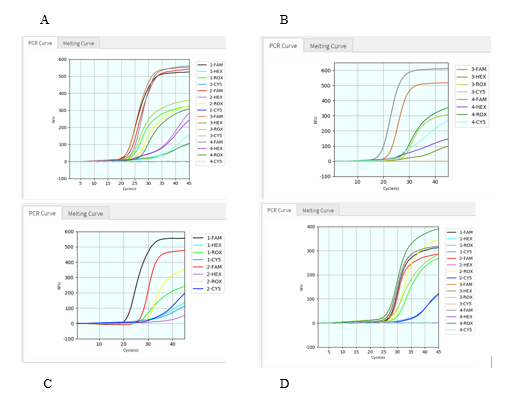
To further test if this protocol applies well in variant detection of clinical samples, we conducted the virus RNA extraction from 15 SARS-CoV-2 positive patient samples, then tested them with this assay on Coyote’s Flash20 PCR. All 15 samples were found to be detectable for virus ORF1ab gene and human RNase P gene (Figure 4, S. Table 1). Among them, 10 patient samples were identified as SARS-CoV-2 Delta variant (T478K detected, K417N undetected); meanwhile, the other five patient samples were identified as Delta plus variant (both T478K and K417N detected). The results showed that these exemplary SARS-CoV-2 variants could be facilely detected by this POCT PCR assay.
Figure 4: Validation of clinical samples with viral RNA extraction. The graphs above (a – d) displayed RT-qPCR results of totally 15 patient samples (n = 4 for each of 4 graphs), in a multiplex PCR setting with four dyes for every sample: FAM detection channel is for viral ORF1ab gene, HEX for viral K417N mutation, CY5 for viral T478K mutation, and ROX for human RNase P gene.
3.3 Validation of SARS-CoV-2 Variant Detection Directly by Coyote’s Flash20 without Viral RNA Extraction
After confirming that these SARS-CoV-2 mutants can be detected by our kit with RNA extraction step in Coyote instrument Flash20, we then try to detect SARS-CoV-2 variants directly without viral RNA extraction step. First, we added Coyote Bioscience’s Sample Dilution Buffer (namely lysis buffer) to the patient sample at 1:1 volume ratio, and then directly use the mixed sample for RT-qPCR on Flash20. Total turnaround time is about 50 min from the sample to the report result. All 11 patient samples were found detectable for both virus ORF1ab gene and human RNase P gene (Figure 5, S. Table 2). Among them, 7 patient samples showed T478K positive but K417N negative, thus identified as the Delta variant; other 4 patient samples showed both T478K positive and K417N positive, thus identified as the Delta-plus variant. Figure 5 and S. Table 2 demonstrate that all these eleven SARS-CoV-2 positive patients got infected by either Delta variant or Delta-plus variant. Compared with variant detection with viral extraction, the rapid SARS-Co-V-2 variant detection assay without viral RNA extraction has a similar result (Table 3). Concordance is 100% for both methods.
Table 3: Comparison of POCT Assays with and without Viral RNA Extraction at RT-qPCR to Identify SARS-CoV-2 Delta Variant and Delta-plus Variant.
|
Sample ID |
Flash 20 with Viral RNA Extraction |
Flash 20 without Viral RNA Extraction |
Final Call |
||||||
|
FAM |
HEX |
ROX |
CY5 |
FAM |
HEX |
ROX |
CY5 |
||
|
ORF |
K417N |
RNP |
T478K |
ORF |
K417N |
RNP |
T478K |
||
|
1 |
22.37 |
44.82 |
29.62 |
24.53 |
21.08 |
30.98 |
27.95 |
32.65 |
Delta-plus |
|
3 |
23.2 |
N/A |
30.86 |
25.59 |
26.58 |
N/A |
29.76 |
31.91 |
Delta |
|
8 |
18.75 |
N/A |
30.32 |
21.54 |
27.81 |
N/A |
30.53 |
34.25 |
Delta |
|
10 |
20.13 |
N/A |
28.44 |
22.65 |
28.16 |
N/A |
28.89 |
34.31 |
Delta |
|
13 |
21.4 |
N/A |
32.24 |
23.83 |
27.24 |
N/A |
29.28 |
33.08 |
Delta |
|
34 |
16.04 |
20.1 |
23.15 |
17.12 |
23.94 |
28.85 |
27.28 |
30.71 |
Delta-plus |
|
89 |
14.53 |
N/A |
20.94 |
16.26 |
21.91 |
N/A |
23.86 |
33.98 |
Delta |
|
162 |
14.94 |
19.33 |
22.43 |
16.88 |
22.98 |
28.91 |
26.49 |
39.2 |
Delta-plus |
|
214 |
17.69 |
22.38 |
24.07 |
19.42 |
21.58 |
32.63 |
28.03 |
33.41 |
Delta-plus |
|
219 |
13.96 |
N/A |
20.03 |
16.06 |
21.57 |
N/A |
24.48 |
34.68 |
Delta |
Figure 5: Validation of clinical samples without viral RNA extraction. The graphs (a – d) above displayed RT-qPCR results of totally 11 patient samples (n = 4, 2, 2, 4 respectively), in a multiplex PCR setting with four dyes for every sample: FAM detection channel is for viral ORF1ab gene, HEX for viral K417N mutation, CY5 for viral T478K mutation, and ROX for human RNase P gene.
4. Discussion
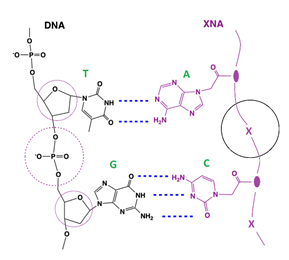
Xenonucleic acid, or XNA, is a class of artificially-made genetic polymers containing the identical natural bases (A/T/C/G) of nucleic acids on side chains and a variety type of backbones. Based on the backbone type, XNAs have various sub-groups including LNA (locked nucleic acid), HNA (hexitol nucleic acid), GNA (glycerol nucleic acid), PNA (peptide nucleic acid) and others [8]. Some of them like peptide nucleic acids or analogs have neutrally-charged backbone thus leading to its even stronger hybridization with the negatively-charged DNA chain compared to the hybridization in the natural DNA-DNA duplex, due to the lack of electrostatic repulsion between the two chains of formed XNA-DNA duplex. (Figure 6) Thus the Tm of XNA-DNA duplex is significantly higher than their DNA-DNA duplex counterpart. Even more importantly, when hybridizing with XNA here, even if a single-base mutation/mismatch at the center of the DNA target sequence can result in a substantial Tm drop of 10 - 18 °C compared to the wildtype DNA sequence. [9] Thus XNA clamping can be used in PCR to selectively block the amplification of wildtype DNA while allowing the amplification of mutant DNA as normal. Taking advantage of this appealing XNA characteristics, we developed the molecular-clamping multiplex RT-qPCR to detect and identify the SARS-CoV-2 variants. During the COVID-19 pandemic, besides the NGS method to detect the whole sequence of the virus genome mainly for new strain monitoring, PCR method as the “gold standard” has proved its many practical advantages on detection of SARS-CoV-2 virus in clinical samples: accuracy, speed, efficacy and cost, thus numerous RT-qPCR diagnostic kits have been approved by FDA as EUA applications [10]. Many of these FDA EUA-approved PCR kits choose a combination of multiple characteristic targets in the virus genome to assure the reliable detection of SARS-CoV-2 in clinical samples. For instance, our QuantiVirus™ SARS-CoV-2 Multiplex Test Kit detects ORF1ab gene, N gene, and E gene simultaneously [6]. Using the same methodological strategy above, we chose the unique combination of multiple proper targets including some characteristic mutations for a specific variant of SARS-CoV-2, so as to reliably detect this certain variant by RT-qPCR method (see Table 2). Of particular note, our PCR method for variant detection is based on our proprietary XNA clamping technology, in which XNA is used to completely block the amplification of wildtype virus cDNA (reverse-transcribed from virus RNA) if existing in the clinical samples, while letting the viral mutant cDNA be amplified normally. Nevertheless the characteristic pattern of mutation combination (or a “fingerprint”) of this certain variant can be reliably and facilely detected.
The Delta variant was picked in this proof-of-concept study in a POCT assay setting. Prior to Omicron variant, Delta Variant or B.1.617.2 variant, was identified as early as in Oct 2020 and has subsequently spread to approximately 60 countries, quickly becoming as the main variant in mid-July in 2021, replacing the earlier dominating one - Alpha variant (first reported in September 2020 then dominated between April to June in 2021). Delta variant grows more rapidly in the human respiratory tract and was reported to have up to a thousand times higher viral loads than the original strain [11], it was more contagious and associated with a higher degree of mortality than Alpha variant. [12, 13]. Delta variant was characterized by 13 mutations including S-gene mutations D614G, T478K, L452R, D950N, P681R etc; further, it becomes Delta-plus variant when having an additional K417N mutation (Table 2). The Delta Variant kit was quickly developed then at DiaCarta to detect and identify both Delta and Delta-plus variants at regular PCR setting. In this report, this method was applied to POCT setting using a portable PCR instrument. It showed that this RT-qPCR protocol worked well on Flash20 PCR instrument for all three types of samples ranging from DNA oligo standards, commercial reference RNA samples, to the real-world patient samples. (Figures 2-5).
The assay method in this report is an integration of DiaCarta’s XNA technology-based Variant Detection test and Coyote Bioscience’s POCT SARS-CoV-2 test (portable PCR instrument and Sample Dilution Buffer). High sensitivity (100%) and specificity (100%) of each of these two individual kits were proven and reported previously [7, 14]. LOD of DiaCarta’s QuantiVirus™ SARS-CoV-2 Variant Detection test is 100-200 copies/mL for the tested Alpha variant [7]; LOD of Coyote Bioscience’s FlashDetect™ LyocartD SARS-CoV-2 test is about 200 copies/mL [14]. In this study, 7 Delta variant and 4 Delta-plus were identified and confirmed by Sanger Sequencing with its PCR amplicon. COVID-positive patient samples were detected with both PPV 100% and NPV 100%. Sample processing as the first step of PCR workflow oftentimes have significant impact on the assay. Here how did the sample processing methods affect the variant detection in this POCT setting? For the patient sample group selected for RNA extraction processing, 10/15 (67%) are detected as Delta variant, 5/15 (33%) as Delta-plus variant (S. Table 1). For the patient sample group selected for direct processing (without RNA extraction step), 7/11 (64%) are detected as Delta variant, 4/11 (36%) as Delta-plus variant (S. Table 2). Between the two groups, there was a large overlap (10 patient samples). For these ten samples, the two sample-processing options were compared and gave 100% concordance results in variant identification. Interestingly, the simple direct use of patient samples without RNA extraction did work with the POCT PCR workflow as demonstrated here. This has great significance in that: 1) as anticipated, the Coyote Bioscience’s Sample Dilution Buffer accompanying with Flash20 device is well-compatible with our Variant Detection kit for XNA-based RT-qPCR protocol; 2) the skipping of RNA-extraction step can further save 60-90 minutes in the PCR turn-around time (TAT), making this integrated POCT PCR technique even faster (less than one hour), appealing, and highly suitable for point-of-care applications in SARS-CoV-2 variant detection.
Worthy of note, the POCT characteristics of this reported technique include: 1) portable, durable and easy-to-operate PCR device; 2) no RNA-extraction needed for sample processing thus saving time/labor and reducing exposure risk; 3) the fast turn-around time of < 1 hour from adding sample to reporting results; 4) the lyophilizable PCR reagents could be stored and transported at room temperature; 5) simple POCT experimental set-up yet still allowing a 4-plex assay for multiple samples (n =4). Besides providing a super-fast, economic, accurate, and POCT-intended method for SARS-CoV-2 variant detection/identification with our innovative XNA-based PCR technique, this study has several limitations: 1) this was an early proof-of-concept study in 2021 with a focus solely on then-dominating Delta/Delta+ variant, not covering the current dominating Omicron variant and its many sub-variants; 2) relatively small number of clinical samples were used in response to the urgent Delta variant spreading worldwide in the first half year of 2021; 3) limited replicates were tested due to the inherent limitation of small-size portable POCT instruments (e.g. only four sample wells/lots available each session for Flash20 PCR). However, this could be potentially improved by running an array of small Flash20 PCR devices parallelly.
Of note, even if this POCT study has only dealt with detection and identification of SARS-CoV-2 Delta variant, we believe this technique can be similarly applied to other new emerging variants without difficulty when needed, after re-designing the sequences of PCR components (new mutation targets, primers/probe, and XNA clamper) accordingly [7]. Taken together, we conducted a proof-of-concept study of successfully applying the XNA-clamping PCR technique to fast detect and identify then-concerned major variants of SARS-CoV-2, the Delta/Delta+ variant here as a typical example, within 1-hour on a small portable PCR device to simulate the real-world point-of-care testing applications.
References
- Point of care diagnostics market by product, platform, purchase, sample, end user and region - Global Forecast to (2027).
- "Global COVID-19 Rapid Antigen Test Kits Market Research Report 2022". Market Reports World. 10 June (2022).
- Kumar A, Parihar A, Panda U, Parihar DS. Microfluidics-Based Point-of-Care Testing (POCT) Devices in Dealing with Waves of COVID-19 Pandemic: The Emerging Solution. ACS Appl Bio Mater 5 (2022): 2046-2068.
- Dakhave M, Gadekar S, Malekar A, Wankhede G. CoviSwift TM, A point-of-care RT-PCR device for SARS-CoV-2 and its variant detection. J Virol Methods 315 (2023): 114714.
- Cooper BS, Evans S, Jafari Y, et al.The burden and dynamics of hospital-acquired SARS-CoV-2 in England. Nature 623 (2023): 132–138.
- Sun Q, Li J, Ren H, Pastor L, Loginova Y, et al. Saliva as a testing specimen with or without pooling for SARS-CoV-2 detection by multiplex RT-PCR test. PLOS ONE 16 (2021): e0243183.
- Shen S, Fu A, Jamba M, Li J, Powell M, Zhang A, et al. A Rapid SARS-CoV-2 Variant Detection by Molecular-Clamping Based RT-qPCR, medRxiv (2021).
- Wang Q, Chen L, Long Y, Tian H & Wu J. Molecular Beacons of Xeno-Nucleic Acid for Detecting Nucleic Acid. Theranostics 3 (2013) 395–408.
- Kyger EM, Krevolin MD & Powell MJ. Detection of the Hereditary Hemochromatosis Gene Mutation by Real-Time Fluorescence Polymerase Chain Reaction and Peptide Nucleic Acid Clamping. Anal. Biochem 260 (1998): 142–148.
- https://www.fda.gov/medical-devices/covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas-molecular-diagnostic-tests-sars-cov-2
- Li B. et al. Viral infection and transmission in a large, well-traced outbreak caused by the SARS-CoV-2 Delta variant (2021).
- Singh et al. SARS-CoV-2 variants of concern are emerging in India. Nature Medicine 27 (2021): 1126–1134.
- Expert reaction to cases of variant B.1.617 (the ‘Indian variant’) being investigated in the UK | Science Media Centre.
- https://en.coyotebio.com/reagent_detail/id/96.html
Supplementary
Table 1: Summary of SARS-CoV-2 Variants Detection with RNA Extraction
|
Sample |
FAM |
HEX |
ROX |
CY5 |
FAM |
HEX |
ROX |
CY5 |
Final Call |
|
ID |
ORF |
K417N |
RNP |
T478K |
ORF |
K417N |
RNP |
T478K |
|
|
1 |
22.37 |
44.82 |
29.62 |
24.53 |
+ |
+ |
+ |
+ |
Delta-plus |
|
3 |
23.2 |
N/A |
30.86 |
25.59 |
+ |
- |
+ |
+ |
Delta |
|
4 |
21.57 |
44.04 |
31.21 |
23.77 |
+ |
+ |
+ |
+ |
Delta-plus |
|
5 |
22.54 |
N/A |
29.24 |
25.11 |
+ |
- |
+ |
+ |
Delta |
|
6 |
21.34 |
N/A |
29.74 |
24.33 |
+ |
- |
+ |
+ |
Delta |
|
8 |
18.75 |
N/A |
30.32 |
21.54 |
+ |
- |
+ |
+ |
Delta |
|
10 |
20.13 |
N/A |
28.44 |
22.65 |
+ |
- |
+ |
+ |
Delta |
|
13 |
21.4 |
N/A |
32.24 |
23.83 |
+ |
- |
+ |
+ |
Delta |
|
30 |
21.13 |
N/A |
29.14 |
23.72 |
+ |
- |
+ |
+ |
Delta |
|
31 |
20.6 |
N/A |
34.45 |
23.47 |
+ |
- |
+ |
+ |
Delta |
|
34 |
16.04 |
20.1 |
23.15 |
17.12 |
+ |
+ |
+ |
+ |
Delta-plus |
|
89 |
14.53 |
N/A |
20.94 |
16.26 |
+ |
- |
+ |
+ |
Delta |
|
162 |
14.94 |
19.33 |
22.43 |
16.88 |
+ |
+ |
+ |
+ |
Delta-plus |
|
214 |
17.69 |
22.38 |
24.07 |
19.42 |
+ |
+ |
+ |
+ |
Delta-plus |
|
219 |
13.96 |
N/A |
20.03 |
16.06 |
+ |
- |
+ |
+ |
Delta |
Table 2: Summary of SARS-CoV-2 Variants Detection without RNA Extraction.*
|
Sample |
FAM |
HEX |
ROX |
CY5 |
FAM |
HEX |
ROX |
CY5 |
Final Call |
|
ID |
ORF |
K417N |
RNP |
T478K |
ORF |
K417N |
RNP |
T478K |
|
|
1 |
21.08 |
30.98 |
27.95 |
32.65 |
+ |
+ |
+ |
+ |
Delta-plus |
|
3 |
26.58 |
N/A |
29.76 |
31.91 |
+ |
- |
+ |
+ |
Delta |
|
8 |
27.81 |
N/A |
30.53 |
34.25 |
+ |
- |
+ |
+ |
Delta |
|
10 |
28.16 |
N/A |
28.89 |
34.31 |
+ |
- |
+ |
+ |
Delta |
|
13 |
27.24 |
N/A |
29.28 |
33.08 |
+ |
- |
+ |
+ |
Delta |
|
14 |
27.23 |
N/A |
26.07 |
34.11 |
+ |
- |
+ |
+ |
Delta |
|
34 |
23.94 |
28.85 |
27.28 |
30.71 |
+ |
+ |
+ |
+ |
Delta-plus |
|
89 |
21.91 |
N/A |
23.86 |
33.98 |
+ |
- |
+ |
+ |
Delta |
|
162 |
22.98 |
28.91 |
26.49 |
39.2 |
+ |
+ |
+ |
+ |
Delta-plus |
|
214 |
21.58 |
32.63 |
28.03 |
33.41 |
+ |
+ |
+ |
+ |
Delta-plus |
|
219 |
21.57 |
N/A |
24.48 |
34.68 |
+ |
- |
+ |
+ |
Delta |
*Note: Clinical samples were directly mixed (1:1) with Coyote’s VTM/Lysis buffer for PCR


 Impact Factor: * 3.5
Impact Factor: * 3.5 Acceptance Rate: 71.36%
Acceptance Rate: 71.36%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
