Performance of GBTsol ICA System: A New Point-of-Care Testing Instrument for HIV Monitoring Through CD4 and CD8 Enumeration in Resource-Poor Settings
Stephen Sung1, Hyon-Suk Kim2, Jae-Woo Song2, Jae-Young Lee1, Shabnam Aghamoradi*, 1, Seung-Lim Back1
1Glory Biotechnologies Corp., Seoul, Republic of Korea.
2Department of Laboratory Medicine, College of Medicine, Yonsei University, Seoul, Republic of Korea.
*Corresponding author: Shabnam Aghamoradi, Glory Biotechnologies Corp., Seoul, Republic of Korea.
Received: 30 January 2023; Accepted: 06 February 2023; Published: 22 February 2024
Article Information
Citation:
Stephen Sung, Hyon-Suk Kim, Jae-Woo Song, Jae-Young Lee, Shabnam Aghamoradi, Seung- Lim Back. Performance of GBTsol ICA System: A New Point-of-Care Testing Instrument for HIV Monitoring Through CD4 and CD8 Enumeration in Resource-Poor Settings. Archives of Microbiology and Immunology. 8 (2024): 35-43.
View / Download Pdf Share at FacebookAbstract
This study demonstrates the application of a POCT microchip platform for enumerating leukocytes, CD4+ T-lymphocytes, and CD8+ T-lymphocytes in whole blood, using fluorochrome-conjugated primary antibodies as a detection method. Specifically, we used phycoerythrin conjugated primary antibodies specific to CD4 and CD8 antigens to enumerate CD4+ and CD8+ T cells. Comparative analysis was performed using Fluorescence-Activated Cell Sorting (FACS) count to evaluate total leukocytes, CD4+ T cell count, and CD8+ T cell count in whole blood samples, aiming at monitoring the immune systems of patients with human immunodeficiency virus infection. Statistical analyses for precision, correlation, and agreement were performed. Coefficients of variation ranging from 0.67% to 12.78%, 0.81% to 13.68%, and 0.29% to 8.33% were obtained for CD4, CD8, and leukocyte recovery, respectively. A significant correlation was identified between the two assays for CD4 and CD8 counts, exhibiting correlation coefficients of 0.90 and 0.91, respectively. Using Bland–Altman plots, we determined a mean bias of 23, 38, and 490 cells/µ L (95% CI, n = 113) for CD4, CD8, and total leukocyte counts, respectively. These findings affirm that the GBTsol ICA (Immune Cell Analyzer) is comparable to the FACS count platform method, providing a cost-effective, user-friendly, and expeditious approach for quantifying CD4 T cells, CD8 T cells, and total leukocytes in blood samples, facilitating the monitoring of HIV/AIDS patients.
Keywords
<p>CD marker, Point-of-care Testing (POCT), Leukocyte, Acquired Immune Deficiency Syndrome, CD4/CD8, immune cell counting, AIDS, HIV</p>
Article Details
1. Introduction
Importance of CD4 and CD8 enumeration monitoring in patients with HIV/AIDS
As of December 2022, an estimated 39.0 million (33.1–45.7 million) people worldwide were living with HIV infection, with a majority residing in resource-limited areas. Monitoring the health of HIV patients requires methods such as CD4 counting and assessing the CD4/CD8 ratio. However, many available laboratory tests are technically complex, time-consuming, expensive, and impractical for use in resource-limited settings. The CD4 cell count has played a central role in the care of both HIV-infected children and adults, serving as a crucial measure of immunosuppression and guiding decisions regarding antiretroviral therapy (ART). The routine collection of CD4 data at the time of diagnosis remains essential in shaping treatment priorities and plays a critical role in identifying instances of late diagnosis [1-3]. One reason for the underutilization of CD4 results is the limited accessibility of clinical centers and centralized laboratories, where the current gold standard technology, flow cytometry, has been in use for 40 years. However, there are many limitations to traditional flow cytometry, including the ability of the laser to analyze only one cell at a time. Cells must be in suspension to be analyzed, highly trained technicians are required, intensive quality control measures are needed, and cells must be viable to be analyzed. These requirements have constrained CD4 testing in several resource-constrained settings, particularly in rural areas where clinical laboratories are not easily accessible [4-8]. In addition to the above-mentioned limitations, flow cytometry measurements lack standardization, and reproducible protocols for sample preparation, including red blood cell (RBC) lysis, cell staining, gating strategies, and acquisition protocols, have proven challenging to maintain consistently across multicenter clinical studies [9]. Furthermore, patients in many countries lack a unique patient number, making it difficult to trace test results from different laboratories, especially for patients referred between hospitals and clinics. Long turnaround times for tests sent to central laboratories delay clinical decision-making and impose a significant strain on patients.
Conventional tests involve transporting samples in complex and insufficient transport networks over lengthy, rough highways. These transport networks are restricted, costly, and often compounded by short sample stability problems. A study in Mozambique, Malawi, and South Africa reported that as many as 50% of CD4 test results did not return to clinics for follow-up [10]. To address this drawback, healthcare programs are using POCT for CD4 results to facilitate individualized care, leading to higher retention in care and reduced loss to follow-up [11]. This commercial tool of the 21st century with a peculiar CD marker measurement method within multiple maladies provides a solution when and where the absence of similarities in others cannot. Hence, in this study, we focused on the accuracy, reproducibility, and time effectiveness of the POCT technology (GBTsol ICA) compared with the results of FACS count data. The POCT technology (GBTsol ICA) is based on leukocyte separation from whole blood, labeling of all leukocytes with DNA fluorescent stains, and CD marker enumeration (CD4+ and CD8+) using primary antibodies conjugated with fluorescent markers.
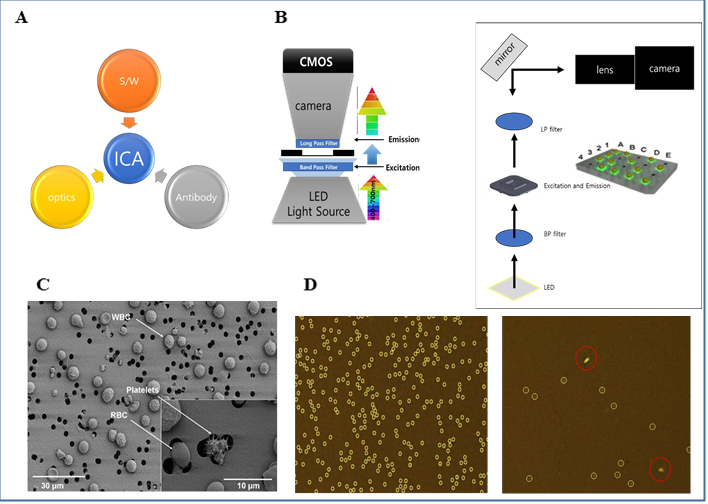
GBTsol ICA system
The GBTsol ICA system comprises three main components. The first is an ICA reader machine, the second is a cartridge, and the third part involves reagents and an optimized phycoerythrin (PE) conjugated CD marker antibody. Each component is meticulously designed and plays a crucial role in the results of the experiments. Various parts of the ICA are shown in Fig. 1. The cartridge is disposable and made of three parts with a specified height and a filter membrane with 3 μm pores; it plays a key role in separating WBCs, eliminating the need for RBC lysis or the use of a centrifuge for cell separation, which can result in cell loss during the experiment. As a CD marker antibody, we used mouse IgG anti-human CD4-PE from clone RPA-T4 for CD4 marker detection and mouse IgG anti-human CD8-PE from clone OKT-8 for CD8 marker detection. These antibodies are optimized with highly purified antibodies and can react with whole blood, without the need to lyse RBCs, and the reaction is completed in five minutes.
1) ICA imaging system consist of optic part (Cool LED, Filter, CMOS CAMERA), Software part (graphical user interface, GUI, Image Capture, Analyzing), Biotechnology part (CD and WBC antibody Fluorescence Dye).
2) In optic parts LPF (Long-Pass Filter) filtering out unwanted wavelengths of fluorescence.
BPF (Band-Pass Filter) allows signals within a selected range of frequencies to be detect, while preventing signals at unwanted frequencies from getting through. Target cells will be visible by immunofluorescence antibody markers.
3) Cells more than 3um cannot pass through filter membrane but filter will let the excess dye pass through it during the washing.
Software part counts signal and filters the signal with different size or shape from target cell.
2. Methods and Materials
PE-conjugated CD4 and CD8 primary mouse antibodies were briefly centrifuged before use. Analytical-grade chemicals and reagent-grade solvents were used for all buffers and solutions. Dimethyl sulfoxide (DMSO), 0.5% trypsin-ethylenediaminetetraacetic acid (EDTA) solution (10X), and phosphate-buffered saline (PBS, pH 7.4) were procured from Sigma Chemical Co. (St. Louis, MO, USA). Fixative and flow cytometry (FC) reagents were purchased from Beckman Coulter (Fullerton, CA). Rainbow microsphere beads were purchased from BD Biosciences (San Diego, USA). Whole blood samples were obtained by a phlebotomist from anonymous donors at Severance Hospital (Yonsei University, Seoul) in accordance with the Institutional Review Board (IRB, 1-2018-009). Procedures involved using a standard venipuncture technique with EDTA as an anticoagulant in 5 mL tubes. Samples were evaluated within 24 hours of collection. Data were obtained from April 16, 2018, to August 15. 2018.
Methodology development for a biomarker detection kit
Typically, when a patient with HIV/AIDS visits a doctor, blood is collected and sent to the clinical laboratory for CD4 and CD4/CD8 tests with WBC count. Test results may be submitted to the doctor within 24–48 hours or up to one week, depending on the number of technicians in the laboratory, distance, resources available, and the chosen biomarker test. The device presented here is a POC detection kit designed to be used by a healthcare provider to readily identify and diagnose CD4, CD8, and leukocyte counts.
The GBTsol ICA detection kit consists of the following:
1) A microseparation filter for the entrapment of leukocytes.
2) A tube containing a leukocyte- specific antibody for a specific test.
3) A tube containing reaction buffer for specific reactions (i.e., CD4, CD8, leukocyte counting).
4) A tube containing washing solution to remove other proteins nonspecifically bound to the microseparation filter.
Each test used a different kit: GBTsol ICA001 for CD4 count, GBTsol ICA002 for CD8 count, and GBTsol ICA 003 for leukocyte count.
The GBTsol ICA procedure involves five steps:
1) Incubation of the sample and respective antibody depending on the test, i.e., CD4 test, CD8 test, or leukocyte test. 2) Wetting step for the removal of dust particles or any other unwanted substance from the Cartridge to lower background noise.
3) Sampling step for dispensing the incubated sample to the GBTsol ICA.
4) Washing step for the removal of extra antibody or unconjugated antibody from the cartridge.
5) Processing step for quantitative results for CD4, CD8, and leukocytes.
After the sample is added, the user injects a washing solution, following which the instrument processes the sample automatically. Results can be printed or sent to a storage device.
Sensitivity assessment
To assess assay sensitivity, various concentrations of microsphere rainbow beads were dispersed in phosphate-buffered saline (pH 7.0) at levels of 100, 250, 500, 1000, 1500, 2000, 2500, 3000, 3500, 4000, 4500, and 5000 microsphere rainbow beads per microliter. Bead counts were performed using GBTsol ICA, following the cell counting method. The counting process was repeated three times to determine the average number of microsphere rainbow beads.
Identification of leukocytes and enumeration of specific blood cell subsets
Blood samples were collected in EDTA tubes, and the antibodies used in these studies were stored at 4–8°C. Each blood sample (5µ L) was combined with a cell-staining solution (5µ L) (either GBTsol ICA-001 or GBTsol ICA-002) containing fluorescently labeled antibodies targeting CD antigens. This mixture was then incubated at room temperature for 5 minutes. After incubation, 990 µ L of reaction buffer was added. Subsequently, the samples were directly dispensed onto the GBTsol ICA cartridge for the enumeration of CD markers. The GBTsol ICA is a semiautomated and fully quantitative device. After the injection of samples, the tests were executed automatically to generate results.
Flow cytometry analysis
A FACSCount flow cytometer (NAVIOUS Ex Flow Cytometer Leukocyte Count) was used as the reference method. The protocol provided by the manufacturer was followed for conducting the flow cytometry analysis. True count tubes from BD Biosciences were used to determine absolute counts for leukocytes, CD4+ cells, and CD8+ cells.
Data management and analysis
All study data were collected and managed using electronic data capture tools based on our algorithm, hosted at Glory Biotechnologies Corp. The platform is a secure, password- protected, web-based application designed to facilitate data capture for research studies. All data were entered once, and each entry underwent thorough checks for accuracy. Access to the entered data was restricted solely to the staff directly involved in either data entry or analysis. Leukocyte, CD8, and CD4 results were exported directly from the GBTsol ICA device and analyzed using Microsoft Excel to determine the number of tests performed, rates of invalid tests, and types of invalid tests. Error assessment was conducted through scatterplot and best- line analyses using linear regression to determine the coefficient of determination (R2). Additionally, Bland–Altman analysis was performed to determine systematic bias, limit of agreement (LOA), and the imprecision of the GBTsol ICA.
Image analysis of leukocytes and specific blood cell subsets
Each blood sample underwent staining with fluorescently labeled antibodies targeting specific CD antigens for 5 minutes. Fluorescence intensities, cell size, and shape were used to identify leukocytes. Specific surface markers were used to classify leukocyte subpopulations, with a specific fluorescent dye conjugated to antibodies against each CD marker.
3. Results
The need for rapid, reproducible, sensitive, accurate, and user-friendly tests is crucial in healthcare. Such a test holds obvious advantages over existing tests and other instrument-based systems that use microfluidic technology to count cells on cartridges.
Specimen collection and GBTsol ICA performance
A total of 113 patients from Severance Hospital were tested over a 4-month period, with patient ages ranging from 8 to 65 years. Overall, there were minimal issues with specimen compromise during blood collection. Out of the 113 CD4 and CD8 tests performed using GBTsol ICA, 2 tests (1.76%) for CD4 and 2 tests (1.76%) for CD8 failed due to pipetting errors or improper sample handling. Additionally, 1 (0.88%) each for CD4 and CD8 showed clotting before conducting the respective tests (Tables 1 and 2). Another one (0.88%) of both CD4 and CD8 tests was aborted before completion. GBTsol ICA instrument errors were primarily associated with improper loading, either due to insufficient or excessive volume (Tables 1 and 2). However, GBTsol ICA CD4 and CD8 test cartridges or analyzers might play a role in some invalid results. Operation abortion of GBTsol ICA CD4 and CD8 tests can occur due to mechanical or technical problems with the analyzer itself or due to testing procedure errors by research personnel. We analyzed invalid GBTsol ICA test rates to determine if a particular problem was more responsible for invalid GBTsol ICA tests. The analysis indicated that pipetting errors or improper sample handling were the major contributors to invalid tests. However, no tests out of 113 conducted were deemed invalid due to error messages reported by GBTsol ICA. Further investigation revealed that CD4 counts less than 100 and CD8 counts greater than 9000 were not suitable for analysis with GBTsol ICA (data not shown). Many existing laboratory tests are time-consuming, tedious, and require trained personnel and expensive equipment [12]. POC-CD4 testing is valuable in the scale-up of ART for HIV care and linkage to treatment, especially in high-risk, resource-restricted settings, as conventional flow cytometry measurement of CD4 count often involves sending samples to a central research facility, causing delayed of 24–72 hours or even up to 7 days [13]. POC innovations can significantly reduce such delays, enabling faster and more immediate care. To improve testing capabilities in resource-restricted settings, it is crucial to have access to high-caliber and low-cost CD4 assays [12,14]. These POC instruments and assays should provide precise and reliable CD4 results while being simple to use. Therefore, in this study, a novel immunofluorescence test based on biomarker-specific antibodies was used to detect specific CD markers and validated for CD4 and CD8 monitoring in patients with HIV/AIDS. This study demonstrated high accuracy and reproducibility in predicting CD4 and CD8 counts using the GBTsol ICA device compared with flow cytometry conducted at Severance Hospital, Yonsei University, Seoul, South Korea.
Table 1: Reason for CD4 results recording failure out of 113 tests performed.
|
GBTsol ICA reading error |
0 |
0% |
|
Pipetting/Human error |
2 |
1.76% |
|
Blood Clotted |
1 |
0.88% |
|
Test Aborted |
1 |
0.88% |
|
Total |
4 |
3.53% |
Table 2: Reason for CD8 results recording failure out of 113 tests performed
|
GBTsol ICA reading error |
0 |
0% |
|
Pipetting/Human error |
2 |
1.76% |
|
Blood Clotted |
1 |
0.88% |
|
Test Aborted |
1 |
0.88% |
|
Total |
4 |
3.53% |
Reproducibility and efficiency of the GBTsol ICA
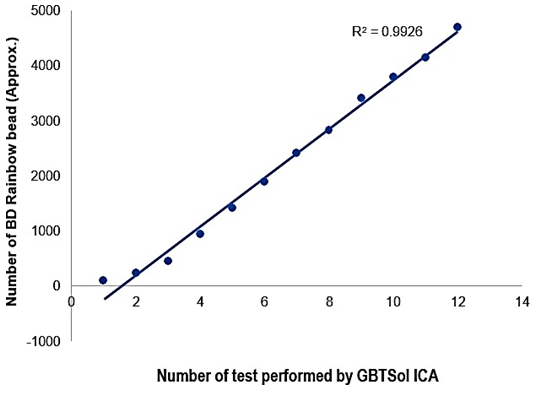
To assess the detection sensitivity of GBTsol ICA, rainbow beads at concentrations of 100, 250, 500, 1000, 1500, 2000, 2500, 3000, 3500, 4000, 4500, and 5000 beads per microliter were subjected to GBTsol ICA and detected using the device. The calculated detection efficiency consistently exceeded 90% when 100 to 5000 beads were present per 200 µ L of the rainbow bead sample (Fig. 2). The CV for various bead concentrations ranged from 2.21% to 5.38%, aligning with the WHO recommendation of < 15% for counts up to 500 and < 10% for counts up to 5000. Therefore, GBTsol ICA had excellent detection efficiency and reproducibility, facilitating accurate detection of CD4, CD8, and leukocytes (Fig. 4 (A, B), 5 (A, B), 6 (A, B)). The GBTsol ICA POC analyzer demonstrated high reproducibility with CV consistently below 5% when assessing both normal and low concentrations of beads. In this study, the GBTsol ICA POC analyzer overestimated counts compared with the BD FACSCount method in CD4+ T-cell enumeration, which is in agreement with most studies using capillary or venous blood [15-18]. However, this overestimation was minimal and deemed not clinically significant. Discrepancies have been reported in conventional CD4 testing platforms between BD FACSCount and BD FACSCalibur, with a mean bias of −76 cells/mm3 (95% CI, LOA - 316.0–163.0) [19]. Adequate correlation for CD4, CD8, and leukocyte counts between the GBTsol ICA analyzer and FACSCalibur (CVs of 0.90, 0.91, and 0.90, respectively) corroborate similar findings [13]. Although a correlation exceeding 0.90 was observed for CD4 enumeration between the two platforms, differences were due to variability in instrument settings, antibodies and fluorochromes used, sample volume inputs, and assay procedures and methods.
Figure 2: Evaluation of the sensitivity of GBTsol ICA at various target concentrations. A known number of rainbow microsphere beads (100-5000 beads) was added into 1 mL phosphate buffered saline (pH.7.0) and detected using the device integrated with a size-selective micro cavity array. The plot represents the number of cells recovered and the number of tests performed (n=12). CV ranged from 2.28 to 5.38%, and the correlation coefficient (R2) was 0.99.
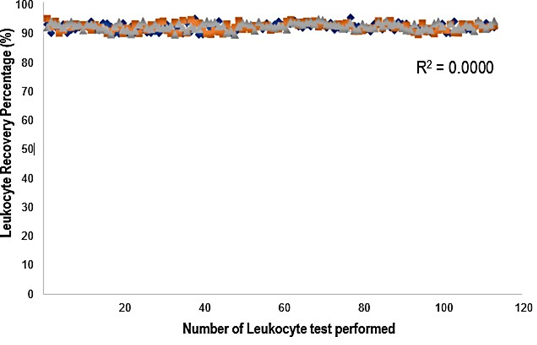
WBC counting by GBTsol ICA
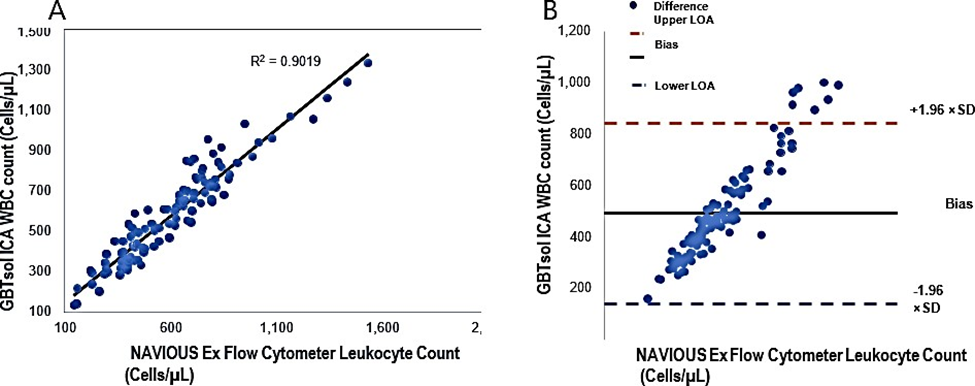
The leukocyte count was determined using GBTsol ICA for all 113 samples collected within 24 hours and was compared with FACSCount. The recovery of leukocytes exceeded 92% when compared with FACSCount (Fig. 3). The coefficient of determination (R2) was 0.90. The mean bias between the two platforms was 490 cells/µ L (95% CI LoA 139.25–840.84, n = 113), and the standard deviation of the mean was 178.97 (Fig. 4A). These results closely align with the FACSCount results, demonstrating high accuracy.
Figure 3: Leukocyte recovery rate from 113 blood samples using the GBTsol ICA device. Blood samples containing 1uL of whole blood were introduced into the leukocyte- counting device GBTsol ICA. Total leukocytes recovered by the GBTsol ICA were enumerated by an internal computer algorithm using RGB values. Leukocytes were incubated with DNA staining solution from the kit (GBTsol ICA-001). All counts with GBTsol ICA were repeated three times. The average number of leukocytes per microliter of whole blood was calculated. The correlation value was 0.98, the precision value was <10% (CV%), and the Leukocyte recovery percent was > 92%. Method comparison and correlation studies for total leukocyte counts of 113 human blood samples.
Figure 4: Linear regression and Bland-Altman analysis of GBTsol ICA and FACS count for WBC. In a comparison using venous blood for GBTsol ICA and FACScount, the coefficient of determination (R2) was 0.90 while the mean bias between these two platforms was 490 cells/µL (CI-95%, LoA 139.25 to 840.84, n=113) and the SD mean was 178.97. In the Bland Altman plot (Right), the difference between the 0 line and the black line indicates the bias of GBTsol ICA minus the NAVIOUS Ex Flow cytometer.
CD4 enumeration using GBTsol ICA
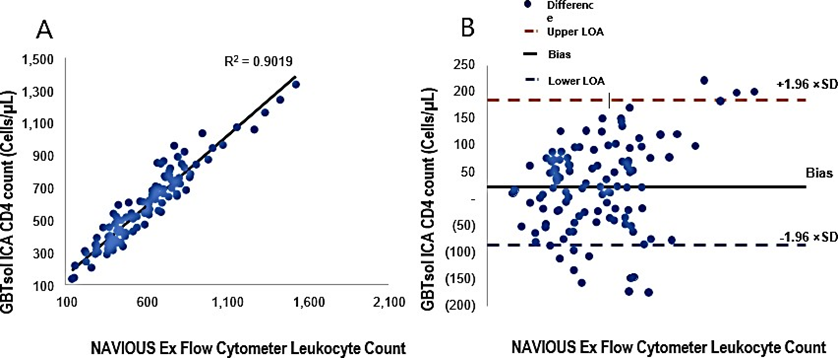
A total of 113 patient samples were collected over a 4-month period under the supervision of Yonsei University IRB board members. For each sample, 200 µ L containing 1 µL of blood was processed using GBTsol ICA. Subsequently, human whole blood samples with different known CD4+ T-cell counts, as determined by flow cytometry, were tested using GBTsol ICA. After subtracting the count due to nonspecific binding, the number of captured cells was plotted against the CD4 count, resulting in a correlation coefficient of 0.90 between our assay and standard flow cytometry (Fig. 5A). Despite the initial intention to use fluorescence exclusively for examining cell capture, challenges arose in counting individual T cells within clumps. The CD4 count exhibited CVs ranging from 0.67% to 12.78%. However, the Bland–Altman analysis revealed a mean bias of 23, with a 95% CI LoA between −86.47 and 186.16 (n = 113) (Fig. 5B). A recent report indicated that same-day POC CD4 testing did not show significant benefits on health outcomes [27]. In alignment with findings from other studies [20,21,25,26], we propose the feasibility of using an existing framework to establish a small-scale POC research facility to offer tests for the staging and pathology of ART. The overall accuracy of the GBTsol ICA test in samples was thoroughly assessed, and the correlation coefficient was 0.99, validating the reliability of GBTsol ICA. However, in the CD4 T-cell count, an overestimation was observed. This phenomenon of misclassification, overestimation, or underestimation has been documented in various studies using the PIMA POC analyzer [12,17,20-24].
Figure 5: Linear regression and Bland-Altman analysis of GBTsol ICA and FACS count for CD4. In a comparison using venous blood for GBTsol ICA and FACScount, the coefficient of determination (R2) was 0.90 while the mean bias between these two platforms was 23 cells/µL (CI-95%, LoA -86.47 to 186.16, n=113) and the SD mean was 83.23. In the Bland Altman plot (Right), the difference between the 0 line and the black line indicates the bias of GBTsol ICA minus the NAVIOUS Ex Flow Cytometer.
Enumeration of CD8 by GBTsol ICA
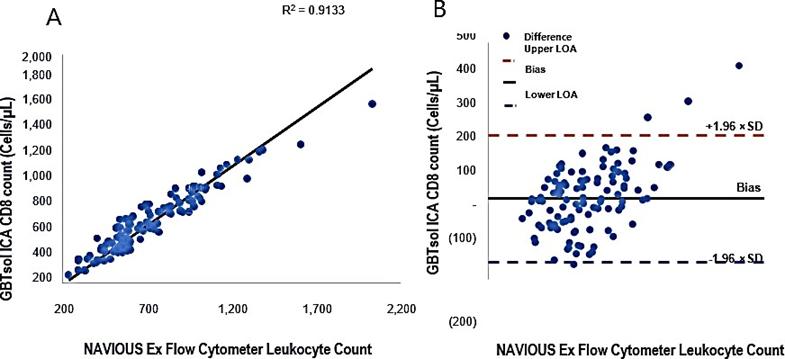
All samples used for CD4 enumeration underwent GBTsol ICA for the enumeration of CD8+ T cells. The assay demonstrated a robust correlation coefficient of 0.91 when compared with standard flow cytometry. Moreover, the CD8 count exhibited CVs ranging from 0.81% to 13.68% (Fig. 6A). However, upon conducting the Bland–Altman analysis, a mean bias of 38 was observed, with a 95% CI of LoA between −149.84 and 226.13 (n = 113) (Fig. 6B).
From an operational perspective, the use of the GBTsol ICA POC analyzer yielded results consistent with other studies using venous or capillary blood [12,13,20,22,27]. In our experience, test aborted and pipetting/human errors of 0.88% and 1.76% for CD4 and CD8, respectively. The operator in our study was a trained researcher rather than a health professional such as a nurse or counselor. Our studies indicated that the GBTsol ICA POC analyzer is interchangeable with conventional platforms and provides results comparable to those obtained with PIMA POC technologies [12,16,17,19,20,22,27]. To further validate these results, additional testing is warranted in communities with HIV/AIDS infection, performed by various health professionals, technicians, and hospitals. This additional testing can verify the CV of repeatability and assess misclassification, particularly in the context of under treatment compared with FACSCalibur [19]. Researchers in South Africa observed that the median time for patients to return for their CD4 results was 8 days for those with ≤ 200 cells/mm3, and 7 days for others, with a median of 49 days from CD4 testing to ART initiation. The use of POC technologies plays a crucial role in expediting the process, saving time, and aiding in the effective management of HIV infection, care, and monitoring [28].
Figure 6: Linear regression and Bland-Altman analysis of GBTsol ICA and FACS count for CD8. In a comparison using venous blood for GBTsol ICA and FACScount, the coefficient of determination (R2) was 0.91 while the mean bias between these two platforms was 38 cells/µL (CI-95%, LoA -149.84 to 226.13, n=113) and the SD mean was 95.91. In the Bland Altman plot (Right), the difference between the 0 line and the black line indicates the bias of GBTsol ICA minus the NAVIOUS Ex Flow Cytometer.
Conclusions
Recent systematic reviews [29] have shown that the establishment of such a resource should streamline administration by minimizing patient visits to centers [30]. It should address psychosocial issues and barriers to healthcare [31], enhance counseling and peer support to patients in need [32], optimize the significance of starting and continuing ART if eligible [33], and provide positive well-being coaching and encouragement for patients. A family-focused model of coordinated human services, consolidating the majority of the previously mentioned healthcare framework, has recently shown success in a similar population, resulting in high adherence (94%) and retention of maintenance of HIV-1-positive individuals [34,35]. In this study, we used a device that enabled highly efficient separation of leukocytes from small amounts of whole blood (less than that required by conventional techniques) and provided highly accurate enumeration of total numbers of leukocytes and their subsets. We demonstrated that CD4 and CD8 counts from a few microliters of whole blood using our method exhibited a good correlation with flow cytometry analysis.
Therefore, our device holds promise as an affordable yet effective tool for rapid leukocyte counting, enabling more comprehensive studies and monitoring of HIV/AIDS patients in decentralized sites. The accuracy and simplicity of absolute leukocyte counting provide numerous potential applications in single-cell analysis, particularly in POC diagnostic systems such as human CD marker enumerations. Previous studies have shown that the immediate provision of CD4+ T-cell counts increased the number of patients receiving care and reduced patient failure to obtain treatment (33%) [11,25,33,36,37]. In conclusion, the overall agreement between FACSCount and the GBTsol ICA analyzer for CD4+ T-cell enumeration was deemed acceptable, with a clinically nonsignificant mean bias and high accuracy and reproducibility. No significant differences were observed in the results obtained from GBTsol ICA compared with those of FACSCount. The GBTsol ICA POC CD4 test demonstrated potential in CD4 and CD8 T-cell enumeration, offering an avenue to broaden access to CD4 testing, particularly in rural settings where current needs are unmet by existing laboratory testing networks.
Acknowledgment
This study received approval from the Institutional Review Board (IRB) of Severance Hospital (Seoul, Republic of Korea), an affiliated hospital of Yonsei University Health System (Approval Number: IRB, 1-2018-009). Clinical blood samples used in this study were residual samples obtained and processed in clinical laboratories. The requirement for obtaining written consent forms from patients was waived for the use of the remaining blood samples after routine clinical laboratory tests, as per the approval of the IRB. All experiments were conducted in accordance with relevant guidelines and regulations.
References
- Antinori T, Coenen D, Costagiola N, Dedes M, Ellefson J, Gatell E, et al. De Wolf, HIV medicine 12 (2011): 61-64.
- Rice BD, Yin Z, Brown AE, Croxford S, Conti S, De Angelis D, et al. AIDS and Behavior 21 (2017): 83-90.
- Mocroft A, Lundgren JD, Sabin ML, Brockmeyer N, Casabona J, Castagna A, et al. PLoS Med 10 (2013): e1001510.
- Habiyambere V, Ford N, Low-Beer D, Nkengasong J, Sands A, Pérez González M, et al. PLoS medicine 13 (2016): e1002088.
- Drain PK, Hyle EP, Noubary F, Freedberg KA, Wilson D, Bishai WR, et al. IV. Bassett, Lancet Infect Dis 14 (2014): 239-49.
- Bassett IV, Wang B, Chetty S, Mazibuko M, Bearnot B, Giddy J, et al. Acquir. Immune. Defic. Syndr 51 (2009): 135-139.
- Drain PK, Rousseau C, Curr. Opin. HIV AIDS 12 (2017): 175-181.
- Vogt F, Tayler-Smith K, Bernasconi A, Makondo E, Taziwa F, Moyo B, et al. Reid PLoS One 10 (2015): e0129166.
- Matson A, Soni N, Sheldon J, Anaesth. Intensive Care 19 (1991): 182-186.
- Initiative CHA. in CHAI data from Mozambique, Malawi and South Africa. Data presented at AIDS conference (2016).
- Jani IV, Sitoe NE, Alfai ER, Chongo PL, Quevedo JI, Rocha BM, et al. Peter, Lancet 378 (2011): 1572-1579.
- Zachariah R, Reid SD, Chaillet P, Massaquoi M, Schouten EJ, Harries AD, Trop. Med. Int. Health 16 (2011): 37-41.
- Harris AD, Schouten EJ, Libamba E. The Lancet 367 (2006): 1870-1872.
- Wynberg E, Cooke G, Shroufi A, Reid SD, Ford N, Int. AIDS Soc 17 (2014): 18809.
- Glencross DK, Janossy G, Coetzee LM, Lawrie D, Aggett HM, Scott LE, et al. Cytometry B Clin Cytom 74 (2008): S40-S51.
- Sukapirom K, Onlamoon C. Thepthai, K. Polsrila, B. Tassaneetrithep, K. Pattanapanyasat, J. Acquir. Immune Defic. Syndr 58 (2011): 141-147.
- Mtapuri-Zinyowera S, Chideme M, Mangwanya D, Mugurungi O, Gudukeya S, Hatzold K, et al. Acquir. Immune Defic. Syndr 55 (2010): 1-7.
- Schaik N, Kranzer K, Wood R, Bekker LG. 18th Conference on Retroviruses and Opportunistic Infections (2010).
- Herbert S, Edwards S, Carrick G, Copas A, Sandford C, Amphlett M, Benn P. Sex Transm. Infect 88 (2012): 413-417.
- Glencross DK, Coetzee LM, Cassim N, PLoS One 9 (2014): e114727.
- Cassim N, Coetzee LM, Schnippel K, Glencross DK. PLoS One 9 (2014): e115420.
- Diaw PA, Daneau G, Coly AA, Ndiaye BP, Wade D, Camara M, et al. Acquir. Immune Defic. Syndr 58 (2011): e103-11.
- Galiwango RM, Lubyayi L, Musoke R, Kalibbala S, Buwembo M, Kasule J, et al. PLoS One 9 (2014): e88928.
- Wade D, Diaw PA, Daneau G, Camara M, Dieye, S. Mboup, L. Kestens, PLoS One 8 (2013): e75484.
- Faal M, Naidoo N, Glencross DK, Venter WD, Osih R, Acquir J. Immune Defic. Syndr 58 (2011): e54-9.
- Glencross DK, Coetzee LM, Faal M, Masango M, Stevens WS, Venter WF, Osih R. Int. AIDS Soc 15 (2012): 1-3.
- Thakar M, Mahajan B, Shaikh N, Bagwan S, Sane S, Kabra S, et al. Paranjape, AIDS Res. Ther 9 (2012): 1-7.
- Skhosana M, Reddy S, Reddy T, Ntoyanto S, Spooner E, Ramjee G, et al. South Afr. J HIV Med 17 (2016): 444.
- Govindasamy D, Meghij J, Kebede Negussi E, Clare Baggaley R, Ford N, et al. J. Int. AIDS Soc 17 (2014): 19032.
- Myer L, Daskilewicz K, McIntyre J, Bekker LG, J Int. AIDS Soc 16 (2013): 18649.
- Govindasamy D, Ford N, Kranzer K, AIDS 26 (2012): 2059-2067.
- Muhamadi L, Tumwesigye NM, Kadobera D, Marrone G, Wabwire-Mangen F, Pariyo G, et al. Ekström, Trials 12 (2011): 1-11.
- Patten GE, Wilkinson L, Conradie K, Isaakidis P, Harries AD, Edginton ME, et al, J. Int. AIDS Soc 16 (2013): 18518.
- Kwaan L, Kindra G, Mdutyana L, Coutsoudis A, Southern African Journal of HIV Medicine 11 (2010): 8-10.
- Coutsoudis A, Spooner E, Kindra G, Ndlovu N. Department of Paediatrics and Child Health, University of KwaZulu-Natal (2013).
- Larson BA, Schnippel K, Ndibongo B, Xulu T, Brennan A, Long L, et al. Acquir. Immune. Defic. Syndr 61 (2012): e13-7.
- Kranzer K, Govindasamy D, Ford N, Johnston V, Lawn SD, J. Int. AIDS Soc 15 (2012): 17383.


 Impact Factor: * 3.5
Impact Factor: * 3.5 Acceptance Rate: 71.36%
Acceptance Rate: 71.36%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
