Inferring the Hoxa1 Gene Regulatory Network in Mouse Embryonic Stem Cells: Time-Series RNA-seq Data and Computational Modeling Approach
Ugochi Emelogu1, Bryan Rogers1, Yaser Banad2, Candice Cavalier1, Anna Wilson1, Xiaoping Yi1, Oswald D’Auvergne1, Samire Almeida de Oliveira1, Nana Akwaboa1, and Eduardo Martinez-Ceballos1, *
1Departmena of Biological Sciences and Chemistry, Southern University, Baton Rouge, LA, 70813, USA
2School of Electrical and Computer Engineering, University of Oklahoma, Norman, OK, 73019, USA
*Corresponding author: Eduardo Martinez-Ceballos, Department of Biological Sciences and Chemistry, Southern University, Baton Rouge, LA, 70813, USA.
Received: 06 December 2023 Accepted: 11 December 2023 Published: 15 December 2023
Article Information
Citation: Ugochi Emelogu, Bryan Rogers, Yaser Banad, Candice Cavalier, Anna Wilson1, Xiaoping Yi, Oswald D’Auvergne, Samire Almeida de Oliveira, Nana Akwaboa, and Eduardo Martinez- Ceballos. Inferring the Hoxa1 Gene Regulatory Network in Mouse Embryonic Stem Cells: Time-Series RNA-seq Data and Computational Modeling Approach. Archives of Microbiology and Immunology. 7 (2023): 456-467
View / Download Pdf Share at FacebookAbstract
The homeotic gene Hoxa1 plays a pivotal role in regulating embryonic pattern formation and morphogenesis during mouse embryogenesis. However, despite the identification of a number of putative Hoxa1 target genes, the intricate regulatory relationships between these targets remain largely elusive. Leveraging the advancements in high-throughput technologies and sophisticated computational methods, we aimed to infer the Gene Regulatory Networks (GRNs) governed by Hoxa1 that direct cellular function, morphology, and/or differentiation. To achieve this, we generated time-series RNA-seq data from Retinoic Acid (RA)-treated Wild Type versus Hoxa1-null mouse ES cells, enabling the construction of the Hoxa1 GRN. To create this GRN, we employed NARROMI, a published technique known for its noise reduction capabilities and improved accuracy in gene-regulation inference. Using this technique, we identified putative direct and indirect connections between Hoxa1 and a set of genes with known relevance in embryonic development. Validation through qPCR confirmed the Hoxa1-dependence on mRNA expression for selected genes, both within the immediate vicinity (direct) and in secondary interactions (indirect). Furthermore, by mapping the candidate genes to relevant Gene Ontology (GO) networks, we verified their involvement in processes likely regulated by Hoxa1. Our findings provide compelling evidence supporting the accuracy of the NARROMI analysis in generating a hierarchical network of genes under the transcriptional control of the Hoxa1 Transcription Factor (TF), specifically in mouse ES cells. This network reveals a pool of promising candidate genes that may function as direct targets of Hoxa1. However, further investigations, including the characterization of Hoxa1 protein interactions with target loci DNA, are necessary to confirm their direct regulatory relationship with this TF. Moreover, the time-series RNA-seq data from Wild Type ES cells, coupled with the methodology employed in this study, hold the potential for constructing GRNs for additional TFs activated by RA. This comprehensive approach can shed further light on the intricate regulatory networks governing cellular function, morphology, and differentiation, advancing our understanding of embryonic development and gene regulation processes.
Keywords
<p>RNA-seq; ES cells; Hoxa1; Retinoic Acid; Gene Regulatory Network</p>
Article Details
1. Introduction
The homeobox (Hox) family of transcription factors comprises important regulators of embryonic patterning and organogenesis [1-6]. In mammals, the Hox genes are located in four separate chromosome clusters, Hoxa, Hoxb, Hoxc and Hoxd. The expression of Hox genes during development depends upon their position in the chromosomal cluster; genes positioned at the 3' end are expressed earlier and more anteriorly, whereas 5' end genes are expressed at later times and more posteriorly [1-2]. Treatment of ES cells or teratocarcinoma cells with Retinoic Acid (RA, a Vitamin A derivative) results in the sequential activation of several Hox genes in a manner that resembles their position in the clusters, e.g. 3' genes are activated before 5' genes [7-9]. In fact, a retinoic acid response element (RARE) was discovered in the 3’ enhancer of Hoxa1, the most 3’ end gene in the Hoxa cluster [9-12]. The spatially restricted patterns of Hox gene expression within the developing central nervous system (CNS) in vertebrates, both along the anteroposterior and dorsoventral axes of the embryo, suggest a regulatory role in patterning of the CNS, as well as in cell specification [13-14]. Inactivation of both alleles of the Hoxa1 gene in mice results in numerous developmental defects, including hindbrain deficiencies and abnormal skull ossification, and ultimately, in neonatal death [15-18]. The abnormalities of the mouse Hoxa1 null embryos emphasize the importance of this gene in the proper development of the early brain and neural crest-derived structures [19-21]. Furthermore, although endoderm is relatively normal in Hoxa1 null embryos, the presence of Hoxa1 expression in a number of endodermal-derived structures in wild type embryos suggests a role for Hoxa1 during the normal endodermal development of vertebrate embryos [22-27]. In humans, mutations in this gene have been described in association with various Central Nervous System (CNS) disorders [28-29] and its overexpression has been associated with cancer development [30-32]. Thus, these observations highlight the importance of Hoxa1 for proper embryonic development and the onset of human disorders and cancer, which suggest a complex regulatory mechanism that orchestrates the actions of the Hoxa1 signaling pathway in cells. Although the relevance of Hoxa1 during embryonic development has been recognized for over two decades, little is known about the mechanism of action of this important TF. An important barrier to the complete understanding of the mechanism of Hoxa1 action may be the lack of information about its downstream pathway. In this regard, different approaches employing cDNA microarray analyses have been taken in order to elucidate the Hoxa1 pathway through the identification of Hoxa1 targets genes. For instance, Hoxa1 target genes identified from microarray analyses were first reported in a study that employed teratocarcinoma cells [33], then in mouse Embryonic Stem (ES) cells [34], and also by researchers that compared embryonic tissues from Wt vs. Hoxa1-null embryos [35]. These studies identified different sets of putative Hoxa1 target genes but, due to the nature of the technologies employed, the findings did not provide information about the regulatory interactions between the putative Hoxa1 targets identified. In this work, we have performed time-series RNA-seq analyses in an attempt to construct the Hoxa1 Gene Regulatory Pathway (GRN) activated during the RA-induced differentiation of mouse ES cells in vitro. To construct the Hoxa1 GRN, we used NARROMI, a noise and redundancy reduction technique shown to improve accuracy of GRN inference [36], which was shown to significantly outperform other methods [37] such as LP [38], LASSO [39, 40], ARACNE [41], and GENIE3 [42].
2. Materials and Methods
2.1 Cells and drug treatments
For this work, we used mouse J1 Wild-type ES cells (American Type Culture Collection or ATCC), and a Hoxa1-null ES cell line, EMC-ES-Hoxa1–15, whose generation and characterization we previously described [34]. Undifferentiated J1 and Hoxa1–15 ES cells were maintained on gelatin-coated dishes in ES medium (Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% ES qualified fetal bovine serum (Atlanta Biologicals), 100 mM MEM nonessential Amino acids, 0.1 mM β-mercaptoethanol, Penicillin and Streptomycin (Invitrogen), 1 mM sodium pyruvate, and 1X103 U/ml leukemia inhibitory factor (LIF, Esgro, Millipore), as previously described [43]. To avoid spontaneous differentiation, the ES cells were passaged every two days for up to three passages before drug treatments. For all experiments, ES cells were treated 24 hours after plating with 1 μM all-trans-Retinoic Acid (RA) for different periods of time. Control cells were treated with 0.1% ethanol (vehicle). After various times of drug treatment, cells were harvested for total RNA extraction for either RNA sequencing (RNA-seq) or qPCR analyses.
2.2. RNA-seq
For time-series RNA-seq, ES cells cultured as monolayers were treated with RA for 12, 24, 36, 48, and 60 hours (RA_12, RA_24, RA_36, RA_48, and RA_60, respectively). Control cells were treated with vehicle only for 48 hours (Ctl_48). To identify Hoxa1 target genes, J1 (Wild type or Wt) and Hoxa1-null (knockout or KO), were treated with RA or vehicle only for 48 hours. Total RNA was extracted from four biological replicates for each treatment and time point combination. Library preparation and sequencing was performed by Novogene (Novogene Inc.).
2.3. Data Analysis
Downstream analysis was performed using a combination of programs including STAR, HTseq, Cufflink and Novogene’s wrapped scripts. Alignments were parsed using STAR program and differential expressions were determined through DESeq2/edgeR.
2.4. Reads mapping to the reference genome
Reference genome and gene model annotation files were downloaded from genome website browser (NCBI/UCSC/Ensembl) directly. Indexes of the reference genome was built using STAR and paired-end clean reads were aligned to the reference genome using STAR (v2.5). STAR used the method of Maximal Mappable Prefix (MMP) which can generate a precise mapping result for junction reads.
2.5. Quantification of gene expression level
STAR will count number reads per gene while mapping. The counts coincide with those produced by HTseq-count with default parameters. And then FPKM of each gene was calculated based on the length of the gene and reads count mapped to this gene. FPKM, Reads Per Kilobase of exon model per Million mapped reads, considers the effect of sequencing depth and gene length for the reads count at the same time, and is currently the most commonly used method for estimating gene expression levels [44].
2.6. Network construction
NARROMI, a noise and redundancy reduction technique [36], was employed to construct the Hoxa1 GRN. This method requires two data sets: a Target Gene (TG) set, and a Transcription Factor (TF) set, where TF is comprised of relevant Transcription Factors present within TG. To obtain the TG set, time-series RNA-seq was performed and data was filtered, based on levels of expression and significant differential expression among treatment, using MATLAB & Simulink software (MathWorks, Natick, Mass.). To construct the TF set for NARROMI, we first obtained a list of all Transcription Factors that were upregulated in RA-treated Wt versus KO cells by at least 2-fold. This list was then compared to the TG set in order to obtain a time-series subset comprised of Hoxa1 dependent Transcription Factors. Subsequently, the Hoxa1 GRN was identified by implementing NARROMI on MATLAB using threshold default conditions as indicated by Zhang et al. (2013) [36]. The resulting network was visualized using Cytoscape [45].
2.7. qPCR analyses
Total RNA was isolated using TRIzol reagent (Thermo Fisher Scientific), according to standard procedures. The concentration of RNA was determined using a NanoDrop (Thermo Scientific). Real-time PCR with SYBR green detection was performed on a QuantStudio 3 thermocycler (Applied Biosystems) using the following primers: Rplp0 (internal control),
forward 5′-AGAACAACCCAGCTCTGGAGAAA-3′,
reverse 5′-ACACCCTCCAGAAAGCGAGAGT-3′;
Nanog forward 5′-GCGGACTGTGTTCTCTCAGGC-3′,
reverse 5′-TTCCAGATGCGTTCACCAGATAG-3′;
Nes, forward 5′-CAGAGAGGCGCTGGAACAGAGATT-3′,
reverse 5′-AGACATAGGTGGGATGGGAGTGCT-3′;
Tubb3, forward 5′-TCAGCGATGAGCACGGCATA-3′,
reverse 5′-CACTCTTTCCGCACGACATC-3′;
Mest, forward 5′-GCTGCCGCGGTCCACAGTGTCG-3′,
reverse 5′-GTCACCCTTAGAGATGAGGTGGAC-3′;
Sox17, forward 5′-GCTGATCAGCCAGGATGAGCAGCCCGGATGCGGGATACGCC-3′,
reverse 5′-ACGAAGGGCCGCTTCTCTGCC-3′;
Zscan4f, forward 5′-GAGGTACCAGGGTTGCAGTC-3′,
reverse 5′-GGATGATTGGCGAAAGCGAC-3′;
Lgals4, forward 5′-CCCGCCCTTTACTCTGCTTCTGTT-3′,
reverse 5′-CTTCCTTGCCCCATTGTCCACTTT-3′,
Col13a1, forward 5′-GGAGAACCAGGTGACGAAGG-3′,
reverse 5′- ACTTGTTCCAGCAGCCTTG-3′;
Coro6, forward 5′-AGCGCCTTCTGTGCCGTCAA-3′,
reverse 5′-CGTCAGGGTGTATGTCGTCCAG-3′.
Reactions were run in triplicate from three independent experiments. Expression data were normalized to the geometric mean of the housekeeping gene Rplp0 (36B4) to control the variability in expression levels and were analyzed using the standard 2 -ΔΔCT method. Expression levels were analyzed by one-way ANOVA with Tukey's post-hoc analysis using GraphPad Prism 5.0 software.
3. Results
3.1 Time-series transcriptomic analysis of Wt ES cells treated with RA
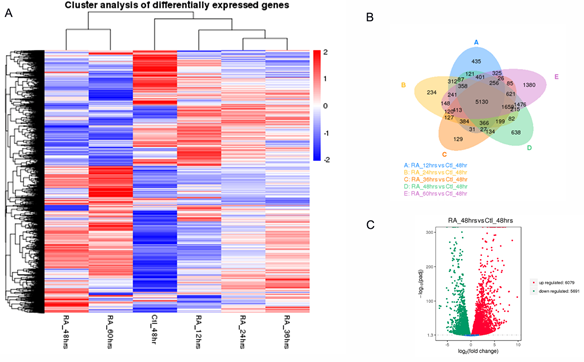
Embryonic Stem Cell (ES) differentiation requires a precise sequence of molecular events that involve specific gene activation and their coordinated regulatory action. This regulatory relationship between genes has traditionally been uncover through biological experimentation [46]. New high-throughput technologies, such as time-series RNA-seq and ChIP-seq, provide ample opportunities to identify novel regulatory gene relationships or Gene Regulatory Networks (GRNs). These technologies, together with the availability of null or transgenic animals and/or ES cells, can allow us to reconstruct GRNs for specific TFs and potentially afford us with a dynamic insight into the transcriptional reprogramming that takes place in differentiating ES cells. In previous work, we had generated mouse Hoxa1-null ES cells and used cDNA microarrays to identify a number of putative Hoxa1 target genes [34]. Thus, for this work, we aimed to construct the Hoxa1 GRN on differentiating mouse ES cells using the NARROMI algorithm. NARROMI requires the time-series RNA-seq data to be input as two separate sets: one is the total number of genes or TG, and the other is the Transcription Factor (TF) data set extracted from the TG set. Our strategy was to first generate TG by performing time-series RNA-seq analyses on D3 (Wt) mouse ES cells. For this purpose, ES cells were treated with 1 mM RA for 0, 12, 23, 36, 48, and 60 hours as described in Materials and Methods. The experiments were done by quadruplicate and whole transcriptome analyses were performed by Novogene, Inc. Filtering for low expression and significant differential expression yielded a preliminary list comprised of 16133 genes. The expression profile for the genes in this preliminary set is shown as a clustered heatmap in Figure 1a, where the average values from four repetitions were used for constructing the heatmap. The Venn diagram in Figure 1b shows the overlaps between differentially-expressed genes (DEGs) among different time-point treatments. The differential expression between RA-treated versus control samples at 48 hours is depicted as a volcano plot in figure 1c. To further trim the TG set for optimal NARROMI analysis, genes were selected if they showed a differential expression of 2-fold or higher between any of the RA-treatment time-points and the untreated control. This produced a working TG set comprised of 4060 genes (Supplementary Data 1).
Figure 1: Time series transcriptomic analysis of Wt ES cells treated with RA. The NARROMI algorithm for GRN construction requires a TG (Total Genes) and a TF (Transcription Factor) data sets. Time series RNA-seq data was used to extract the TG set. A) Clustering heatmap for TG set. B) Venn diagram showing overlap between differentially expressed genes (DEGs) in Wt cells at different times of RA treatment. C) Volcano plot for DEGs at 48 hours of treatment.
Table: Expression of Differentiation Markers in Wt ES cels (in FPKM)
|
Sample ID |
Cti_48hrs |
RA_12hrs |
RA_24hrs |
RA_36hrs |
RA_48hrs |
RA_60hrs |
|
Nanog |
205.56 |
97.34 |
94.38 |
102.82 |
126.41 |
104.43 |
|
Nes |
4.87 |
6.39 |
9.15 |
13.52 |
22.41 |
83.11 |
|
Mest |
48.48 |
77.26 |
207.63 |
459.74 |
650.76 |
502.84 |
|
SOX 17 |
1.36 |
0.94 |
1.26 |
2.24 |
4.01 |
5.04 |
|
Tubb3 |
23.12 |
12.43 |
12.15 |
12.09 |
10.17 |
25.39 |
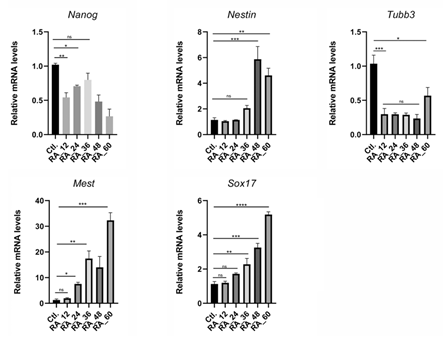
Figure 2: Verification of RNA-seq data by qPCR analysis of ES differentiation markers. The table indicates the genes tested and their FPKM values from RNA-seq. Results from qPCR are shown below. In general, qPCR analyses verified the RNA-seq data for the genes tested. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001 by one-way ANOVA. Means ± SEM are shown.
3.2. Time-series expression of differentiation markers by qPCR
Our RNA-seq results indicate that markers for all three lineages are upregulated in mouse ES cells after RA treatment. Thus, to begin the validation of our time-series RNA-seq results, we performed qPCR analyses for a group of known differentiation markers, including the stem cell marker Nanog, the neuronal precursor Nestin (Nes), Mest (mesodermal marker), and the endodermal marker Sox17. As shown in Table 1 for average RPKM values, RNA-seq results indicate that Nanog expression quickly decreased by more than two-fold after 12 hours of RA treatment, with slight increases at subsequent time-points, but always staying close to the two-fold mark decrease as compared to controls. On the other hand, Nes expression slowly increased after RA treatment followed by a dramatic increase of about 8-fold by 60 hours of treatment. Expression of Tubb3, a neuronal marker also found to be expressed in undifferentiated ES cells, initially decreased after treatment but then increased to reach initial levels of expression at 60 hours of RA treatment. Contrarily, Mest expression was found to increase after RA treatment in a time-dependent manner, reaching a maximum of about 13-fold at 48 hours of treatment versus control. Sox17 expression levels were low in untreated control cells (1.36 FPKM) and increased after RA treatment, reaching a maximum of 5.04 FPKM (about 3.6-fold increase) at 60 hours.
With regards to the qPCR results, Nanog gene expression declined, as expected, after treatment of cells with RA. Nanog mRNA expression decreased by about two-fold after 60 hours of RA treatment. Conversely, Nes expression did not significantly change during the first 36 hours of treatment but increased to about 6-fold at 48 hours as compared to controls. Nes mRNA expression then decreased to about 5-fold, with respect to controls, at 60 hours. As it was the case with the RNA-seq results, Tubb3 mRNA expression was detected by qPCR in untreated controls and levels quickly decreased after RA treatment for 12 to 48 hours, and then presented a small increase at 60 hours whenTubb3 mRNA expression levels were about half of those of controls. In the case of Mest, mRNA expression was found to significantly increase with RA treatment at 24 hours, with a steady increase afterwards, reaching a maximum of about 32-fold with respect to controls at 60 hours of RA treatment. In the case of Sox17, mRNA expression levels also progressively increased with RA treatment, reaching the highest peak of about 5-fold at 60 hours. Thus, the patterns of marker expression obtained from qPCR analyses were similar to those obtained by RNA-seq, which validates our time-series transcriptome analyses.
3.3 Hoxa1 Gene Regulatory Network
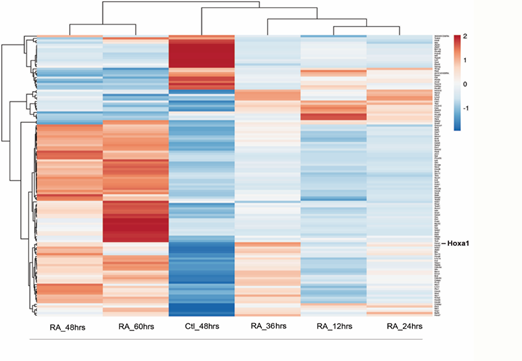
Hoxa1 plays important roles during early embryogenesis and, in vitro, it has been found to be required for the neuronal differentiation of mouse ES cells into neurons. Furthermore, in human cells, HOXA1 has been identified as an important oncogene that drives tumor growth and metastasis when overexpressed. Although we and others have reported the identification of multiple putative Hoxa1-related genes, little is known about the hierarchical relationship between Hoxa1 and its target genes, including those that code for transcription factors. Thus, to infer a Hoxa1-dependent GRN, we next sought to identify transcription factors within the TG set that were differentially expressed in Wt versus Hoxa1-null mouse ES cells as determined by RNA-seq analyses (see Materials and Methods). This analysis identified 124 transcription factors that were differentially expressed in Wt vs Hoxa1 KO Es cells. The resulting TF set used for Narromi is depicted as a heatmap in Fig. 3. The list of genes that constitute the TF set is provided as Supplemental Materials (Supplementary Data 2).
3.4. To verify the relationship between the selected genes for the TF set and the Hoxa1 signaling pathway
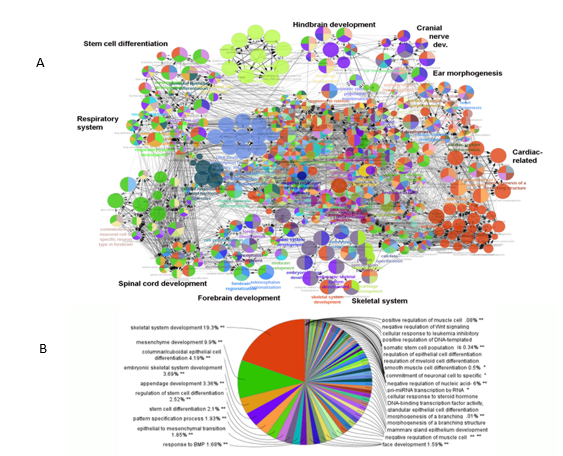
We employed ClueGo, a Cytoscape plugin47, to perform pathway analysis of TF members and to attain their complete Gene Ontological terms (GO). The resulting analysis, depicted in Figure 4, indicated that the genes selected for the TF set are involved in processes found to be defective in Hoxa1 knockout mice. These processes include hindbrain and cranial nerve development, ear morphogenesis, cardiovascular development, and respiration, among others [15-18,48]. Thus, pathway analyses using ClueGo verified the relationship between genes that integrate the TF set and the Hoxa1 signaling pathway.
Figure 4: Pathway analysis using ClueGo. Functional analysis indicated that selected TFs interconnected to pathways related to observed deficiencies in Hoxa1 knockout mice. A) Representation of GO terms enriched in the TF gene set (p<0.005). Processes with observed deficiencies in Hoxa1 knockout mice are shown in bold. B) Pie-chart indicating the prevalence of GO terms among TF genes.
3.5. Construction of the Hoxa1 GRN
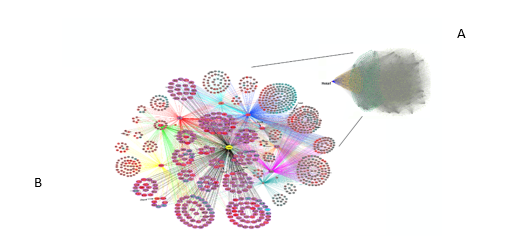
Following the verification of the Hoxa1-related TF set by Gene Ontology, we then conducted the NARROMI analysis on MATLAB as described in Materials and Methods. The analysis from NARROMI resulted in a network containing a total of 27497 connections among genes or nodes. Those connections were visualized using Cytoscape (Figure 5, top right). An amplified partial view of the GRN is also shown for Hoxa1 and its first- and second-level connections (first- and second- neighbors). In this amplified view, the nodes are colored based on the differential expression of genes at 48 hours of RA treatment versus untreated control. Red color indicates increase of expression, and green indicates decrease of expression after treatment. In this view, the size of the nodes indicates whether the genes are first-Hoxa1 neighbors (larger nodes) or second neighbors (smaller nodes). In this network, NARROMI identified 281 direct connections between Hoxa1 and its putative targets (see Supplementary Data 3). Thus, using NARROMI, we were able to construct the Hoxa1 GRN and identified a set of putative direct targets of this TF, along with its indirect second-tier neighbors. A high-resolution image for figure 5 is provided as Supplementary Data 4.
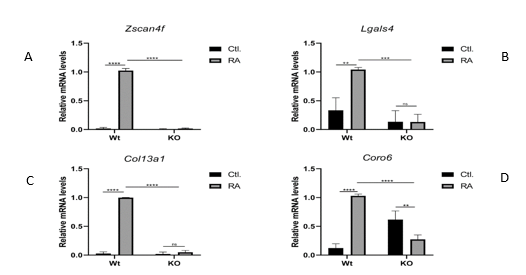
3.6. To verify the gene connections identified by NARROMI, and to test the Hoxa1-dependence of first and second-level neighbors identified in the GRN, we examined the expression of selected genes by qPCR in Wt vs Hoxa1-null mouse ES cells cultured in the presence or absence of 1 mM RA for 48 hours. To accomplish this goal, we examined two genes that are directly connected to Hoxa1, Lgals and Zscan4f, and two genes that are indirectly connected to Hoxa1 through Hoxa10, Col13a1 and Coro6. As observed in figure 6, the mRNA expression of both Zscan4f and Lgals4 significantly increased in Wt ES cells after RA treatment, but no increase by RA was observed in Hoxa1-null cells (panels A and B). Thus, these results indicate that Hoxa1 is required for the expression of both Zscan4f and Lgals4. With respect to the qPCR analysis of indirect Hoxa1 target genes expression, we found that both Col13a1 and Coro6 mRNA levels were significantly upregulated by treatment with RA (figure 6, panels C and D) after 48 hours. As it was the case with Zscan4f and Lgals4, RA treatment failed to upregulate the mRNA expression levels of these two genes in Hoxa1-null cells. Interestingly, the levels of Coro6 mRNA expression were found to be significantly higher in untreated Hoxa1-null than in untreated Wt ES cells, which indicates that small basal differences exist between these two cell lines even in the absence of RA treatment. All together, our qPCR results indicate that the examined genes, both first- and second-level neighbors, are all dependent on Hoxa1 for their expression, which constitutes an initial validation of the Hoxa1 GRN constructed using NARROMI with our time-series RNA-seq data.
Figure 5: Hoxa1 Gene Regulatory Network. The full network, top right panel (A), contains 27497 connections among genes or nodes, where Hoxa1 is displayed as a red dot towards the left of the network. In the full network, the nodes for the Hoxa1-first neighbors are colored yellow, while the second neighbor nodes are shown in green. All other nodes are gray. (B) Close-up view of the Hoxa1 GRN showing the first- and second-level neighbors of Hoxa1 only. Bigger ovals indicate putative Hoxa1 direct targets and smaller ovals indicate indirect targets or second Hoxa1 neighbors. Node colors indicate whether target genes are upregulated (red) or downregulated (blue) by Hoxa1. The indicated genes were selected for verification of the network by qPCR.
Figure 6: Verification of Hoxa1 GRN. Expression of selected putative direct (Zscan4f and Lgals4) and indirect (Col12a1 and Coro6) target genes was examined by qPCR on Wild Type versus Hoxa1-null mouse ES cells grown in the presence (RA) or absence (Ctl.) of Retinoic Acid treatment. These results demonstrated that the genes tested depend on Hoxa 1 for transcriptional activation, which correlates with the findings from the constructed GRN.
* p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001 by one-way ANOVA. Means ± SEM are shown.
4. Discussion
Embryonic stem cells are critical in regenerative medicine due to their ability to differentiate into all cell types of the body [49]. Retinoic acid, a potent inducer of ES cell differentiation, acts by activating the expression of genes that code for important developmental proteins, including the members of the Hox family of Transcription Factors [50, 51]. Hoxa1, the most 3’ member of the Hoxa cluster, is a direct RA target and a master regulator of mouse embryogenesis [52]. In humans, its overexpression correlates with tumor cell proliferation in different types of cancer [30-32, 53].
Although, extensive research has shed light into the molecular mechanism of Hoxa1 action through the identification of putative Hoxa1 target genes [33-35], the regulatory relationship between these targets remains largely unknown. In this study, we sought to shed light on this gap of knowledge by constructing a Hoxa1 Gene Regulatory Network (GRN) using RNA-seq time series data analyzed by the NARROMI algorithm. Our base information for NARROMI consisted in filtered RNA-seq data from Wild-type ES cells treated with 1 mM RA for different times: 0, 12, 24, 36, 48, and 60 hours. These data constituted the TG set required for running the algorithm. To generate a GRN specific for Hoxa1, RNA-seq from mouse Hoxa1-null ES cells treated with RA for 48 hours was employed to identify TF-coding genes differentially-expressed between Wt and Hoxa1-null ES cells treated with RA for 48 hours. This time point was selected for the differential expression analysis because this is when we observed the highest expression levels of Hoxa1 mRNA in Wt ES cells treated with RA [34]. From the NARROMI analysis, we found a group of 281 genes that were directly connected to Hoxa1 (Figure 5). This group included Hoxa1 itself, which suggest an autoregulatory feed-back loop for this gene (see Supplementary Data 3). Among the primary Hoxa1 neighbors, Zscan4f and Lgals were selected for qPCR verification, and were confirmed to be differentially expressed between Wt and Hoxa1-null ES cells treated with RA for 48 hours. Importantly, these genes have been found to be key regulators of various developmental processes. Zscan4f plays a role in the regulation of telomere length and genomic stability in ES cells, as well as in the regulation of pluripotency and early embryonic development [54]. Similarly, Lgals is involved in a wide range of biological processes, including cell adhesion, signaling, differentiation, apoptosis, and immune modulation [55]. Our qPCR analyses also verified the differential expression of the second-level Hoxa1 neighbors Col13a1 and Coro6, who were indirectly connected to Hoxa1 through Hoxa10. Research suggests that Col13a1 plays a role in the development and maintenance of neuromuscular junctions [56], while Coro6 was found to be differentially regulated in muscle after denervation [57]. Similarly, the protein expression of Coronin 6 was upregulated in skeletal muscle during development and after denervation [58]. Interestingly, Hoxa10, which connects Hoxa1 to Col13a1 and Coro6 in our GRN, has been found to play a role during murine smooth muscle development [58] and during the regeneration of skeletal muscle in adult mice [59, 60], which hints to a role for Hoxa1 during the development and regeneration of murine muscles. Although qPCR was used as one validation method for the Hoxa1 GRN, there are limitations that must be considered when using GRNs. One limitation of using a gene regulatory network (GRN) is that they are often constructed based on data from a single cell type or developmental stage, which may not accurately reflect the complex interactions that occur between genes and signaling pathways in vivo [61]. The GRN constructed by our team addresses this issue by employing RNA-seq data from both Wt and Hoxa1-null mouse ES cells. Nevertheless, as the accuracy of GRN predictions are known to vary widely depending on the algorithm and modeling approach used, which highlights the challenges of constructing reliable GRNs [61], these limitations underscore the need for rigorous experimental validation of GRN predictions and the use of complementary approaches, such as genetic perturbations and single-cell transcriptomics, to refine and improve GRN models. To refine our GRN model, we are currently performing proteomic analyses on Wt vs Hoxa1-null mouse ES cells treated with RA for various periods of time. Taken together, our approach is the first step on the elucidation of the regulatory interactions among Hoxa1 and its targets. Furthermore, our TG data can be used to construct a GRN for other TFs if a list of differentially expressed transcription factors is generated or obtained from published data for mouse ES cells.
Contributions
EMC conceptualized the study. EMC and UE conducted most of the experiments and performed NARROMI analyses. YB handed bioinformatics data. BR wrote sections of the manuscript and provided advice on the interpretation of genomic data, XY, AW, and OD wrote sections of the manuscript and provided editorial advice. CC submitted the RNA-seq data to the database. SAO and NA performed some qPCR experiments and performed the ClueGo analyses.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Funding
This work was supported by a grant from the National Science Foundation (Grant number 1901346 to EMC).
Data Availability
Transcriptome data has been deposited in NCBI's BioProject accession number PRJNA770639.
References
- Deschamps J and Meijlink F. Mammalian homeobox genes in normal development and neoplasia. Crit Rev Oncog 3 (1992): 117-173.
- Duboule D and Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet 10 (1994): 358-364.
- Means AL, Gudas LJ. The roles of retinoids in vertebrate development. Ann Rev Biochem 64 (1995): 201-233.
- Capecchi MR. Hox genes and mammalian development. Cold Spring Harb Symp Quant Biol 62 (1997): 273-281.
- Cillo C, Cantile M, Faiella A and Boncinelli E. Homeobox genes in normal and malignant cells. J Cell Physiol 188(2001): 161-169.
- Mavilio F. Regulation of vertebrate homeobox-containing genes by morphogens. Eur J Biochem 212 (1993): 273-288.
- Simeone A, Acampora D, Arcioni L, Andrews PW, Boncinelli E, Mavilio F. Sequential activation of HOX2 homeobox genes by retinoic acid in human embryonal carcinoma cells. Nature 346 (1990): 763-766.
- Stornaiuolo A, Acampora D, Pannese M, D'Esposito M, Morelli F, Migliaccio E, et al. Human HOX genes are differentially activated by retinoic acid in embryonal carcinoma cells according to their position within the four loci. Cell Differ Dev 31 (1990): 119-127.
- Langston AW, and Gudas LJ. Identification of a retinoic acid responsive enhancer 3' of the murine homeobox gene Hox-1.6. Mech Dev 38 (1992): 217-227.
- Langston AW, Thompson JR, and Gudas LJ. Retinoic acid-responsive enhancers located 3' of the HoxA and HoxB homeobox gene clusters. J Biol Chem 272 (1997): 2167-2175.
- LaRosa GJ and Gudas LJ. The early retinoic acid-induced F9 teratocarcinoma stem cell gene ERA-1: alternate splicing creates both a transcript for a homoebox-containing protein and one lacking the homeobox. Mol Cell Biol 8 (1988): 3906-3917.
- LaRosa GJ and Gudas LJ. An early effect of retinoic acid: cloning of an mRNA (Era-1) exhibiting rapid and protein synthesis-independent induction during teratocarcinoma stem cell differentiation. Proc Natl Acad Sci 85 (1988): 329-333.
- Akin ZN and Nazarali AJ. Hox genes and their candidate downstream targets in the developing central nervous system. Cell Mol Neurobiol 25 (2005): 697-741.
- Lichtneckert R and Reichert H. Insights into the urbilaterian brain: conserved genetic patterning mechanisms in insect and vertebrate brain development. Heredity 94 (2005): 465-477.
- Carpenter EM, Goddard JM, Chisaka O, Manley NR and Capecchi MR. Loss of Hox-A1 (Hox-1.6) function results in the reorganization of the murine hindbrain. Development 118 (1993): 1063-1075.
- Chisaka O, Musci TS and Capecchi MR. Developmental defects of the ear, cranial nerves and hindbrain resulting from targeted disruption of the mouse homeobox gene Hox-1.6. Nature 355 (1992): 516-520.
- Lufkin T, Dierich A, Le Meur M, Mark M and Chambon P. Disruption of the Hox-1.6 homeobox gene results in defects in a region corresponding to its rostral domain of expression. Cell 66 (1991): 1105-1119.
- Mark M, Lufkin T, Vonesch JL, Ruberte E, Olivo JC, Dolle P, et al. Two rhombomeres are altered in Hoxa-1 mutant mice. Development 119 (1993): 319-338.
- Gavalas A, Trainor P, Ariza-McNaughton L, and Krumlauf R. Synergy between Hoxa1 and Hoxb1: the relationship between arch patterning and the generation of cranial neural crest. Development 128 (2001): 3017-3027.
- Hunt P, Wilkinson D and Krumlauf R. Patterning the vertebrate head: murine Hox 2 genes mark distinct subpopulations of premigratory and migrating neural crest. Development 112 (1991): 43-51.
- Murphy P and Hill RE. Expression of the mouse labial-like homeobox-containing genes, Hox 2.9 and Hox 1.6, during segmentation of the hindbrain. Development 111 (1991): 61-74.
- Technau U and Scholz CB. Origin and evolution of endoderm and mesoderm. Int J Dev Biol 47 (2003): 531-539.
- Fukuda K and Kikuchi Y. Endoderm development in vertebrates: fate mapping, induction and regional specification. Dev Growth Differ 47 (2005): 343-355.
- Graham A, M Okabe, et al. The role of the endoderm in the development and evolution of the pharyngeal arches. J Anat 207 (2005): 479-487.
- Kaestner KH. The making of the liver: developmental competence in foregut endoderm and induction of the hepatogenic program. Cell Cycle 4 (2005): 1146-1148.
- Familari M. Characteristics of the endoderm: embryonic and extraembryonic in mouse. Scientific World Journal 6 (2006): 1815-1827.
- Lewis SL. and Tam PP. Definitive endoderm of the mouse embryo: formation, cell fates, and morphogenetic function. Dev Dyn 235 (2006): 2315-2329.
- Ingram JL, Stodgell CJ, Hyman SL, Figlewicz DA, Weitkamp LR, Rodier PM. Discovery of allelic variants of HOXA1 and HOXB1: genetic susceptibility to autism spectrum disorders. Teratology 62 (2000): 393–405.
- Bosley TM, Alorainy IA, Salih MA, Aldhalaan HM, Abu-Amero KK, Oystreck DT, et al. The clinical spectrum of homozygous HOXA1 mutations. Am J Med Genet, A 146A (2008): 1235–40.
- Liu LJ, Sun XY, Yang CX, & Zou XY. MiR-10a-5p restrains the aggressive phenotypes of ovarian cancer cells by inhibiting HOXA1. The Kaohsiung Journal of Medical Sciences 37 (2021): 276-285.
- de Bessa Garcia SA, Araújo M, Pereira T, Mouta J & Freitas R. HOX genes function in breast cancer development. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer 1873 (2020): 188358.
- Mohankumar KM, Xu XQ, Zhu T, Kannan N, Miller LD, Liu ET, et al. HOXA1-stimulated oncogenicity is mediated by selective upregulation of components of the p44/42 MAP kinase pathway in human mammary carcinoma cells. Oncogene 26 (2007): 3998–4008.
- She J, Wu H and Gudas LJ. Molecular cloning and analysis of a group of genes differentially expressed in cells which overexpress the Hoxa-1 homeobox gene. Exp Cell Res 259 (2000): 274-83.
- Martinez-Ceballos E, Chambon P, and Gudas LJ. Differences in gene expression between Wild type and Hoxa1 knockout embryonic stem cells after retinoic acid treatment or leukemia inhibitory factor (LIF) removal. J Biol Chem 280 (2005): 16484-16498.
- Makki N and Capecchi MR. Identification of novel Hoxa1 downstream targets regulating hindbrain, neural crest and inner ear development. Dev Biol 357 (2011): 295-304.
- Zhang X, Liu K, Liu ZP, Duval B, Richer JM, Zhao XM, et al. NARROMI: a noise and redundancy reduction technique improves accuracy of gene regulatory network inference. Bioinformatics 29 (2013): 106-113.
- Yang B, Xu Y, Maxwell A, Koh W, Gong P, and Zhang C. MICRAT: a novel algorithm for inferring gene regulatory networks using time series gene expression data. BMC systems biology 12 (2018): 19-29.
- Wang Y, Joshi, T, Zhang XS, Xu D and Chen L. Inferring gene regulatory networks from multiple microarray datasets. Bioinformatics 22 (2006): 2413-2420.
- Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society: Series B (Methodological) 58 (1996): 267-288.
- Geeven G, van Kesteren RE Smit AB and de Gunst MC. Identification of context-specific gene regulatory networks with GEMULA—gene expression modeling using LAsso. Bioinformatics 28 (2012): 214-221.
- Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R and Califano A. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. In BMC bioinformatics 7 (2006): 1-15.
- Irrthum A, Wehenkel L and Geurts P. Inferring regulatory networks from expression data using tree-based methods. PloS one 5 (2010): e12776.
- Addae C, Yi X, Gernapudi R, Cheng H, Musto A, Martinez-Ceballos E. All-trans-retinoid acid induces the differentiation of encapsulated mouse embryonic stem cells into GABAergic neurons. Differentiation 83 (2012): 233–241.
- Mortazavi, Ali, Brian A. Williams, Kenneth McCue, Lorian Schaeffer, and Barbara Wold. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods 5 (7): 621-628.
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13 (2003): 2498–2504.
- Lemmens K, Dhollander T, De Bie T, Monsieurs P, Engelen K, Smets B, et al. Inferring transcriptional modules from ChIP-chip, motif and microarray data. Genome biology 7 (2006): 1-14.
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25 (2009): 1091-1093.
- Makki N and Capecchi MR. Cardiovascular defects in a mouse model of HOXA1 syndrome. Human molecular genetics 21 (2012): 26-31.
- Baker DE, Harrison NJ, Maltby E, Smith K, Moore HD, Shaw PJ, et al. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nature 45 (2007): 1355-1361.
- García-Carpizo V, Ruiz-Llorente L, Menéndez-Gutiérrez MP, Lafarga V, Ibáñez de Cáceres I, Lazo PA, et al. Retinoic acid activation of Hoxa1 gene expression in embryonic stem cells involves retinoid receptor-mediate and active DNA demethylation. BMC Developmental Biology 11 (2011): 1-16.
- Gudas LJ and Wagner JA. Retinoids regulate stem cell differentiation. Journal of cellular physiology 226 (2011): 322-330.
- De Kumar B, Parker HJ, Paulson A, Parrish ME, Zeitlinger J and Krumlauf R. Hoxa1 targets signaling pathways during neural differentiation of ES cells and mouse embryogenesis. Developmental Biology 432 (2017): 151-164.
- Shenoy US, Adiga D, Kabekkodu SP, Hunter KD and Radhakrishnan R. Molecular implications of HOX genes targeting multiple signaling pathways in cancer. Cell Biology and Toxicology (2022): 1-30.
- Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL and Golub TR. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 464 (2010): 858-863.
- Rabinovich GA and Toscano MA. Turning 'sweet' on immunity: galectin–glycan interactions in immune tolerance and inflammation. Nature Reviews Immunology 9 (2009): 338-352.
- Kowal K, Plóciennikowska A, Klimaszewska-Wisniewska A and Rychlowski M. Type XIII collagen: a new actor in healthy and diseased tissues. International Journal of Biochemistry & Cell Biology 112 (2019): 66-70.
- Tang H, Cheung WM, Ip FC and Ip NY. Identification and characterization of differentially expressed genes in denervated muscle. Molecular and Cellular Neuroscience 16 (2000): 127-140.
- Chen Y, Ip FC, Shi L, Zhang Z, Tang H, Ng YP, et al. Coronin 6 regulates acetylcholine receptor clustering through modulating receptor anchorage to actin cytoskeleton. Journal of Neuroscience 34 (2014): 2413-2421.
- El-Mounayri O, Triplett JW, Yates CW and Herring BP. Regulation of Smooth Muscle-specific Gene Expression by Homeodomain Proteins, Hoxa10 and Hoxb8. Journal of Biological Chemistry 280 (2005): 25854-25863.
- Yoshioka K, Nagahisa H, Miura F, Araki H, Kamei Y, Kitajima Y, et al. Hoxa10 mediates positional memory to govern stem cell function in adult skeletal muscle. Science Advances 7 (2021): 7924.
- Gitter A, Bar-Joseph Z and Berman J. Challenges and opportunities in computational developmental biology. BMC Developmental Biology 19 (2019): 1-9.


 Impact Factor: * 3.5
Impact Factor: * 3.5 Acceptance Rate: 71.36%
Acceptance Rate: 71.36%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
