Multi-Matrix Real Time PCR in 108 Patients with Polymorphic Signs Suggestive of Fibromyalgia or Related to A Tick Bite
Marie Mas4, Alexis Lacout5, Véronique Perronne2, Yannick Lequette1, Yves Gadiolet7, Béatrice Rambeaud3, Paul Trouillas6, Michel Franck1, Christian Perronne8
1ADNucleis 3 route des Pierres Blanches, 69290 Grézieu la Varenne
2Raymond Poincaré University Hospital, Public Assistance – Paris Hospitals, Department of Infectiology. University of Versailles – Saint Quentin, Paris-Saclay
3General Medicine Center, 16 boulevard de Savoie 74200 Thonon les bains
4Clinique Convert, General Medicine, Emergency Department, 62 avenue de Jasseron, 01000 Bourg en Bresse, France
5ELSAN diagnostic center, Medical-surgical center 83 avenue Charles de Gaulle 15000 Aurillac, France
6Department of Neurology, Claude Bernard University, 8 avenue Rockefeller, 69008 LYON, France
7North West Tarare Hospital, 69170 Tarare
8Infectious Diseases, Paris
*Corresponding author: Alexis Lacout, Centre de diagnostic ELSAN, Centre médico-chirurgical 83 avenue Charles de Gaulle 15000 Aurillac, France.
Received: 27 September 2023 Accepted: 10 October 2023 Published: 01 November 2023
Article Information
Citation: Marie Mas, Alexis Lacout, Véronique Perronne, Yannick Lequette, Yves Gadiolet, Béatrice Rambeaud, Paul Trouillas, Michel Franck, Christian Perronne. Multi-Matrix Real Time PCR in 108 Patients with Polymorphic Signs Suggestive of Fibromyalgia or Related to A Tick Bite. Archives of Microbiology and Immunology. 7 (2023): 250-270.
View / Download Pdf Share at FacebookAbstract
Introduction: Ticks are frequently polyinfected and can thus transmit numerous micro-organisms. A large number of bacteria, parasites and viruses are transmitted by tick bites and could cause different signs and symptoms in patients. The main goal of this study was to search for these numerous micro-organisms in patients presenting with persistent polymorphic syndrome possibly due to a tick bite.
Patients and Methods: The following micro-organisms were searched for in saliva, urine, venous and capillary blood by using real time PCR : Borrelia burgdorferi sensu lato, Borrelia hermsii, Borrelia garinii, Borrelia afzelii, Borrelia miyamotoi, Bartonella spp., Bartonella quintana, Bartonella henselae, Ehrlichia spp., Anaplasma spp., Rickettsia spp., Coxiella burnetii, Brucella spp., Francisella tularensis, Mycoplasma spp., Chlamydia spp., Babesia spp., Theileria spp., EBV, HHV-6, CMV, VZV, Tick-borne encephalitis virus (TBEV), Bourbon virus, West Nile virus, Chikungunya virus, Dengue virus, Zika virus, Powassan virus, Eyach virus.
Results: 108 patients were included. 65.7% of the patients were poly-infected, and 23.1 % harboured at least three different micro-organisms. Borrelia spp. were not the most frequent bacteria observed, far behind Mycoplasma spp., Rickettsia spp. and Theileria spp. which were the most frequent micro-organisms observed. The most sensitive matrix was saliva, followed by urine, capillary blood and venous blood.
Conclusion: Our prospective study has shown that patients with SPPT with polymorphic signs suggestive of fibromyalgia or related to a tick bite, could harbor several tick-borne micro-organisms.
Keywords
<p>Lyme, Borrelia, Babesia, PCR, PTLDS, SPPT, capillary blood, saliva</p>
Article Details
1. Introduction
Lyme disease is a tick-borne infectious disease caused by Borrelia burgdorferi sensu lato (including B. burgdorferi sensu stricto, B. afzelii, B. garinii and B. hermsii). The prevalence seems to be increasing in many countries around the world, as in France. In addition, some experts make the assumption that Lyme disease is not the only factor to explain the persistent polymorphic syndrome possibly due to a tick bite / Syndrome persistant polymorphe après une possible piqûre de tique (SPPT), a syndrome close to the post-treatment Lyme disease syndrome (PTLDS) [1]. SPPT is officially recognized by the French High Authority for Health (HAS) (a governmental institution https://www.has-sante.fr). SPPT includes post-treatment Lyme disease syndrome (PTLDS), but also addresses other hidden infections (crypto-infections). A large number of bacteria (other than Borrelia), parasites and viruses are transmitted by tick bites and could cause different signs and symptoms in patients, the so-called co-infections. In addition, the efficiency of Lyme serology is controversial with a tough scientific debate and it seems that the recommended two-tier testing for antibody detection (ELISA test followed, if positive, by a Western blot test) lacks of sensitivity [2-8]. Above all, this serology only looks for some species of Borrelia. Another controversial issue is the persistence of the bacterium after antibiotic treatment.
Indeed, ticks are frequently polyinfected and can transmit numerous micro-organisms [9-12], whether bacteria (other species of Borrelia, Bartonella spp., Ehrlichia spp., Rickettsia spp....), viruses (tick-borne encephalitis virus (TBEV), Bourbon virus, Powassan virus, Eyach virus), or parasites, first and foremost piroplasms (Babesia). Lyme disease is due to Borrelia burgdorferi sensu lato, other species of Borrelia are responsible for relapsing fevers (transmitted by ticks or lice according to the species. Borrelia miyamotoi is now found in symptomatic patients, either with acute or chronic disease [13, 14]. The best-known species of Bartonella are Bartonella henselae, responsible for cat scratch disease and Bartonella quintana, transmitted by lice and the agent of trench fever. They are usually investigated by serology [15-17]. In fact, there are many other species of Bartonella which may be transmitted by ticks and may cause neurological damage: radiculitis, myelitis, neurocognitive disorders for example. Bartonella have also been identified from patients with psychiatric diseases [17].
Other species of Borrelia, known in veterinary medicine has been found in the blood of patients with chronic syndromes [18]. Ehrlichia are responsible for ehrlichiosis, a tick-borne disease, which may be a Lyme disease co-infection [19]. Rickettsia species are numerous and are responsible for different groups of diseases, such as spotted fevers and typhus. Piroplasms are tick-borne monocellular parasites infecting red blood cells. Piroplasms include Babesia and Theileria species. Babesiosis is mainly known as an acute severe disease in splenectomised or deeply immunosuppressed persons, but the chronic form of the disease has not been much studied and is not well known. At the present time, Theileria, well known in veterinary medicine, have been identified for the first time in humans in our previous study [20]. A number of viruses are known to be transmitted by tick bites, and some others could reactivate in these patients, due to an immune deficiency related to Lyme disease. Many co-infections due to bacteria, parasites or viruses have been described in these persistent disorders. These observations are leading to the concept of stealth infections, denominated by experts as “crypto-infections” [21].
This study is a continuation of the one we carried out in the past, to which we added other microorganisms, in particular viruses [20]. The main goal of this study was to look for the different micro-organisms transmitted or not by ticks in patients suffering from polymorphic signs and symptoms (SPPT/PTLDS), using real time quantitative polymerase chain reaction (real time qPCR), which is a direct diagnostic method amplifying the DNA of the micro-organisms sought. As the yield of PCRs looking for Borrelia or co-infections in the venous blood is low [22, 23], another aim was to compare the results obtained by real time qPCR from several matrices (venous blood, urine, saliva and capillary blood). The last aim was to evaluate the relevance to draw two samples, at Day 0 and Day 3.
2. Patients and Methods
This is a prospective multi-centre observational study associating 9 clinical centres and one laboratory performing PCR analyses (AdNucleis) including patients with persistent polymorphic syndrome possibly due to a tick bite (SPPT).
SPPT is defined by a clinical triad associating several times a week, for more than 6 months: a polyalgic syndrome (musculoskeletal pain and/or neuropathic pain and/or headaches) ; persistent fatigue with reduced physical capacities ; cognitive complaints. The difference between SPPT and PTLDS is that a diagnosis of Lyme disease has not to be proven and patients may have not been treated [24, 25].
2.1 Patients
The criteria for inclusion in this prospective study were: SPPT patients of at least 18 years of age, of either sex, presenting with the following signs or symptoms evolving for more than 6 months:
1) Neurocognitive disorders.
2) More than 2 of the following chronic sign or symptom categories:
- Musculoskeletal: muscle pain, arthritis or arthralgia;
- Neurological: facial paralysis, central or peripheral involvement, myelitis, root pain, paresthesias, dysesthesias, radiculopathy.
3) Abnormal asthenia.
4) Lack of etiology.
5) Have given their signed informed consent.
The following patients were excluded from the study:
1) Patients under 18 years of age, a context of acute intercurrent infectious pathology.
2) Patients who had received prior systemic antibiotic or antiparasitic treatment within two months prior to blood collection.
3) The protocol was approved by the ethical committee (Comité de protection des personnes CPP SUD EST VI Clermont Ferrand, France; document number : Ref. CPP AU 1396 Ref. ID-RCB 2017-A02705-48). All patients and control persons signed an informed consent in accordance with the Declaration of Helsinki.
2.2 Micro-Organisms Searched
The following micro-organisms were searched for by using real time PCR (Tables 1, 2) : Borrelia burgdorferi sensu lato (s.l.) (including B. afzelii, B. garinii and B. hermsii), Borrelia miyamotoi, Borrelia garinii, Borrelia afzelii, Borrelia hermsii, Bartonella spp., Bartonella quintana, Bartonella henselae, Ehrlichia spp., Anaplasma spp., Rickettsia spp., Coxiella burnetii, Brucella spp., Francisella tularensis, Mycoplasma spp., Chlamydia spp., Babesia spp., Theileria spp., some herpesviruses (EBV, HHV-6, CMV, VZV), tick-borne encephalitis virus (TBEV), Bourbon virus, West Nile virus, Chikungunya virus, Dengue virus, Zika Virus, Powassan virus, Eyach virus.
Table 1: Real Time Multiplex Polymerase chain reaction (PCR)
|
Samples |
Urine and saliva were collected in dry bottles, five milliliters of blood were collected by venous puncture and around 500µl of capillary blood were collected by finger prick in tubes with EDTA as anti-coagulant, before any antibiotic treatment and were sent in Vacutainer® K2 tubes. Samples (venous blood, urine, saliva, capillary blood) were drawn twice at Day 0 (D0) and Day 3 (D3). |
|
Selection of Primers |
To allow the detection of bacteria and parasites, primers targeting specific genes of each micro-organism were used to amplify DNA by qPCR. Details of qPCR kits used is listed in Table 2. |
|
Robustness of PCR Mixes |
The portion of target genes were synthesized and introduced into a plasmid to obtain a control DNA and facilitate its multiplication. This control DNA was used to validate the amplification mixes. Serial dilution of the plasmid was performed and amplified to determine the robustness parameters of each PCR kit: the limit of detection (LOD), the limit of quantification (LOQ), the repeatability and the reproducibility. |
|
DNA Extraction and Purification |
The DNA was extracted without any prior treatment using 300 μl of whole blood with an equal volume of ADNucleis extraction buffer (5 M guanidium thiocyanate, 500 mM TrisHCL, 50 mM EDTA, 20% Tween 20, 20% Triton X-100, 750 μg proteinase K). After incubation for 20 min at 56°C and 15 min at 80°C, the extracted DNA was purified by means of silica magnetic beads and eluted in 250 μl of elution buffer (10 mM TrisHCl, pH 8.5). |
|
Control of the Extraction |
Human glyceraldehyde-3-phosphate dehydrogenase (hGAPDH) was used as a housekeeping gene as an internal control for PCR extraction and inhibition. The extracted samples were first checked with a PCR targeting the GAPDH gene. If the results of this PCR were consistent (Ct of GAPDH below 32), the samples were then analyzed for the other pathogens. The sequence of interest of GAPDH was inserted into a plasmid and this plasmid was used as a positive DNA for the validation of GAPDH primers and PCR mix as well as a positive control for subsequent PCRs. The primers used for GAPDH are described in Table 2. |
|
Real-Time PCR (real time PCR) |
Real-time PCR was carried out in a total volume of 50 μl with a PCR mix containing ADNucleis PCR buffer (20 mM Tris-HCl, 10 mM NH4SO4, 10 mM KCl, 2 mM Mg2+, 0.1% TritonX-100, pH 8.8), 2 mM of each dNTP, 600 nM of each primer, 1 μl of Evagreen and 5 units of Taq polymerase ADNucleis. Twelve μl of extracted samples were amplified. An initial denaturation step of 5 min at 95°C was followed by 42 cycles of 15 s at 95°C and 40 s at 60°C (hybridization-elongation). The dissociation curves were generated by a last step of 10 min with temperature increments from 75 to 95°C for qPCR kits using Sybr green technology. |
|
Quantification |
Positive samples were quantified using a standard curve obtained by amplifying known and calibrated concentrations of control DNA of the desired targets. Quantification was obtained using the standard curve equation (Ct = a (Log10 [DNA]) + b) where “a” is the slope and “b” the intercept of the curve. The results were expressed in genome units (UG) per ml of sample. |
LOD: limit of detection
LOQ: limit of quantification
UG: Genome Units
Ct : Cycle threshold.
LOD limit of detection and LOQ limit of quantification: the LOD is calculated by comparing the response of the PCR kit with respect to a reference method, which is most often a method for cultivating the microbial population. Once this microbial population has been cultured, it is stopped when the population is most abundant (eg 10E9); a count is carried out and the microbial population is subjected to successive dilutions in order to be able to have samples from 10E9 to 0, passing through all the intermediates (10E8, 10E7, etc.); these samples constitute the reference and the Borrelia analysis kit is evaluated for each dilution; we look for the sensitivities of the PCR kits making it possible to detect at least 10E2 DNA copies / PCR reaction volume, at best 10 copies / PCR reaction volume or less. LOD is therefore expressed in DNA copies (or RNA for most viruses) detected per PCR reaction volume; and when we evaluate the regression of the response of the kit (in Ct with respect to each dilution) we must obtain a straight line of which we evaluate the linearity (equation) and the slope (a of the equation y = ax + b, y being the log value of the concentration of the bacterial population, x the value of the response of the kit in Ct); most often, this linearity is not complete, in particular for low concentrations of the microbial population; Then comes the LOQ (limit of quantification) which is the lowest detection value evaluated on the linear part of the regression line. The LOQ is therefore always greater than the LOD, if the latter is of the order of 5 copies / PCR reaction volume, the LOQ can be between 40 and 100 copies / PCR reaction volume; these values are always carefully assessed by the manufacturer before placing the PCR reaction kit on the market.
Table 2: List of desired targets and details of PCR kits
Sybr: Syber Green fluorophore
The choice of the PCR technique (Sybr green or Taqman) is essentially linked to the sensitivity of each of the techniques; contrary to what is usually said, the two techniques are roughly equivalent, one being better than the other for certain targets and vice versa. And since we are looking for the best sensitivity in all cases, the laboratory has kept both techniques.
FAM: 6-carboxyfluorescéine (fluorophore)
LOD: limit of detection
LOQ: limit of quantification
UG: Genome Units
The sensitivity of each kit, in particular the limit of detection (LOD) and the limit of quantification (LOQ), are the subject of an analytical evaluation calculated according to the recommendations of Regulation (EU) 2017/746.
LOD limit of detection and LOQ limit of quantification
the LOD is calculated by comparing the response of the PCR kit with respect to a reference method, which is most often a method for cultivating the microbial population.
Once this microbial population has been cultured, it is stopped when the population is most abundant (eg 10E9); a count is carried out and the microbial population is subjected to successive dilutions in order to be able to have samples from 10E9 to 0, passing through all the intermediates (10E8, 10E7, etc.); these samples constitute the reference and the analysis kit is evaluated for each dilution; we look for the sensitivities of the PCR kits making it possible to detect at least 10E2 DNA copies or genome unit (UG) / PCR reaction volume, at best 10 copies / PCR reaction volume or less.
LOD is therefore expressed in DNA copies (Genome Units UG) detected per PCR reaction volume; and when we evaluate the regression of the response of the kit (in Ct with respect to each dilution) we must obtain a straight line of which we evaluate the linearity (equation) and the slope (a of the equation y = ax + b, y being the log value of the concentration of the bacterial population, x the value of the response of the kit in Ct); most often, this linearity is not complete, in particular for low concentrations of the microbial population; Then comes the LOQ (limit of quantification) which is the lowest detection value evaluated on the linear part of the regression line. The LOQ is therefore always greater than the LOD, if the latter is of the order of 5 copies / PCR reaction volume, the LOQ can be between 40 and 100 copies / PCR reaction volume; these values are always carefully assessed by the manufacturer before placing the PCR reaction kit on the market.
2.2.1 Sample collection and nucleic acid sample preparation
Five ml of blood were collected by venous puncture in tubes with EDTA as anti-coagulant, capillary blood from the tip of the fingers was collected in tubes with EDTA, urine was collected in sterile bottle and saliva was collected using swab. 300 µl of venous and capillary blood were used to extract nucleic acid. 10 ml of urine were first centrifuged 10 min at 4500 rpm in swinging bucket rotor, then the supernatant was discarded, and the pellet was resuspended in 300 µl of RNase DNase free water. Saliva swab sample was resuspended in 1 ml of Tris HCl 10 mM EDTA 1 mM buffer. 300 µl of the resuspension was used to extract nucleic acid. The ADNucleis DNA extraction-purification bead kit (ref ADNPVG300+BM) was used to extract DNA directly from 300 μl of blood samples or from processed sample as described above (urine and saliva).
2.2.2 Real time PCR method
Control DNA plasmids containing the amplified fragment was constructed to validate the amplification mixes, to be used as positive control. Serial dilution of the plasmid from 108 copies/PCR to 1-10 copies/PCRwas made and used to determine the limit of detection (LOD), the limit of quantification (LOQ) of each target, Tm for the sybr mixes (listed in table 2). The primer and probes are listed in the table 2. Primes and probes were from the literature or designed for this study. Each target was tested as individual monoplex. The real time PCR was performed using TaqMan or sybr technologies. Mixes was validated with specific PCR components used and developed by ADNucleis (including the Taq polymerase). The thermal cycling conditions of real time qPCR were as following: 95°C 5 min, 42 cycles of 95°C 10s and 60°C 40s for the TaqMan real time qPCR. The sybr PCR thermal cycling condition were the same the TaqMan one, expect the addition of the melting curve step (95°C 15s, 75°C 15s increasing temperature to 95°C and 95°C 15s). The reaction volume was 40 µl of the master mix and 12 µl of DNA samples. A Ct cycle thermic (Ct) higher than 38 was considered as negative for the TaqMan mixes. A Tm within the range TM +/-1 °C was considered positive for the sybr mixes. Mixes was validated with specific PCR components used and developed by AD Nucleis (including the Taq polymerase).
3. Results
3.1 Lack of detection of micro-organisms in healthy persons using venous blood qPCR
In a group of 24 healthy asymptomatic students, Borrelia burgdorferi s.l., Borrelia miyamotoi, Ehrlichia spp., Babesia spp., Borrelia hermsii, Bartonella henselae, Bartonella quintana were searched by PCR in venous blood. All the PCR results were negative (Table 3).
Table 3: Lack of detection of micro-organisms in the healthy persons in venous blood qPCR
|
PCR Inhibition |
Ct GAPDH values |
Detection |
|
|
FDC071 |
No |
29.44 |
Not Detected |
|
MCM072 |
No |
24.46 |
Not Detected |
|
MGA073 |
No |
28.57 |
Not Detected |
|
MFA074 |
No |
28.47 |
Not Detected |
|
FBF075 |
No |
27.7 |
Not Detected |
|
MDW076 |
No |
27.76 |
Not Detected |
|
FDT077 |
No |
30.97 |
Not Detected |
|
MAJ078 |
No |
28.29 |
Not Detected |
|
MMC079 |
No |
28.81 |
Not Detected |
|
FMS081 |
No |
28.08 |
Not Detected |
|
MSL082 |
No |
31.28 |
Not Detected |
|
MMD085 |
No |
31.55 |
Not Detected |
|
MPA088 |
No |
30.57 |
Not Detected |
|
FVA089 |
No |
29.98 |
Not Detected |
|
MGW092 |
No |
31.17 |
Not Detected |
|
FDN093 |
No |
29.15 |
Not Detected |
|
MBA094 |
No |
31.87 |
Not Detected |
|
FFS095 |
No |
28.32 |
Not Detected |
|
FBA096 |
No |
28.7 |
Not Detected |
|
FGA098 |
No |
28.89 |
Not Detected |
|
MACA101 |
No |
26.54 |
Not Detected |
|
MLS103 |
No |
30.83 |
Not Detected |
|
FLH105 |
No |
31.77 |
Not Detected |
|
FLL106 |
No |
28.44 |
Not Detected |
|
Positive control |
No |
22.83 |
Detected |
|
Negative control |
No |
0 |
Not Detected |
3.2 PCR and Patients
In this study, 108 patients were included. A total of 864 samples were analyzed. At D0 and D3, 236 samples and 237 samples were respectively positive for at least one micro-organism. A total (D0+D3) of 473 (54,7%) samples were positive. 474 micro-organisms were found including 455 bacteria and parasites, and 19 viruses.
3.3 Real time qPCR at Day 0 (D0) and Day 3 (D3)
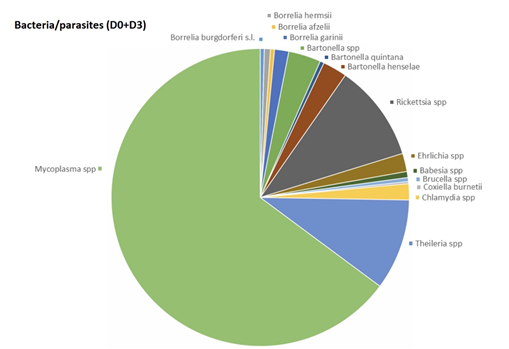
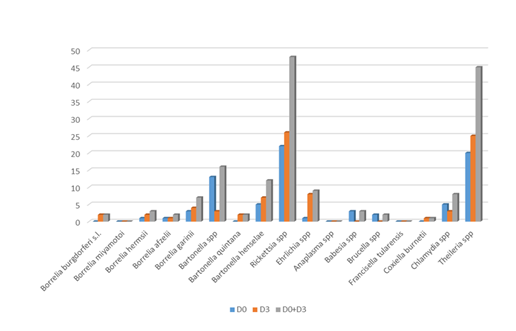
No micro-organisms were found in 5 (4.6%) out of 108 patients. One hundred and three patients out of 108 (95.4%) were positive for a least one micro-organism (Figures 1-5) (Table 4). Seventy-four (68.5%) patients were positive for at least one micro-organism excluding Mycoplasma spp (Mycoplasma spp. were excluded from this analysis, since these bacteria could be commensal).
- Bacteria and parasites:
The most frequent bacteria found were Mycoplasma spp., following by Rickettsia spp. and Theileria spp. For two bacterial genera, Borrelia and Bartonella, specific species of the same genus were identified. Fourteen PCRs (5 at D0 and 9 at D3) in 10 patients (9.3%) were positive for Borrelia: 2 (1 at D0 and 1 at D3) in 2 patients (1.9%) for B. afzelii, 0 for B. miyamotoi, 3 (1 to D0 and 2 at D3) in 2 patients (1.9%) for B. hermsii, 7 (3 to D0 and 4 at D3) in 7 patients (6.5%) for B. garinii.
Thirty PCRs (18 at D0 and 12 at D3) in 22 patients (20.4%) were positive for Bartonella, 2 (0 at D0 and 2 at D3) in 2 patients (1.9%) for B. quintana, 12 (5 at D0 and 7 at D3) in 11 patients (10.2%) for B. henselae, and 16 (13 at D0 and 3 at D3) in 12 patients (11.1%) for Bartonella spp.
Three PCRs (3 at D0 and 0 at D3) in 3 patients (2,8%) were positive for Babesia spp.
45 PCRs (20 at D0 and 25 at D3) in 30 patients (27,7%) were positive for Theileria spp.
48 PCRs (22 at D0 and 26 at D3) in 31 patients (28.7%) were positive for Rickettsia spp.
1 PCR (0 at D0 and 1 at D3) in 1 patient (0.9%) was positive for Coxiella burnetii.
9 PCRs (1 at D0 and 8 at D3) in 7 patients (6.5%) were positive for Ehrlichia spp.
2 PCRs (2 at D0 and 0 at D3) in 2 patients (1.9%) was positive for Brucella spp.
8 PCRs (5 at D0 and 3 at D3) in 7 patients (6.5%) were positive for Chlamydia spp.
295 PCR (153 at D0 and 142 at D3) in 101 patients (93.6%) were positive for Mycoplasma spp.
Anaplasma spp. and Francisella tularensis have not been detected in this study.
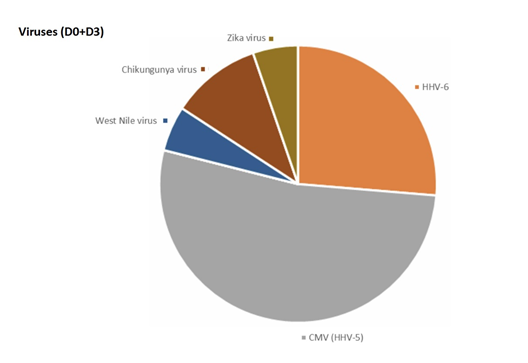
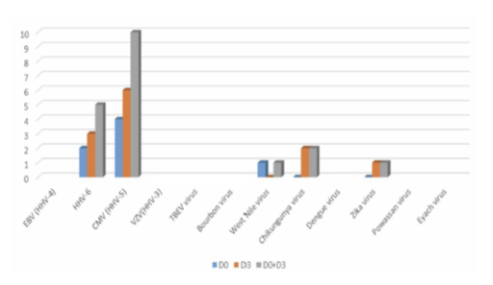
-Viruses:
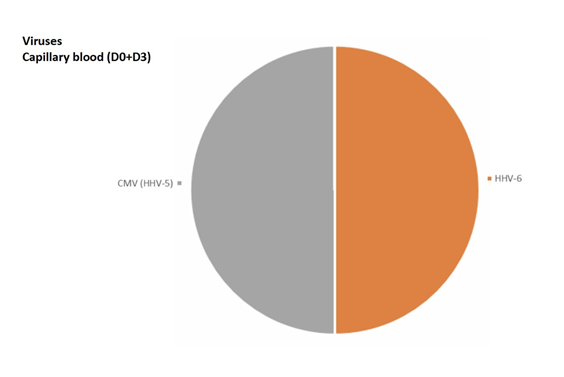
The most frequent viruses found were HHV6 and CMV.
5 PCR (2 at D0 and 3 at D3) in 3 patients (2.8%) were positive for HHV6.
10 PCR (4 at D0 and 6 at D3) in 3 patients (2.8%) were positive for CMV.
1 PCR (0 at D0 and 1 at D3) in 1 patient (0.9%) was positive for West Nile virus.
2 PCR (0 at D0 and 2 at D3) in 2 patients (1.9%) were positive for Chikungunya virus.
1 PCR (0 at D0 and 1 at D3) in 1 patient (0.9%) was positive for Zika virus.
EBV, VZV, TBEV virus, Bourbon virus, Dengue virus, Powassan virus and Eyach virus have not been detected in this study.
3.3.1 Day 0 (D0) versus Day 3 (D3)
Globally the results are similar between D0, D3 and D0 + D3. Individually, in 1 patient where real time PCR had isolated no micro-organisms at D0, the second analysis found micro-organisms. Six patients negative in blood at D0 were positive at D3; fifteen patients negative in capillary blood at D0 were positive at D3; two patients negative in saliva at D0 were positive at D3; fourteen patients negative in urine at D0 were positive at D3. In some cases, the second analysis showed different micro-organisms (Figure 6) (Table 4).
Table 4: PCRs results
|
Results at D0+D3 |
D0+D3 |
D0+D3 |
D0+D3 |
D0+D3 |
|||||
|
D0 |
D3 |
D0+D3 |
Blood (108) |
Urine (108) |
Saliva (108) |
Capillary Blood (108) |
Patients |
% of patients |
|
|
Borrelia burgdorferi s.l. |
0 |
2 |
2 |
0 |
1 |
0 |
1 |
2 |
1,9 |
|
Borrelia miyamotoi |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Borrelia hermsii |
1 |
2 |
3 |
0 |
1 |
2 |
0 |
2 |
1,9 |
|
Borrelia afzelii |
1 |
1 |
2 |
0 |
0 |
1 |
1 |
2 |
1,9 |
|
Borrelia garinii |
3 |
4 |
7 |
0 |
1 |
6 |
0 |
7 |
6,5 |
|
Bartonella spp. |
13 |
3 |
16 |
5 |
2 |
4 |
5 |
12 |
11,1 |
|
Bartonella quintana |
0 |
2 |
2 |
0 |
1 |
1 |
0 |
2 |
1,9 |
|
Bartonella henselae |
5 |
7 |
12 |
3 |
6 |
1 |
2 |
11 |
10,2 |
|
Rickettsia spp. |
22 |
26 |
48 |
1 |
16 |
29 |
2 |
31 |
28,7 |
|
Ehrlichia spp. |
1 |
8 |
9 |
2 |
1 |
2 |
4 |
7 |
6,5 |
|
Anaplasma spp. |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Babesia spp |
3 |
0 |
3 |
1 |
1 |
1 |
0 |
3 |
2,8 |
|
Theileria spp. |
20 |
25 |
45 |
4 |
3 |
17 |
21 |
30 |
27,7 |
|
Brucella spp. |
2 |
0 |
2 |
1 |
0 |
1 |
0 |
2 |
1,9 |
|
Francisella tularensis |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Coxiella burnetii |
0 |
1 |
1 |
0 |
1 |
0 |
0 |
1 |
0,9 |
|
Chlamydia spp. |
5 |
3 |
8 |
0 |
0 |
8 |
0 |
7 |
6,5 |
|
Mycoplasma spp. |
153 |
142 |
295 |
3 |
76 |
187 |
29 |
101 |
93,6 |
|
EBV (HHV-4) |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
HHV-6 |
2 |
3 |
5 |
0 |
1 |
2 |
2 |
3 |
2,8 |
|
CMV (HHV-5) |
4 |
6 |
10 |
0 |
3 |
5 |
2 |
3 |
2,8 |
|
VZV (HHV-3) |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
TBEV |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Bourbon virus |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
West Nile virus |
1 |
0 |
1 |
0 |
1 |
0 |
0 |
1 |
0,9 |
|
Chikungunya virus |
0 |
2 |
2 |
0 |
2 |
0 |
0 |
2 |
1,9 |
|
Dengue virus |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Zika virus |
0 |
1 |
1 |
0 |
1 |
0 |
0 |
1 |
0,9 |
|
Powassan virus |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Eyach virus |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
Spp: species plurimae (all species in the genus).
TBEV: Tick-borne encephalitis virus
Table 5: Number of positive matrices per micro-organism for a given patient
|
Detected in 1 matrix |
Detected in 2 matrix |
Detected in 3 matrix |
Detected in 4 matrix |
|
|
Borrelia burgdorferi s.l. |
2 |
0 |
0 |
0 |
|
Borrelia miyamotoi |
0 |
0 |
0 |
0 |
|
Borrelia hermsii |
2 |
0 |
0 |
0 |
|
Borrelia afzelii |
2 |
0 |
0 |
0 |
|
Borrelia garinii |
7 |
0 |
0 |
0 |
|
Bartonella spp. |
9 |
2 |
1 |
0 |
|
Bartonella quintana |
2 |
0 |
0 |
0 |
|
Bartonella henselae |
10 |
1 |
0 |
0 |
|
Rickettsia spp. |
26 |
5 |
0 |
0 |
|
Ehrlichia spp. |
4 |
2 |
0 |
0 |
|
Anaplasma spp. |
0 |
0 |
0 |
0 |
|
Babesia spp. |
3 |
0 |
0 |
0 |
|
Theileria spp. |
23 |
5 |
1 |
1 |
|
Brucella spp. |
2 |
0 |
0 |
0 |
|
Francisella tularensis |
0 |
0 |
0 |
0 |
|
Coxiella burnetii |
1 |
0 |
0 |
0 |
|
Chlamydia spp. |
7 |
0 |
0 |
0 |
|
Mycoplasma spp. |
43 |
44 |
13 |
1 |
|
EBV (HHV-4) |
0 |
0 |
0 |
0 |
|
HHV-6 |
3 |
0 |
0 |
0 |
|
CMV (HHV-5) |
1 |
1 |
1 |
0 |
|
VZV (HHV-3) |
0 |
0 |
0 |
0 |
|
TBEV virus |
0 |
0 |
0 |
0 |
|
Bourbon virus |
0 |
0 |
0 |
0 |
|
West Nile virus |
1 |
0 |
0 |
0 |
|
Chikungunya virus |
2 |
0 |
0 |
0 |
|
Dengue virus |
0 |
0 |
0 |
0 |
|
Zika virus |
1 |
0 |
0 |
0 |
|
Powassan virus |
0 |
0 |
0 |
0 |
|
Eyach virus |
0 |
0 |
0 |
0 |
3.4 Matrices
3.4.1 Micro-organisms found by matrix
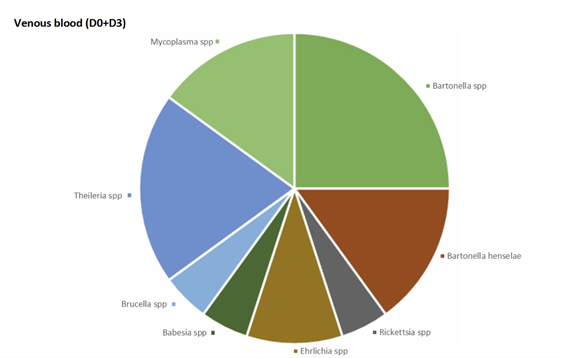
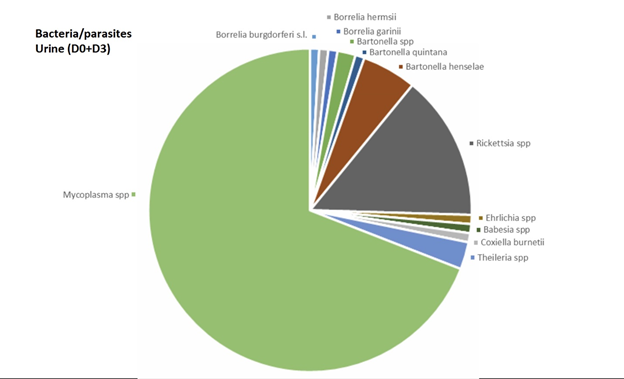
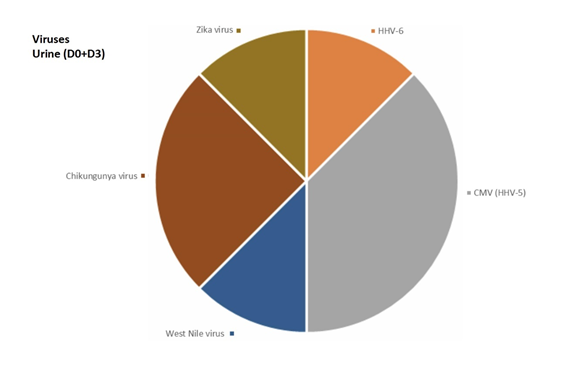
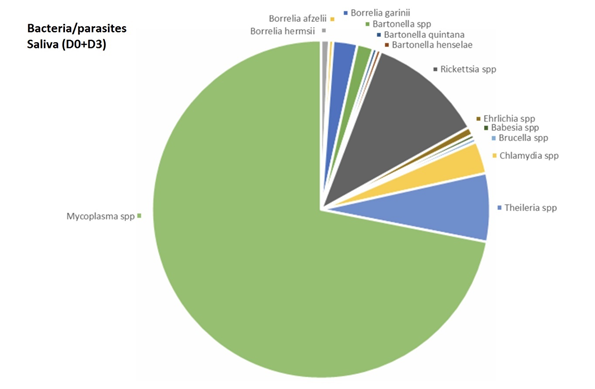
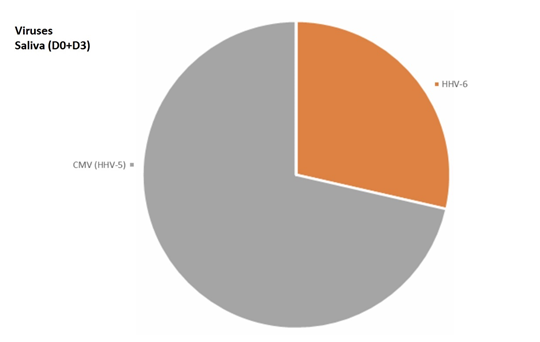
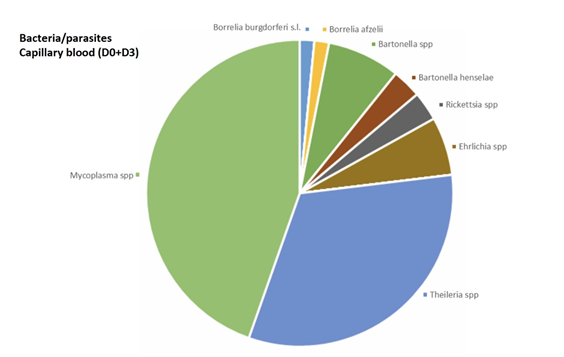
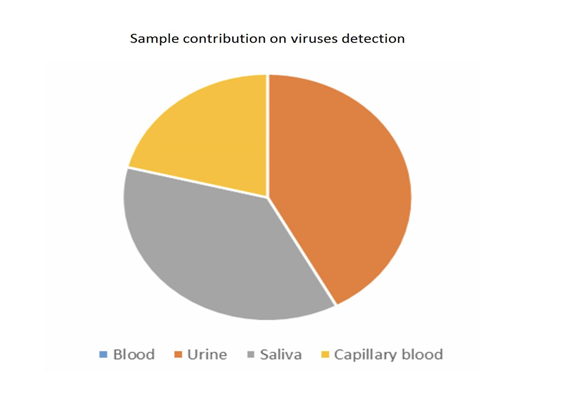
In venous blood, the main micro-organism found was Bartonella (B. quintana, B. henselae, Bartonella spp.) (found 8 times out of a total of 30 positive results, 26.7%) followed by Theileria spp. (found 4 times out of a total of 45 positive results, 8.9 %) (Figure 2). In urine, the main micro-organism found was Mycoplasma spp. (found 76 times out of a total of 295 positive results, 25.8%) followed by Rickettsia spp. (found 16 times out of a total of 48 positive results, 33.3%) (Figure 3). In saliva, the main micro-organism found was Mycoplasma spp. (found 187 times out of a total of 295 positive results, 63.4%) followed by Rickettsia spp. (found 29 times out of a total of 48 positive results, 60.4%) (Figure 4). In capillary blood, the main micro-organism found was Mycoplasma spp. (found 29 times out of a total of 295 positive results, 9.8%) followed by Theileria spp. (found 21 times out of a total of 45 positive results, 46.7%) (Figure 5).
3.4.2 Matrix performance - Overall results
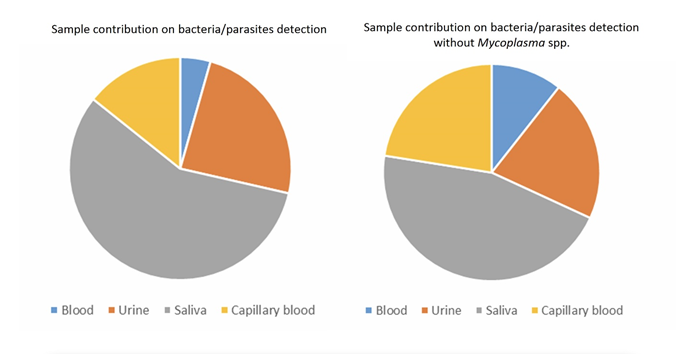
Overall, the most informative matrix was saliva (267 micro-organisms found, 56.3%), followed by urine (118 micro-organisms found, 24.9%), capillary blood (69 micro-organisms found, 14.6%), and venous blood (20 micro-organisms found, 4.2%) (Figure 7).
3.4.3 Matrix performance - Results by micro-organism:
- Bacteria and parasites:
Mycoplasma spp. were the most frequently identified micro-organism. Mycoplasma spp. was found mainly in saliva. Mycoplasma spp. was found 3 times in venous blood (1.2%), 29 times in capillary blood (9.8%), 76 times in urine (25.8%) and 187 times in saliva (63.4%).
For Borrelia, the informative matrices were urine (21.4%) and saliva (64.3%) and capillary blood (14.3%). Borrelia was not found in venous blood.
Bartonella spp. was found in all 4 matrices. Bartonella quintana was found once in urine (50 %) and once in saliva (50%). Bartonella henselae was found thrice in venous blood (25%), twice in capillary blood (16.7%), 6 times in urine (50%) and once in saliva (8.3%).
Bartonella spp. was found 5 times in venous blood (31.2%), 5 times in capillary blood (31.2%), twice in urine (12.5%) and 4 times in saliva (25%).
Babesia spp. were found once in venous blood (33.3%), not found in capillary blood, found once in urine (33.3%) and once in saliva (33.3%).
Theileria spp. was found 4 times in venous blood (8.9%), 21 times in capillary blood (46.7%), three times in urine (6.7%), and 17 times in saliva (37.8%).
Rickettsia spp. were found once in venous blood (2.1%), twice in capillary blood (4.2%), 16 times in urine (33.3%) and 29 times in saliva (60.4%).
Coxiella burnetii was found once in urine.
Ehrlichia spp. was found twice in venous blood (22.2%), 4 times in capillary blood (44.4%), once in urine (11.1%), and twice in saliva (22.2%).
Brucella was found once in blood (50%) and once in urine (50%).
Chlamydia spp. was found 8 times in saliva (100%).
For a given patient, bacteria and parasites were most often found in one matrix only (62.4%), then in two matrices (26.9%), three matrices (6.8%), and even four matrices (0.9%).
Mycoplasma spp. and Theileria spp. were the only micro-organisms observed in all 4 matrices, both once (Table 5).
Viruses:
HHV-6 was not found in venous blood, found twice in capillary blood (40%), once in urine (20%), and twice in saliva (40%).
CMV was not found in venous blood, found twice in capillary blood (20%), 3 times in urine (30%), and 5 times in saliva (50%).
West Nile virus was found once in urine.
Chikungunya virus was found twice in urine.
Zika virus was found once in urine.
For a given patient, viruses were most often found in one matrix only (80%), then in two matrices (10%), or three matrices (10%). No virus was found in all the four matrices.
3.5 Poly-infection
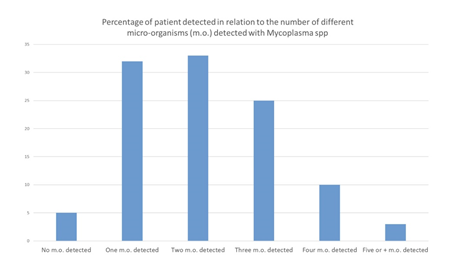
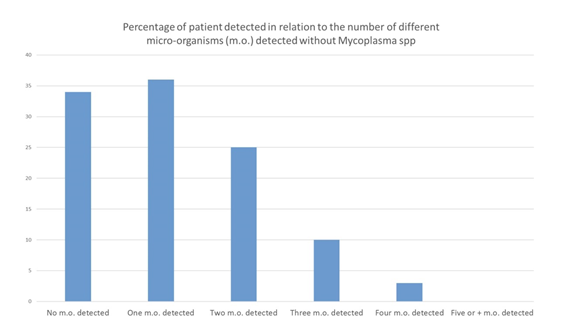
Among the 108 patients included in our study, no micro-organism was found in 5 patients (4,6%), only one micro-organism in 32 patients (29.6%), two different micro-organisms in 33 patients (30,6%), three different micro-organisms in 25 patients (23.1%), four micro-organisms in 10 patients (9.3%), five or more different micro-organisms in 3 patients (2.8%) (Figures 8) (Table 6). 71 patients (65.7%) were poly-infected. The same analysis was performed without taking into account Mycoplasma spp., that could be considered as commensal bacteria. In this analysis, no micro-organism was found in 34 patients (31.5%), one single micro-organism in 36 patients (33.3%), two different micro-organisms in 25 patients (23.1%), three micro-organisms in 10 patients (9.3%), four micro-organisms in 3 patients (2.8%). Excluding Mycoplasma spp., 38 patients (35.2%) patients were poly-infected.
Table 6: Poly-infection: number of micro-organisms detected per patient
|
Number of patients |
|||||||
|
Number of micro-organism(s) detected per patient |
0 |
1 |
2 |
3 |
4 |
5 and more |
Total |
|
With Mycoplasma spp. |
5 |
32 |
33 |
25 |
10 |
3 |
108 |
|
Without Mycoplasma spp. |
34 |
36 |
25 |
10 |
3 |
0 |
108 |
|
Percentage of patients based on the number of different micro-organisms detected |
||||||
|
Number of micro-organism(s) detected per patient |
0 |
1 |
2 |
3 |
4 |
5 and more |
|
With Mycoplasma spp. |
4.63 |
29.63 |
30.56 |
23.15 |
9.26 |
2.78 |
|
Without Mycoplasma spp. |
31.48 |
33.33 |
23.15 |
9.26 |
2.78 |
0 |
Spp : species plurimae
4. Discussion
This study showed that most patients with a combination of signs and symptoms, e.g. joint, muscle and tendon pain, neurological pain, tingling, sweats, hot flashes, fatigue, sleep difficulty, focusing/concentrating, neck pain, headache, difficulty finding words, irritability, low back pain, joint swelling, double vision, drooping facial muscle, drooping eyelid, tinnitus, heart palpitations, consistent with the diagnosis of SPPT/PTLDS harbour micro-organisms that are well detected using real time qPCR. As a matter of fact, Borrelia spp., as described in our previous article, were not the most frequent bacteria isolated. Rickettsia spp. and Theileria spp. were the most frequent micro-organisms identified (Figure 1). Borrelia could be less frequent than estimated, or more difficult to detect despite the use of several matrices, in the hypothesis that this bacteria could be particularly localized in organs such as brain, or deep tissues. In this case, PCRs performed on biopsies would be more relevant [26]. In our study, piroplasms (Babesia spp. and Theileria spp.) were found in a significant number of patients. A surprising result is that we isolated much more Theileria spp. than Babesia spp. In our previous study, we performed a search for Theileria spp. in 33 patients, which was found 17 times in total, 5 times in capillary blood (29.4%), once in urine (5.9%) and 11 times in saliva (64.7%) [20]. The present study confirms that this parasite is frequently encountered in patients with SPPT/PTLDS (28.7% of patients positive for Theileria spp.), far ahead of Babesia (2.8% were positive for Babesia spp. int the present study 8.7% in our previous study) [20].
Before our two studies, Theileria pathogen-specific PCR sequences, well known in veterinary medicine as it usually infests horses [27], were never isolated from humans. There is a high genetic proximity between Babesia microti and Theileria microti and our PCR primers are studied to specifically distinguish the genus Babesia from the genus Theileria. These preliminary results suggest that Theileria could be a significant pathogen for humans and should be confirmed by additional studies. Babesia are most commonly known to cause severe infection with shock, mainly in patients who have undergone splenectomy. However, some authentic Babesia spp. infections may occur in immunocompetent patients with a torpid, chronic presentation [28-30]. Thus, our study confirms that piroplasm infections are most likely underestimated [31]. As previously suggested by Muriel Vayssier Taussat et al [18], various species of Bartonella, including species which were only known in veterinary medicine, could be involved in SPPT/PTLDS. Some bacteria such as Mycoplasma spp. are not usually transmitted by ticks and could be commensal bacteria or transmitted by sexual contamination. However, some mycoplasmas such as Mycoplasma fermentans can be transmitted by ticks and be pathogenic [32]. Surprisingly, a case of West Nile virus was observed. This virus is not normally transmitted by tick, but by mosquitoes, and possibly by horseflies (we have observed the presence of this virus in a not yet published series looking for different micro-organisms in horseflies). It seems that this virus, potentially at the origin of flue-like syndrome or acute encephalitis, is increasing in France and in Europe [33].
It is both surprising and interesting to find in these chronically ill patients viruses such as Zika or Chikungunya, which are known to cause acute illness. This raises the question of whether or not these viruses can persist in the body and cause chronic signs and symptoms in some patients with fibromyalgia/SPPT/ PTLDS. Some other viruses found in our series, such as CMV or HHV 6, which have never been described in ticks, could reactivate following a depression of the immune system due to Lyme disease, and may qualify as opportunistic infections [34]. It might be thus more accurate to change the paradigm, and consider the term “crypto-infections” rather than exclusively “tick-borne infections” [21]. The study showed that nearly 65 % of the patients were poly-infected, 35 % of them harboured at least three different micro-organisms, and some patients had up to 5 micro-organisms, which could play a role in the polymorphism and severity of symptoms.
However, at this preliminary stage of the research, it is not possible to distinguish between latent carriage and active infection. As a matter of fact, the isolation of micro-organisms in saliva may be difficult to interpret. As the mouth is an open area, it could be a contamination from the outside. However, salivary secretion of micro-organisms exists. Salivary glands are holomerocrine: secretion needs disruption of the apex of the acini cells which must multiply rapidly, this could enhance the tropism of some micro-organisms. Some micro-organisms such as rabies, HHV6 or Epstein Barr virus have a tropism for salivary glands. For Mycoplasma, a high level of isolation from saliva and urine was observed. There was a superiority of capillary blood samples. For the Mycoplasma urine results, a possible contamination from the genital tract cannot be excluded, and the Mycoplasma found in saliva could be commensal bacteria: a precise characterization of the Mycoplasma species should be further studied. For Borrelia, saliva samples seemed the most interesting (6 Borrelia garinii found in saliva), before urine. Globally, few micro-organisms were found in venous blood, significantly more in capillary blood, and in two matrices which were most frequently positive: saliva and urine. This comparison has never been described before (except in our previous study) [20]. These various samples are easy and non-invasive to collect. Theileria is most frequently found in capillary blood, although almost as frequently in saliva. This is not surprising because this parasite infects red blood cells and saliva is derived from blood via a holomerocrine secretion. Thus, this study showed that venous blood was the less sensitive matrix, far behind saliva, urine and capillary blood. In our study, Borrelia was found a few times in urine and saliva, but not in blood. Previous studies have indeed showed a low interest of venous blood PCR for the detection of Borrelia. As a matter of fact, PCR sensitivities and specificities are heterogeneous [35]. Some studies used PCR to identify Borrelia burgdorferi in early Lyme disease, while we studied it in a chronic stage [36, 37]. Liveris et al reported a sensitivity of 40.6 % (36) while in Eshoo et al's study, the sensitivity was 62% and the specificity 100% [37]. In these studies, on early Lyme disease, the two-tier serology sensitivity was higher than PCR direct detection. However, in Bil-Lula's study, 3% of patients with negative ELISA IgM test, 2.8% with negative Line blot IgM test, 3.1% and 2.7% with negative ELISA IgG and Line blot IgG tests, respectively, were positive in real time PCR [22]. Few studies looked for Borrelia in urine by PCR. In one study, detection rate was 91% in patients with Lyme disease skin lesions [38]. In another study, results were disappointing [39].
Our study showed the advantage of a double sample for PCR at Day 0 and Day 3: individually, in one case where real time PCR had isolated no micro-organism at D0, the second analysis found micro-organisms. We have observed many cases where a matrix was negative at D0 but positive at D3. In some cases, the second analysis showed different micro-organisms. Therefore, performing the PCR assays several times (in this case at D0 and D3), could increase the sensitivity of detection and improve the diagnosis of patient infections. Some have speculated that SPPT/PTLDS clinical signes have a psychiatric cause [40]. May we put foward that infections could play a role, even if dysimmunitary phenomenons have to be taken into account [40]. Futures studies are necessary to show that healthy individuals do not exhibit a similar infection rate. In patients presenting with SPPT/PTLDS, a precise evaluation of appropriate anti-infective treatments, and real time qPCR tests after treatment should be considered. SPPT/PTLDS is very similar and difficult to dissociate from fibromyalgia syndromes [24, 25, 41, 42]. These syndromes associate three major signs: disabling fatigue, neuro-psychic disorders (memory, sleep, concentration disorders) and various somatic signs; pain remains a predominant symptom [24, 25]. As all patients suffered from signs and symptoms, it is possible that the isolated micro-organisms were actually responsible for the disease, or at least a cofactor. Further studies on large populations, including healthy control persons, should look at the possibility of asymptomatic carriage.
In conclusion, our prospective study has shown that patients with persistent polymorphic syndrome possibly due to a tick bite (SPPT) / post-treatment Lyme disease syndrome (PTLDS), a syndrome close to fibromyalgia, could harbour several micro-organisms, some of them transmitted by tick bites. Microbiologic analyses should not be limited to Borrelia's research alone, nor just look for micro-organisms in venous blood. Taking several successive samples on different days could improve the detection of these micro-organisms. Our results confirm, as in our previous study, that capillary blood is a more informative matrix than venous blood, which seems to be less sensitive than saliva and urine. Thus, in the case of SPPT/PTLDS, qPCR analyses should be performed on these different matrices to ensure optimal cost-effectiveness diagnostic. Due to the strong Lyme controversy, the absence of gold standards, the limited results published on co-infections identification, the low sensitivity of blood PCRs, it is difficult to rely on the literature to try to establish comparisons. Further studies are needed. Finally, these micro-organisms are difficult to detect. This may justify protocols using antibiotic therapy or probabilistic anti-parasitic drugs, without the germ having been detected, provided that the patient's condition is accurately assessed throughout the course of treatment. Prolonged antibiotic therapy could be useful [43].
Conflict of Interest
Michel Franck is CEO of ADNucleis
Yannick Lequette is employed by ADNucleis
The others authors do not declare any conflict of interest
Statement
The study has been approved by an appropriate ethics committee and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. National laws have been observed. The protocol was approved by the ethical committee (Comité de protection des personnes CPP SUD EST VI Clermont Ferrand, France; document number: Ref. CPP AU 1396 Ref. ID-RCB 2017-A02705-48).
Key points
Our prospective real time qPCR (quantitative polymerase chain reaction) study has shown that patients with persistent polymorphic syndrome possibly due to a tick bite (SPPT) / post-treatment Lyme disease syndrome (PTLDS) could harbour several tick-borne micro-organisms. Venous blood seems to be the less sensitive matrix, far behind saliva, urine and capillary blood. The study showed, as our precedent series, that there was a benefit to performing the analyses twice, by increasing the detection of micro-organisms.
Funding
Ramsay santé. The authors declare that this study received funding from Association BonSens.org to cover the publication fees.
References
- https://www.hhs.gov/ash/advisory-committees/tickbornedisease/meetings/2018-07-24/meeting summary/index.html?language=es
- Alby K, Capraro GA. Alternatives to Serologic Testing for Diagnosis of Lyme Disease. Clin Lab Med 35 (2015): 815-25.
- Cook MJ, Puri BK. Commercial test kits for detection of Lyme borreliosis: a meta-analysis of test accuracy. Int J Gen Med 9 (2016): 427-440.
- Leeflang MM, et al., “The diagnostic accuracy of serological tests for Lyme borreliosis in Europe: a systematic review and meta-analysis”, BMC Infectious Diseases 16 (2016): 140.
- Rapport d’activité du Haut Conseil de la Santé Publique HSCP (High Council for Public Health (in French) (2014).
- Schutzer SE, Coyle PK, Belman AL, Golightly MG, Drulle J. Sequestration of antibody to Borrelia burgdorferi in immune complexes in seronegative Lyme disease. Lancet 335 (1990): 312-5.
- Horowitz RI, Lacout A, Marcy PY, Perronne C. To test or not to test? Laboratory support for the diagnosis of Lyme borreliosis. Clin Microbiol Infect 24 (2018): 210.
- Perronne C, Lacout A, Marcy PY, El Hajjam M. Errancy on Lyme Diagnosis. Am J Med 130 (2017): e219.
- Cisak E, Wójcik-Fatla A, Zajac V, Dutkiewicz J. Prevalence of tick-borne pathogens at various workplaces in forest exploitation environment. Med Pr 65 (2014): 575-81.
- Sanchez-Vicente S, Tagliafierro T, Coleman JL, Benach JL, Tokarz R. Polymicrobial Nature of Tick-Borne Diseases. mBio 10 (2019): e02055-19.
- Michelet L, Delannoy S, Devillers E, Umhang G, Aspan A, Juremalm M, et al. High-throughput screening of tick-borne pathogens in Europe. Front Cell Infect Microbiol 29 (2014): 103.
- Nelder MP, Russell CB, Sheehan NJ, Sander B, Moore S, Li Y, et al. Human pathogens associated with the blacklegged tick Ixodes scapularis: a systematic review. Parasit Vectors 9 (2016):265.
- Franck M, Ghozzi R, Pajaud J, Lawson-Hogban NE, Mas M, Lacout A, et al. Borrelia miyamotoi: 43 Cases Diagnosed in France by Real-Time PCR in Patients with Persistent Polymorphic Signs and Symptoms. Front Med (Lausanne) 7 (2020): 55.
- Platonov AE, Karan LS, Kolyasnikova NM, Makhneva NA, Toporkova MG, Maleev VV, et al. Humans infected with relapsing fever spirochete Borrelia miyamotoi, Russia. Emerg Infect Dis 10 (2011): 1816-23.
- Maggi RG, Mozayeni BR, Pultorak EL, Hegarty BC, Bradley JM, Correa M, Breitschwerdt EB. Bartonella spp. bacteremia and rheumatic symptoms in patients from Lyme disease-endemic region. Emerg Infect Dis 18 (2012): 783-91.
- Berghoff W. Chronic Lyme Disease and Co-infections: Differential Diagnosis. Open Neurol J 6 (2012):158-78.
- Breitschwerdt EB, Maggi RG, Nicholson WL, Cherry NA, Woods CW. Bareal time onella Sp. Bacteremia in Patients with Neurological and Neurocognitive Dysfunction. J Clin Microbiol 46 (2008): 2856-61.
- Vayssier-Taussat M, Moutailler S, Féménia F, Raymond P, Croce O, La Scola B, et al. Identification of Novel Zoonotic Activity of Bartonella spp., France. Emerg Infect Dis 22 (2016): 457-62.
- Feder HM Jr, Johnson BJ, O'Connell S, Shapiro ED, Steere AC, Wormser GP, Ad Hoc International Lyme Disease Group, et al. A critical appraisal of "chronic Lyme disease". N Engl J Med 357 (2007): 1422-30.
- Lacout A, Mas M, Pajaud J, Perronne V, Lequette Y, Franck M, et al. Real time micro-organisms PCR in 104 patients with polymorphic signs and symptoms that may be related to a tick bite. Eur J Microbiol Immunol (Bp) 3 (2021).
- Christian Perronne. Crypto-Infections: Denial, Censorship and Suppression the Truth About What Lies Behind. ed. Hammersmith Health Books (2021).
- Bil-Lula I, Matuszek P, Pfeiffer T, Wozniak M. Lyme Borreliosis--the Utility of Improved Real-Time PCR Assay in the Detection of Borrelia burgdorferi Infections. Adv Clin Exp Med 24 (2015): 663-70.
- Ruzic-Sabljic E, Cerar T. Progress in the molecular diagnosis of Lyme disease. Expert Rev Mol Diagn 17 (2017): 19-30.
- Aucott JN. Posttreatment Lyme disease syndrome. Infect Dis Clin North Am 29 (2015): 309-23.
- Haute Autorité de Santé (High Authority for Health). Recommandation de bonne pratique. Borréliose de Lyme et autres maladies vectorielles à tiques (MVT) (in French) (2018).
- Häupl T, Hahn G, Rittig M, Krause A, Schoerner C, Schönherr U, Kalden JR, Burmester GR. Persistence of Borrelia burgdorferi in ligamentous tissue from a patient with chronic Lyme borreliosis. Arthritis Rheum 36 (1993): 1621-6.
- Onyiche TE, Suganuma K, Igarashi I, Yokoyama N, Xuan X, Thekisoe O. A Review on Equine Piroplasmosis: Epidemiology, Vector Ecology, Risk Factors, Host Immunity, Diagnosis and Control. Int J Environ Res Public Health 16 (2019): E1736.
- Vannier E, Gewurz BE, Krause PJ. Human babesiosis. Infect Dis Clin North Am 22 (2008): 469-88
- Krause PJ. Human babesiosis. Int J Parasitol 49 (2019): 165-174.
- Krause PJ, Spielman A, Telford SR 3rd, Sikand VK, McKay K, Christianson D, et al. Persistent parasitemia after acute babesiosis. N Engl J Med 339 (1998): 160-5.
- Martinot M, Paleau A, Greigert V, Brunet J, Hansmann Y, Jouglin M, et al. Babésiose en France et en Europe : une pathologie à redéfinir. Med mal inf 48 (2018): S112.
- Eskow E, Adelson ME, Rao RV, Mordechai E. Evidence for disseminated Mycoplasma fermentans in New Jersey residents with antecedent tick attachment and subsequent musculoskeletal symptoms. J Clin Rheumatol 9 (2003): 77-87.
- Eldin C, Ninove L, Lagier JC. Infection à virus West Nile : une arbovirose émergente en France et en Europe [West Nile virus infection: an emerging arbovirosis in France and Europe] Rev Prat 70 (2020): 333-335.
- Elsner RA, Hastey CJ, Olsen KJ, Baumgarth N. Suppression of Long-Lived Humoral Immunity Following Borrelia burgdorferi Infection. PLoS Pathog 11 (2015): e1004976.
- Waddell LA, Greig J, Mascarenhas M, Harding S, Lindsay R, Ogden N. The Accuracy of Diagnostic Tests for Lyme Disease in Humans, A Systematic Review and Meta-Analysis of North American Research. PLoS One 11 (2016): e0168613.
- Liveris D, Schwartz I, McKenna D, Nowakowski J, Nadelman R, Demarco J, et al. Comparison of five diagnostic modalities for direct detection of Borrelia burgdorferi in patients with early Lyme disease. Diagn Microbiol Infect Dis 73 (2012): 243-5.
- Eshoo MW, Crowder CC, Rebman AW, Rounds MA, Matthews HE, Picuri JM, et al. Direct molecular detection and genotyping of Borrelia burgdorferi from whole blood of patients with early Lyme disease. PLoS One 7 (2012): e36825.
- Schmidt BL, Aberer E, Stockenhuber C, Klade H, Breier F, Luger A. Detection of Borrelia burgdorferi DNA by polymerase chain reaction in the urine and breast milk of patients with Lyme borreliosis. Diagn Microbiol Infect Dis 21 (1995): 121-8.
- Rauter C, Mueller M, Diterich I, Zeller S, Hassler D, Meergans T, et al. Critical evaluation of urine-based PCR assay for diagnosis of Lyme borreliosis. Clin Diagn Lab Immunol 12 (2005): 910-7.
- Kennedy AG. Differential Diagnosis and the Suspension of Judgment. J Med Philos 38 (2013): 487-500.
- Wolfe F, Clauw DJ, Fitzcharles MA, Goldenberg DL, Katz RS, Mease P, et al. The American College of Rheumatology preliminary diagnostic criteria for fibromyalgia and measurement of symptom severity. Arthritis Care Res (Hoboken) 62 (2010): 600-10.
- Rebman AW, Bechtold KT, Yang T, Mihm EA, Soloski MJ, Novak CB, et al. The Clinical, Symptom, and Quality-of-Life Characterization of a Well-Defined Group of Patients with Posttreatment Lyme Disease Syndrome. Front Med (Lausanne) 14 (2017): 224.
- Clarissou J, Song A, Bernede C, Guillemot D, Dinh A, Ader F, et al. Efficacy of a long-term antibiotic treatment in patients with a chronic Tick Associated Poly-organic Syndrome (TAPOS). Med Mal Infect 39 (2009): 108-15.


 Impact Factor: * 3.5
Impact Factor: * 3.5 Acceptance Rate: 71.36%
Acceptance Rate: 71.36%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
