Molecular Study of Rotavirus A Infection in Children with diarrhea, before and after Vaccine Introduction in Brazzaville and Pointe-Noire, Republic of the Congo
Léadisaelle Hosanna LENGUIYA1, Fabien Roch NIAMA*,1,2, PEMBE ISSAMOU MAYENGUE1, 2, Leblanc GAMPOUO GANDZA 2, Cynthia NKOUA BADZI2, Igor Judicael LOUZOLO 2, Nadia Claricelle LOUKABOU MBONGOLO2, Grâce Petula Urielle FILA-FILA1, Sagesse Raïssa Ginelle LOKO1, Louis Régis DOSSOU-YOVO1,2, Félix KOUKOUIKILA-KOSSOUNDA 1, 2
1Faculté des Sciences et Techniques, Université Marien Ngouabi, BP: 69 Brazzaville, Republic of the Congo
2Laboratoire National de Santé Publique, 120 avenue du Général Charles de Gaule, BP: 120 Brazzaville, Republic of the Congo
*Corresponding author: Fabien Roch NIAMA, Laboratoire National de Santé Publique (LNSP), Unité de Biologie Moléculaire, BP: 120, avenue du Général Charles de Gaule, Brazzaville, Republic of the Congo.
Received: 09 January 2022; Accepted: 17 January 2022; Published: 13 February 2023
Article Information
Citation: Léadisaelle Hosanna LENGUIYA, Fabien Roch NIAMA, PEMBE ISSAMOU MAYENGUE, Leblanc GAMPOUO GANDZA, Cynthia NKOUA BADZI, Igor Judicael LOUZOLO, Nadia Claricelle LOUKABOU MBONGOLO, Grâce Petula Urielle FILA-FILA, Sagesse Raïssa Ginelle LOKO, Louis Régis DOSSOU-YOVO, Félix KOUKOUIKILAKOSSOUNDA. Molecular study of Rotavirus A infection in children with diarrhea, before and after vaccine introduction in Brazzaville and Pointe- Noire, Republic of Congo. Archives of Microbiology and Immunology 7 (2023): 10-17.
View / Download Pdf Share at FacebookAbstract
Introduction: Acute gastroenteritis due to Rotavirus A infection is common in both developing and developed countries and is responsible for approximately 215,000 annual deaths especially in developing countries. This study aimed to determine the prevalence of rotavirus and the molecular distribution of stains in congolese children under five years old in Brazzaville and Pointe-Noire before and after the Rotarix vaccine introduction.
Method: From February to September 2013 and from August 2017 to February 2018, stool samples were collected from children under 5 years of age suffering from gastroenteritis in Congolese hospitals before and after vaccine introduction. Rotavirus was detected using the ELISA and the VP7 and VP4 genes were genotyped by multiplex RT-PCR.
Results: Of 154 stool samples analyzed, 84 male and 70 female children were included, ranging in age from 4 days to 59 months. A total of 45.4% (n=49) tested positive before vaccination versus 10.86% (n=5) after vaccination (P<0.0002). Also, a change in genomic profile was observed between the two periods with G1P[8] (77.5%), G9P[8] (2%), G1G2P[6]P[8] (6%) and G1G9P[8] (2%) genotypes before vaccination and G1P[8] (40%), G1P[4]P[6] (20%), G8P[8] (20%) and G12P[8] (20%) genotypes after vaccination.
Conclusion: Although based on a limited number of samples, these results improve our knowledge of rotavirus A circulation in the Republic of Congo, particularly the positive impact of vaccination. In addition, the emergence of recombinant strains after vaccination may suggest a possible change in the molecular profile and therefore requires increased surveillance of these strains and their potential escape from vaccination.
Keywords
<p>Vaccine impact, Rotavirus genotypes, Republic of Congo</p>
Article Details
1. Introduction
In Africa, diarrheal diseases are the leading causes of pediatric consultation and account for about 12% of deaths in the under-five population [1]. Several infectious agents are involved in these diseases including Rotaviruses that are responsible of severe gastroenteritis. It was estimated that, around half of the cases of hospitalization and mortality of infants due to gastroenteritis is attributable to Rotavirus infection [2]. Studies conducted in 2008 have shown that Rotaviruses have claimed the lives of approximately 453,000 children under the age of five and have resulted in hundreds of thousands of hospitalizations worldwide. Most of these cases have been recorded in developing countries, particularly sub-Saharan Africa and the Indian subcontinent [3, 4]. Mortality in developing countries as well as economic consequences in developed countries led the World Health Organization in 2009 to recommend the introduction of Rotavirus vaccines into national immunization programs. The consequence was until 2013 the reduction in this number to about 215,000 deaths [5, 6].
Rotaviruses are non-enveloped, segmented double-stranded RNA viruses approximately 70 nm of diameter and belonging to the family Reoviridae. The 11 segments of the genome encode 6 structural proteins and 6 non-structural proteins. The intermediate protein of the capsid, VP6 intervenes in the subdivision of Rotavirus into eight groups; Rotavirus A to H. Among these groups, only strains of group A (mostly encountered), B and C have been recognized as one of the causes of gastroenteritis in humans [7, 8]. The proteins constituting the outer layer of the capsid, VP7 and VP4 inducing the production of neutralizing antibodies are encoded by genes involved in the classification of Rotavirus to genotypes G (glycoprotein) and P (sensitive protease) [9]. strains, the 5 genotypic combinations G1P[8], G2P[4], G3P[8], G4P[8] and G9P[8] cause the majority of RVA infections throughout the world [10, 11, 12]. In developing countries, rare genotypes such as G1P[6], G2P[6], G3P[6], G1P[4], G2P[8], G9P[4], G12P[8] and G12P[6] are circulating resulting in reassortment between human and animal strains [10, 13]. In 2014, the Republic of Congo joined the immunization program recommended by WHO and introduced Rotarix vaccine (GlaxoSmithKline Biologicals, Belgium) in the [14]. However, information on the diversity of circulating Rotavirus strains in Congo is derived from a single pre-vaccination study conducted in the southern Brazzaville area [15]. In this study, we propose to characterize Rotavirus strains circulating at Brazzaville and Pointe-Noire and evaluate the impact of vaccination on the occurrence of gastroenteritis in the under-five of age population.
2. Materials and Methods
2.1 Study design and setting
This study is part of a sentinel surveillance system on RVA related diarrhea cases in the Republic of Congo. For this purpose, the collection of samples was done randomly from february to july 2013 and to august 2017 to February 2018. samples were collected in 4 different sentinel sites selected for the surveillance of rotavirus gastroenteritis cases during the pre-vaccine period, namely the Marien NGOUABI Pediatric Hospital located in the north part of Brazzaville, Pierre MOBENGO hospital located in the center of the city, Mfilou hospital and the Bacongo hospital located in the south part of Brazzaville.
2.2 Study population
This study was conducted among young children aged from 0 to 59 months.
2.3 Datacollection
From August 2017 to February 2018, samples were collected from 4 sentinel sites selected for rotavirus surveillance. In Pointe-Noire samples were collected in Tié-Tié Based Hospital and in Adolphe Sicé General Hospital and at Brazzaville in Marien NGOUABI Pediatric Hospital and Pierre MOBENGO Hospital. Patients experiencing diarrhea with or without vomiting who had been under 7 days was included in this study. However, patients older than 59 months and with diarrhea lasting for more than fourteen days were not included. For each patient received as outpatient or hospitalized in one the sentinel sites for gastroenteritis, a case notification form was completed by the mother or by the caregiver. The demographic information such as age, sex, city and clinical data such as duration of diarrhea until collection, number of stools per day, vomiting, cough, colds, fatigue, loss of appetite, weight loss, as well as clinical signs of complication of infection such as seizures, lethargy and dehydration have been collected. The information about the immune status related du RVA was collected for all the children included during the post-vaccine period. In addition, a stool sample of approximately 5 ml was collected in a sterile container as soon as the patient arrived at the hospital. For the sites located in Brazzaville, the collected samples were placed in isothermal coolers and immediately transferred to the Laboratoire National de Santé Publique (LNSP). For sites located in Pointe-Noire, the samples were stored at 4°C until shipment to the LNSP where they were stored at -20°C before being analyzed.
2.4 Laboratory analysis
For the identification of the VP6 protein, Rotavirus antigen, the Enzyme linked Immuno-sorbent Assay (ELISA) ProSpectTM Rotavirus Kit (Oxoid Ltd., Basingstoke, Hampshire, UK) was used, according to the manufacturer instructions. The RNA was extracted from 10% fecal suspension using the procedure previously described by the World Health Organization [16]. Briefly, a 10% fecal suspension was prepared by diluting approximately 100 μl of the stool solution in 1 ml of PBS (Phosphate Buffered Saline). The mixture was clarified by centrifugation at 14,000rpm for 3 minutes and 140μl of the mixture was used for RNA extraction with the QIAamp Viral RNA Mini kit (Qiagen, GmbH, Hilden, Germany). The RNA was eluted with 60μl of the elution buffer and stored at -80° C. The amplification of VP4 and VP7 genes was performed using the Superscript III One-Step RT-PCR System Kit with Platinum Taq DNA Polymerase (Invitrogen, USA). Briefly, 5 μL of the RNA extract was denatured for 3 minutes at 97°C and immediately transferred to a refrigerated rack for 2 minutes as previously described [16, 17]. The reaction of the reverse transcription (RT) of RNA and the amplification was done using the following pairs of primers: Beg9-GGCTTTAAAAGAGAGAATTTCCGTCTGG/End9-GGTACACATCATACAATT CTAATCTAAG giving a product of about 1062 bp for the VP7 gene [16, 18] and the Con2-ATTTCGGACCA TTTATAACC / Con3-TGGCTTCGCTCATTTATAGACA couple yielding a product of approximately 876 bp for the VP4 gene [19]. After the initial denaturation step, 20 μl of the reaction mixture containing 1 μl of each primer, 12.5 of the reaction buffer (consisting of 0.4 mM of each dNTP and 2.4 mM of MgSO4), 5 μl of deionized water and 0.5μl of the enzyme mixture (Reverse transcriptase and Taq polymerase) was added to each tube. The RT reaction was run at 45°C for 30 minutes followed by activation of the polymerase at 94°C for 2 minutes and 40 cycles of PCR (15 seconds of denaturation at 94°C, 30 seconds of hybridization at 45°C and 1 minute and 30 seconds elongation at 68°C) and final elongation at 68°C for 5 minutes. The amplified products were analyzed in 1% agarose gel and visualized with UV light. The semi-nested PCR was carried out using 2μl of the RT-PCR product, 1μl of the RVG9 antisense primer and of each sense primer (aAT8, aBT1, aCT2, aDT4, aET3, aFT9, G10 and G12), 23μl of deionized water, 12.5μl of buffer 2x, 1μl of the mixture of dNTPs, 2 μl of MgSO4 and 0.5μl of Taq polymerase.
The PCR conditions are as follows: activation of the polymerase at 94°C for 2 minutes followed by 40 amplification cycles (30 seconds of denaturation at 94°C, 30 seconds of hybridization at 45°C. and 1-minute elongation at 68°C) and final elongation at 68°C for 5 minutes. For the VP4 gene, the second PCR was carried out using 1 μl of the RT-PCR product, 1 μl of the Con3 sense primer and of each antisense primer (1T-ku, 2T-1, 3T-1, 4T-1, 5T -1), 26.5 μl of deionized water, 12.5 μl of the buffer 2x, 1 μl of the dNTP mixture, 2.5 μl of the MgSO4 and 0.5 μl of the Taq polymerase. After homogenization, the tubes were placed in a thermocycler and the reaction was carried according the following conditions: activation of the polymerase at 94°C for 4 minutes followed by 30 amplification cycles (1 minute of denaturation at 94°C, 2 minutes of hybridization at 45°C. and 1 minute elongation at 68°C) and final elongation at 68°C for 5 minutes. The amplified products of both genes (VP7 and VP4) were separated by electrophoretic on 2% agarose gel content ethidium bromide and visualized by UV transillumination. The genotypes were determined based on the size of the result amplicon. The sequences of primers used for amplification and the corresponding genotype have shown in Table 1.
Table 1. Primers used in the RVA multiplex PCR
|
Gene |
Name / strain |
Sequence (5’-3’) |
Position |
Size |
Genotype |
|
VP7 |
RVG9 |
GGTACATCATACAATTCT |
1044-1062 |
||
|
aAT8-69M |
GTCACACCATTTGTAAATTCG |
178–198 |
885 bp |
G8 |
|
|
aBT1-Wa |
CAAGTACTCAAATCAATGATGG |
314–335 |
749 bp |
G1 |
|
|
aCT2-DS-1 |
CAATGATATTAACACATTTTCTGTG |
411–435 |
652 bp |
G2 |
|
|
aDT4-ST-3 |
CGTTTCTGGTGAGGAGTTG |
480–498 |
583 bp |
G4 |
|
|
aET3-P |
CGTTTGAAGAAGTTGCAACAG |
689–709 |
374 bp |
G3 |
|
|
aFT9-W161 |
CTAGATGTAACTACAACTAC |
757–776 |
306 bp |
G9 |
|
|
G10 |
ATGTCAGACTACARATACTGG |
666–687 |
397 bp |
G10 |
|
|
G12 |
CCGATGGACGTAACGTTGTA |
548–567 |
515 bp |
G12 |
|
|
VP4 |
Con3 |
TGGCTTCGCTCATTTATAGACA |
Nov-32 |
||
|
1T-1-KU |
ACTTGGATAACGTGC |
336-356 |
345 bp |
P[8] |
|
|
2T-1- RV5 |
CTATTGTTAGAGGTTAGAGTC |
474-494 |
483 bp |
P[4] |
|
|
3T-1-1076 |
TGTTGATTAGTTGGATTCAA |
259-278 |
267 bp |
P[6] |
|
|
4T-1- K8 |
TGAGACATGCAATTGGAC |
385-402 |
391 bp |
P[9] |
|
|
5T-1-69M |
ATCATAGTTAGTAGTCGG |
575-594 |
594 bp |
P[10] |
With: bp, base pair; Source: [16]
2.5 Statisticalanalysis
All data were entered to Microsoft Office Excel 2013 software and then transferred to R software 3.4 for statistical analysis. The chi-square test was used to test homogeneity and associations between different variable and Student’s t-test for comparison of means. The confidence interval was set at 95% and the significance level at P-values <0.05.
2.6 Ethicalconsiderations
The study was conducted after obtaining clearance from the “Comité d'Ethique pour la Recherche en Sciences de la Santé (CERSSA)” of the Ministry of Research and Innovation Technologies (n°038/CERSSA-2012). The parents whose children were eligible, gave their written informed to participate in the study prior to collection of demographic data and samples. To do this, a statement summarizing the objectives of the investigation was read to each individual, in French or one of the two national languages (Lingala and Kituba). The interviews were conducted in private to ensure the confidentiality of the information collected in accordance with the declaration of Helsinki (Declaration of Helsinki – WMA – The World Medical Association)
3. Results
3.1 Characteristics of study population and Rotavirus A detection
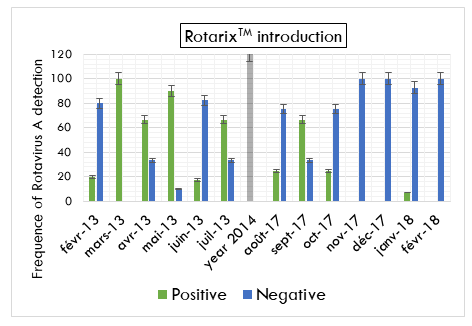
This study included 154 children under five years of age presenting the gastroenteritis symptoms and who were hospitalized or have been visited the sentinel hospitals. A total of 84 male and 70 female children were included in our study with ages ranging from 4 days to 59 months. The study had two groups. The first had constituted by 70% (n=108) patients recruited before vaccine introduction, from February to September 2013 in four hospitals of Brazzaville. The second group included 30% (n=46) patients recruited after vaccine introduction, from August 2017 to February 2018, respectively 21 and 25 patients from two sentinel hospitals at Pointe-Noire and two others at Brazzaville.
Among 108 stool samples collected before vaccine introduction, 45.4% (n=49) were tested positive for RVA. Whereas, 10.86% (n=5) samples collected after vaccination were positive for Rotavirus with the proportions of 16% (n=4) and 4.7% (n=1) respectively in Brazzaville and Pointe-Noire. The difference of the proportion between the two groups was statistically significant (X2=22, P<0.0002). Sixty-five percent of the patients included were recruited during the dry months of the year. The difference of the proportion of RVA was no significant between males (48%) and females (52%) children (X2=0.1, P=0.78) nor in the age group (X2=0.5, P=0.49). Moreover, the proportion of Rotavirus gastroenteritis was high, 52% in the 6-11 months age group and the low prevalence, 9.2% was found in the age group of 24 to 59 months (Table 2).
Table 2: Prevalence of children with gastroenteritis and distribution of RVA into age group
|
Age group |
Children with gastroenteris |
Rotavirus reactive ELISA n (%) |
Rotavirus ELISA |
|
n |
Non-reactive n (%) |
||
|
[0-6] |
26 |
7 (27) |
19 (73.07) |
|
[6-12] |
63 |
28 (44.44) |
35 (55.55) |
|
[12-24] |
52 |
14 (26.92) |
38 (73.07) |
|
[24-59] |
13 |
5 (38.46) |
8 (61.53) |
|
Total |
154 |
54 (35.06) |
100 (64.93) |
With n, number of cases
In the monthly distribution of Rotavirus cases, the proportion of children testing positive for Rotavirus gastroenteritis was more in the dry months (January, February, June, July, August, and September) compared to rainy months (March, April, May, October, November, and December) (X2= 4.6, P=0.03) with one peaked during Mars before vaccine introduction and one other during September in the post vaccine period (Figure 1).
3.2 Rotavirus A genotypes distribution
In the first PCR, among 54 RVA positive samples with ELISA, 52 (96.3%) were successfully amplified with VP4 gene.
For specific PCR, 50 samples were genotyped with VP7 gene (G type) and 52 with VP4 gene (P type).
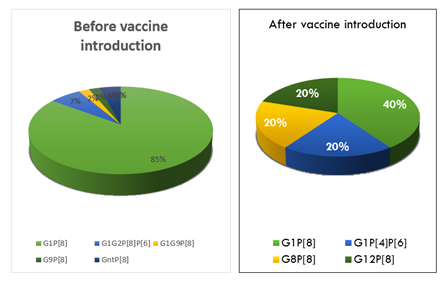
Regarding samples collected before vaccine introduction, 47 out of 49 samples could be amplified and the G1 genotype was more prevalent with 81% followed by G9 genotype with 2% and mix infections between G1G2 and G1G9 were found at 8%. The VP4 genotype showed the P[8] with 79.6% and the mix P[6]P[8] with 6%. Four samples were partially amplified for the VP7 and two for VP4 gene. For samples collected after vaccine introduction, 5 samples could be amplified. We had three G1 genotypes, one G8 and one G12. The P type was P[8] and the mix P[4]P[6] genotype (Table 3). The G12 genotype was found in Pointe-Noire, G1 and G8 in Brazzaville.
Table 3: Distribution of Rotavirus G- and P-types before and after vaccine introduction in under-5 children
|
G-Type |
|||||||||
|
P-Type |
G1 |
G8 |
G9 |
G12 |
G1G2 |
G1G9 |
Gnt |
Total |
Percent (%) |
|
P[8] |
41 |
1 |
1 |
1 |
0 |
1 |
3 |
48 |
88.88% |
|
P[8]P[6] |
0 |
0 |
0 |
0 |
3 |
0 |
0 |
3 |
5.55% |
|
P[4]P[6] |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
1.85% |
|
P[nt] |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
2 |
3.70% |
|
Total |
43 |
1 |
1 |
1 |
3 |
1 |
4 |
54 |
100% |
With nt= untippable strain
For the Rotavirus genotype combination, the G1P[8] were the most Rotavirus genotype detected in these two study groups. The genotypes G9P[8], G1G2P[6]P[8] and G1G9P[8] were detected only before vaccine introduction. In the post vaccine period, the others genotypes detected were the mix G1P[4]P[6], G8P[8] and G12P[8] (Figure 2).
Regarding RVA genotype distribution according season, age group and sex, the predominant genotype G1P[8] was present throughout months of year, age groups and in both sex.
4. Discussion
The propose of this study was to characterize rotavirus strains circulating at Brazzaville and Pointe-Noire and evaluate the impact of vaccination on the occurrence of gastroenteritis in the under-five of age population. Rotaviruses are identified as an important cause of gastroenteritis requiring hospitalization and causing several deaths in Asian and African children [20, 21]. In our study, 54 (35%) out of 154 samples of patients suspected with gastroenteritis were ELISA positives (26 male vs 28 female). This result showed that there was no significant difference in the detection of RVA to sex and implies that RVA affects both male and female children. Conversely, in the age group, we found that 51.8% of RVA positive cases were attributed to the age group of 6 to 11 months in agreement with the studies conducted in surrounding countries such as the Democratic Republic of Congo (DRC) were 54.3% of children were infected in this group [22]; but in contrast to those in Rwanda where it was mostly children aged 12 to 24 months who have been most affected [23].
However, children aged 24 to 59 months were the least infected and accounted for 9.2% of cases. This result can be explained by the role of the immune memory that is built up by repetition of infections and provides a natural immunity that reduces or eliminates severe forms of RVA diarrhea [24]. We also showed that children under 2 years of age were the most RVA infected and represented approximately 90% of cases. These data corroborate those of others sub-Saharan countries that also reported a high incidence of RVA diarrhea in the under-2 age group [22, 25]. Before the introduction of the Rotavirus vaccine into the vaccination program in the Congo, stool samples were collected from children received in 4 hospitals with different localizations (north, center and south) at Brazzaville. About these, 45.4% of pre-vaccination samples were positive. This prevalence is similar to those previously reported in a study conducted in south of Brazzaville and in some neighboring countries such as the Central African Republic (located in northern Congo), with a prevalence of 46.4% and 45% respectively [15], unlike in Gabon, between 2010 and 2011, only 27% of prevalence rate was reported [25].
This high prevalence of Rotavirus infections could be explained by the difficulty, in accordance with the habits in the country, to prevent children from observing certain hygiene measures and access to drinking water recommended to combat diarrhea. Also, the limited size of the sample (154) would not be sufficient to generate representative data of the exact epidemiology of this infection in the country. The number of clinical cases of gastroenteritis may have been underestimated. Nevertheless, a clear decrease in cases was observed after the introduction of Rotavirus vaccination in the country. Indeed, we report a significant decrease of 34.54% between the two periods, thus materializing the effectiveness of the positive impact of Rotavirus A vaccine in the prevention of RVA diarrhea. A clear difference in genotype distribution was observed between the pre-vaccination and post-vaccination periods. Indeed, before the introduction of the vaccine, G1 genotypes of the VP7 gene represented 81% of the isolates, followed by G9 with only 2%. Mixed infection cases were also identified between G1G2 and G1G9 in proportions of 6% and 2%, respectively. Regarding the VP4 gene, the predominant P genotype was P[8] with 79.6% and mixed infections P[6]P[8] with 6% of cases. Rare genotypes such as G8, G10 and G12 previously identified in Brazzaville by Mayindou et al [15] and in other sub-Saharan countries [26, 27] were not reported in the current study. This difference between the work conducted in Brazzaville may be related to the limitations of the two studies, including the duration of sample collection and sample size. After the introduction of the vaccine, G1 genotypes remain predominant; however, there is an emergence of rare genotypes such as G8 and G12. Regarding the P type, P[8] has been detected in the majority of cases. It was characterized as an unusual mixed P[4]P[6] genotype. Overall, the genotypic distribution of the strains we report has also been identified in other studies before or after the introduction of vaccination [22, 28].
The P [6] genotype has been reported in several studies of varicella virus infection, and the P[6] genotype has been associated with a wide range of varicella virus genotypes for both asymptomatic and symptomatic infections [28, 29]. In our study, we report in the pre-vaccine group, 7% of patients with a mixed P[6]P[8] genotype and 20% of P[4]P[6] genotype after vaccine introduction. These genotypes have also been reported in DRC [22]. In this study, we report the predominance of the G1P[8] genotype combination (75.92%). In the previous report in DRC conducted between 2012 and 2013, this genotype was detected in 49.3% of cases [15]. Similarly, G9P[8], G1G2P[8]P[6] and G1G9P[8] genotypes were detected at low prevalence before the introduction of vaccination. The high prevalence of the G1P [8] genotype has also been reported in previous studies in other African countries [30]. In the period following the introduction of the vaccine, a reversal of the genotype distribution was noted with the emergence of G8P[8], G1P[4]P[6] and G12P[8] strains.
However, given the small sample size and the number of positive results obtained after vaccination, it is difficult to compare the distribution of genotypes in the two study groups. Nevertheless, studies in eastern and southern Africa [28] found no difference in the detection of VAR strains before and after the introduction of vaccination. Uncommon genotypes such as G8P[8] and G12P[8] detected in this study have been increasingly identified in several African and Asian countries [31, 32] and suggest greater diversity among Congolese VAR infections. Similarly, mixed infections with the unconventional G1P[4]P[6] strain detected for the first time in the DRC had previously been identified in Kenyan children [33]. These unusual combinations are certainly due to reassortment between human and animal strains of rotavirus [33] or by the fact that rotavirus vaccines do not fully protect children, as shown in several post-vaccination studies [34]. Although these data show a significant positive impact of vaccination, the small number of samples collected between the two periods may underestimate the true prevalence of rotavirus infection in this population and limit the scope of this study.
5. Conclusion
This study on rotaviruses associated with acute gastroenteritis in young children in the Republic of Congo reports similar data to those reported in Africa and other continents in a post-vaccination situation. However, the emergence of strains with a particular molecular profile, notably G8P[8], G1P[4]P[6] and G12P[8], should lead to a genomic follow-up in order to corroborate the potential impact of these strains on vaccine escape.
Acknowledgments
The authors thank the Ministry of Public Health and Population, for permitting and facilitating study activities. We thank also the participants for their time and trust.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
RFN, KKF and PIM designed and coordinated the study, and review the manuscript. HLL, analyzed the samples and data, and write the manuscript. GGL, LI, GPUFF, SRGL, NBC and NCLB supervised samples and data collection; LRD-Y participated in patient’s recruitment, read and approved the final version final manuscript. Talwar GP, Gupta JC, Gupta SK, Wadhwa SN, Frick J, et al. Conjugation to a Carrier Renders a Self-Molecule Immunogenic besides Imparting Immuno-Prophylactic Benefit. Archives of Microbiology & Immunology 1 (2016): 1-11.
References
- Liu L, Johnson HL, Cousens S, et al. Global, regional, and national causes of child mortality: An updated systematic analysis for 2010 with time trends since 2000. The Lancet 379 (2012): 2151–2161.
- Tate JE, Burton AH, Boschi-Pinto C, et al. 2008 estimate of worldwide Rotavirus-associated mortality in children younger than 5 years before the introduction of universal Rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect Dis 12 (2012): 136-41.
- World, Health, Organization. Enfants: réduire la mortalité. Aide-mémoire n°8178.
- Tate JE, Burton AH, Boschi-Pinto C, et al. Estimate of worldwide Rotavirus-associated mortality in children younger than 5 years before the introduction of universal Rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect Dis2012 12 (2008): 136-41.
- Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 385 (2015): 117-71.
- Tate JE, Burton AH, Boschi-Pinto C, et al. Global, Regional, and National Estimates of Rotavirus Mortality in Children <5 Years of Age, 2000-2013. Clin Infect Dis 62 (2016): S96-S105.
- Matthijnssens J, Otto PH, Ciarlet M, et al. VP6-sequence-based cutoff values as a criterion for Rotavirus species demarcation. Archives of virology 157 (2012): 1177–82.
- Mihalov-Kovacs E, Gellert A, Marton S, et al. Candidate new Rotavirus species in sheltered dogs, Hungary. Emerging infectious diseases 21 (2015): 660–3.
- Estes MK. Rotaviruses and their replication. In: Fields BN, Knipe DM, Howley PM, eds. Fields virology. 3rd ed. Vol 2. Philadelphia: Lippincott- Raven (1996): 1625–55
- Santos N, Hoshino Y. Global distribution of Rotavirus serotypes/genotypes and its implications for the development and implementation of an effective Rotavirus vaccine. Rev Med Virol 15 (2005): 29–56.
- Gentsch JR, Laird AR, Bielfelt B, et al. Serotype diversity and reassortment between human and animal Rotavirus strains: implications for Rotavirus vaccine programs. J Infect Dis 192 (2005): S146-S159.
- Sanchez-Fauquier A, Wilhelmi I, Colomina J, et al. Diversity of group A human Rotavirus types circulating over a 4-year period in Madrid, Spain. J Clin Microbiol 42 (2004): 1609-13.
- Global Rotavirus information and surveillance bulletin. Reporting period: January through December 2010. Vol. 4. World Health Organization 4 (2011).
- Base de données de l’OMS sur la vaccination, les vaccins et les produits biologiques, septembre 2016. Organisation mondiale de la Santé, Genève (Suisse) (2016).
- Mayindou G, Ngokana B, Sidibe A, et al, Molecular Epidemiology and Surveillance of Circulating Rotavirus and Adenovirus in Congolese Children with Gastroenteritis, Journal of Medical Virology 88 (2016): 596–605.
- 2009. Manual of Rotavirus detection and characterization methods.
- Donato CM, Ch’ng LS, Boniface KF, et al, Identification of Strains of RotaTeq Rotavirus Vaccine in Infants with Gastroenteritis Following Routine Vaccination, The Journal of Infectious Diseases 206 (2012): 377–83.
- Gouvea V, Glass RI, Woods P, et al. Polymerase chain reaction amplification and typing of Rotavirus nucleic acid from stool specimens. J Clin Microbiol 28 (1990): 276–82.
- Gentsch JR, Glass RI, Woods P, et al. Identification of group A Rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol 30 (1992): 1365–73.
- Parashar UD, Burton A, Lanata C, et al. Global mortality associated with Rotavirus disease among children in 2004. J Infect Dis 200 (2009): S9–15.
- Cunliffe NA, Kilgore PE, Bresee JS, et al. Epidemiology of Rotavirus diarrhoea in Africa: A review to assess the need for Rotavirus immunization. Bull World Health Organ 76 (1998): 525-37.
- Pukuta ES, Esona MD, Nkongolo A, et al. Molecular Surveillance of Rotavirus Infection in the Democratic Republic of the Congo August 2009 to June 2012. Pediatr Infect Dis J 33 (2014): 355–359.
- Uwimana J, Ndishimye P, Bizimana E, et al. Rotavirus gastroenteritis surveillance and prevalence assessment among under five children in Rwanda. Rwanda med. j. (Online)72 (2015): 17-21.
- SCHMITZ J. Vaccinations antiRotavirus. Arch Pediatr 6 (1999): 979-84.
- Lekana-Douki SE, Kombila-Koumavor C, Nkoghe D, et al. Molecular epidemiology of enteric viruses and genotyping of Rotavirus, adenovirus and astrovirus among children under 5 years old in Gabon. Int J Infect Dis 34 (2015): 90-5.
- Tate JE,Ngabo F,Donnen P,et Effectiveness of pentavalent Rotavirus vaccine under conditions of routine use in Rwanda. Clin Infect. Dis 62 (2016): S208-S212. 31.
- Gasparinho C, Piedade J, Mirante MC, et al. Characterization of Rotavirus infection in children with acute gastroenteritis in Bengo province, Northwestern Angola, prior to vaccine introduction. PLoS ONE 12 (2017): e0176046.
- Seheri LM, Magaguna NB, Peenze I, et Rotavirus strain diversity in Eastern and Southern African countries before and after vaccine introduction. Vaccine S0264-410X (17) 31679-1.
- Dennis FE, Fujii Y, Haga K, et Identification of Novel Ghanaian G8P[6] Human-Bovine Reassortant Rotavirus Strain by Next Generation Sequencing. PLoS ONE 9 (2014): e100699.
- Mwenda JM, Ntoto MN, Abebe A, et al. Burden and Epidemiology of Rotavirus Diarrhea in Selected African Countries: Preliminary Results from the African Rotavirus Surveillance Network. J Infect Dis202 (2010): S5-S11.
- Global Rotavirus information and surveillance bulletin. Reporting period: January through December 2010. Vol. 4. World Health Organization (2011).
- Seheri M, Nemarude L, Peenze I, et Update of Rotavirus strains circulating in Africa from 2007 though 2011. Pediatr Infect Dis J 33 (2014): S76-84.
- Wandera A. E. Rotavirus disease epidemiology and molecular characteristics of circulating strains before and after vaccine introduction in KENYA. Program for Nurturing Global Leaders in Tropical and Emerging Communicable Diseases Graduate School of Biomedical Sciences Nagasaki University (2013-2017).
- Jere KC, Sawyerr T, Seheri LM, et al. A first report on the characterization of Rotavirus strains in Sierra Leone. J Med Virol 83 (2011): 540-50.


 Impact Factor: * 3.5
Impact Factor: * 3.5 Acceptance Rate: 71.36%
Acceptance Rate: 71.36%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
