Antimicrobial Resistance and Quorum Sensing Genes Detection among the Biofilm Forming Staphylococcus aureus Isolated from Admitted Patients of Dhaka Medical College Hospital, Dhaka, Bangladesh
Noor-E-Jannat Tania1*, S. M. Shamsuzzaman2, Aminul Islam3
1Lecturer, Department of Microbiology, Dhaka Medical College, Dhaka, Bangladesh
2Professor and Head of the Department, Department of Microbiology, Dhaka Medical College, Dhaka, Bangladesh
3Honorary Medical Officer, Department of Medicine, Dhaka Medical College Hospital, Dhaka, Bangladesh
*Corresponding author:Noor-E-Jannat Tania, Lecturer, Department of Microbiology, Dhaka Medical College, Dhaka, Bangladesh
Received: 06 August 2021; Accepted: 16 August 2021; Published: 21August 2021
Article Information
Citation:
Noor-E-Jannat Tania, S. M. Shamsuzzaman, Aminul Islam. Antimicrobial resistance and quorum sensing genes detection among the biofilm forming Staphylococcus aureus isolated from admitted patients of Dhaka Medical College Hospital, Dhaka, Bangladesh. Fortune Journal of Health Sciences 4 (2021): 441-455.
View / Download Pdf Share at FacebookAbstract
Staphylococcus aureus an infamous human pathogen, is a major cause of the community as well as healthcare associated infections. Staphylococcus aureus has the tendency to form biofilm. Biofilm formation usually increases antimicrobial resistance capacity which shows considerable challenges to successful eradication of infections. The aim of this study was to detect the biofilm forming Staphylococcus aureus phenotypically and to determine its association with drug resistance and also detect the prevalence of different types of quorum sensing genes among them. Out of 275 clinical samples, 75.64% were culture positive among which 21.63% Staphylococcus aureus were isolated by culture and biochemical tests. Biofilm producing Staphylococcus aureus were isolated by tissue culture plate method and tube method. Antimicrobial susceptibility tests were performed by the standard disc diffusion technique. PCR was done to detect agr genes and sequencing of agr-I gene was done. Tissue culture plate method was found superior to tube method for biofilm detection. Tissue culture plate method detected 71.11% and tube method detected 62.22% biofilm producers. Biofilm-producing Staphylococcus aureus showed higher resistance to oxacillin 53.13%, cefoxitin 46.88% and ampicillin 46.88% than non biofilm-producers. Among biofilm-producing Staphylococcus aureus, agr-I gene was predominant (46.88%) than other quorum sensing genes. The study provided insight into the higher proportion of antibiotic resistance among non-biofilm-producing Staphylococcus aureus than biofilm-producers. So, antibiotic resistance is not significantly associated with biofilm-production in Staphylococcus aureus.
Keywords
<p>Antimicrobial resistance; Biofilm; PC;, Quorum sensing; Staphylococcus aureus</p>
Article Details
1. Introduction
Staph. aureus (Staphylococcus aureus) is a leading cause of nosocomial infections worldwide and causative agent of a wide range of diseases. Many of these diseases, including endocarditis, osteomyelitis, and foreign body associated infections, appear to be caused by biofilm-forming Staph. aureus [1,2]. According to some study reports, over 65% of hospital-acquired infections occur by the infecting organisms that have the ability to biofilm production [3].
The biofilm formation is accompanied by the production of extracellular polymer and adhesion matrix and leads to fundamental changes in the bacterial growth and gene expression [4]. There are various definitions for biofilm but all of them consider three major components for it: microbes, slime exopolysaccharide and surface, removing any of them can stop producing biofilm [5].
Susceptibility to antibiotics in bacteria that are protected by biofilm is reduced because antibiotics are prevented to reach the bacteria surrounded by biofilm. Furthermore biofilm keeps bacteria out of reach of host immune defense mechanism and often resulting in persistent infections that are difficult-to-treat and life-threatening due to emergence of multidrug resistance strains and also occurrence of isolates that are able to form strong biofilms [6, 7, 8].
A large number of studies have shown that bacterial quorum sensing (QS) signaling plays important roles in biofilm production [9, 10]. The QS system of the staphylococci is called the accessory gene regulator or “agr” system. QS via agr increases production of virulence factors, which include various enzymes and toxins. Moreover, agr dysfunction is correlated with persistent Staph. aureus bacteremia [11].
There has been continued interest in the agr for the development of anti-staphylococcal drugs [12].Combining use of antibiotic with an anti-QS agent is the most effective clinical strategy for the treatment of bacterial diseases at present [13, 14]. In this study, we investigate the prevalence of agr groups in Staph. aureus isolates obtained from pus, wound swab and blood samples.
This study seeks to determine drug resistance pattern among the biofilm forming Staph. aureus isolated from the patients of Dhaka Medical College Hospital with prevalence of quorum sensing genes among them by PCR assays.
2. Materials and Methods
This Cross sectional study was conducted from 1 January 2019 to 31 December 2019 in the Microbiology laboratory of Dhaka Medical College Hospital, Dhaka, Bangladesh.
2.1. Samples collection and identification
Samples were collected from pus, wound swab and blood of clinically suspected infected patients of inpatient departments of Dhaka Medical College Hospital or were received in the microbiology department for culture and sensitivity after taking informed written consent irrespective of age, sex and antibiotic intake. Patient who did not give consent were excluded from this study. Staph. aureus were identified by Gram staining, catalase test, coagulase test (slide and tube method), colony morphology, hemolytic property, pigment production and mannitol fermentation test in mannitol salt agar media as per standard procedures [15].
2.2. Biofilm experiments
Biofilm formation by these isolates was detected by two in vitro methods: tissue culture plate method and tube method [16, 17].
2.2.1. Tissue culture plate method
The organisms were inoculated in 10 mL of trypticase soya broth (TSB) with 1% glucose at 37°C or 24hours. The cultures were then diluted 1:100 with fresh medium and individual wells of sterile 96 polystyrene tissue culture plates were filled with 200 μl of the diluted cultures. Negative control wells contained inoculated sterile broth. The plates were incubated for 24 hours at 37°C. The wells were gently washed four times with 0.2ml of phosphate buffered saline (PBS) (PH 7.2), fixed by sodium acetate (2%) and stained by 0.1% crystal violet. Excess stain was removed by deionized water, and the plate was kept for drying. The optical density at 570nm (OD570) was determined using a micro-ELISA auto-reader assay. The experiment was performed in triplicate and repeated three times.
2.2.2. Calculation of OD values [18]
The average OD values were calculated for all tested strains and negative controls, since all tests were performed in triplicate and repeated three times. Second, the cut off value (ODc) was established. It was defined as three standards (SD) above the mean OD of the control: ODc = average OD of negative controls + (3 × SD of negative control). Final OD value of a tested strain was expressed as average OD value of the strain reduced by ODc value (OD = average OD of a strain – ODc). ODc value was calculated for each microtiter plate separately. If a negative value obtained, it should be present as zero, while any positive value indicates biofilm.
2.2.3. Interpretation of biofilm production by TCP method
|
Average OD value |
Biofilm production |
|
OD ≤ ODc |
Non biofilm producer |
|
ODc < OD ≤ 2× ODc |
Weak biofilm producer |
|
2 × ODc < OD ≤ 4 × ODc |
Moderate biofilm producer |
|
4 × ODc < OD |
Strong biofilm producer |
2.2.4. Tube method
TSB with 1% glucose (10ml) was inoculated with loopful of microorganism from overnight culture plates and incubated for 24 hours at 37°C. The tubes were decanted and washed with PBS (pH 7.2) and dried. Dried tubes were stained with crystal violet (0.1%). Excess stain was removed and tubes were washed with deionized water. Tubes were than dried in inverted position and observed for biofilm formation. Biofilm formation was considered positive when a visible film lined the wall and bottom of the tube. Experiments were performed in triplicate and repeated three times. The adherence property of biofilm producers was graded as strong, moderate and weak and non-biofilm producer.
2.3. Antimicrobial susceptibility test
Antimicrobial susceptibility tests of the clinical isolates against different antimicrobials were performed in Muller-Hinton agar (MHA) using the standard disk diffusion technique (modified Kirby-Bauer method) [19]. Zones of inhibition of azithromycin (15μg), erythromycin (15μg), clindamycin (2μg), linezolid (30μg), cefoxitin (30μg) were interpreted according to CLSI (2019) and ampicillin (10μg), vancomycin (30μg), novobiocin (30μg), teicoplanin (30μg) according to CLSI (2015) guidelines [20, 21]. Zones of inhibition of amikacin (30μg) and oxacillin (1μg) were interpreted according to EUCAST (2018) and Khan et al. respectively [22, 23]. Antibiotic discs were obtained from commercial source (Oxoid Ltd, Uk).
2.4. Control strain
Staph. aureus ATCC 25923 was used as control strain to assess the performance of the method.
2.5. Statistical Analysis
SPSS software (version 25) was used for data analysis. Chi-square test was used for analysis of categorical data. P value of < 0.05 was considered significant.
2.6. Molecular method [24, 25]
Polymerase chain reaction (PCR) was done for the detection of agr-I, agr-II, agr-III andagr-IV genes in biofilm forming Staph. aureus.
2.6.1. Bacterial pellet formation
A loop full of bacterial colonies from MHA media was inoculated into a microcentrifuge tube containing TSB. After incubation overnight at 37°C, the tubes were centrifuged at 4000 g for 10 minutes at 4°C and the supernatant were discarded and then the tubes containing bacterial pellets were kept at -20°C as pellets until DNA extraction.
2.6.2. DNA extraction
300 µl distilled water was mixed with bacterial pellet and was vortexed until mixed well. The mixture was kept in block heater (DAIHA Scientific, Seoul, Korea) at 100°C for 10 minutes for heating. After heating the tube was immediately kept on ice for further 5 minutes and then centrifuged at 14000 g at 4°C for 10 minutes. Finally, supernatant was taken into another microcentrifuge tube and was used as template DNA for PCR. Extracted DNA was preserved at -20°C for future use.
2.6.3. Mixing of master mix and primer with DNA template
Primers were diluted with Tris-EDTA (TE) buffer according to manufacturer’s instruction. PCR was performed in a final reaction volume of 25 µl in a PCR tube, containing 12.5 µl of master mix (mixture of dNTP, Taq polymerase, MgCl2 and PCR buffer), 1 µl forward primer and 1 µl reverse primer (Promega Corporation, USA) 2 µl extracted DNA and 10.5 µl of nuclease free water. After a brief vortex, the PCR tubes were centrifuged in a microcentrifuge for few seconds.
2.6.4. Amplification through thermal cycler
PCR assays were performed in a DNA thermal cycler (Eppendorf AG, Master cycler gradient, Hamburg, Germany). Each PCR run was comprised of preheat at 94°C for 10 minutes followed by 36 cycles of denaturation at 94°C for 1 minutes, annealing at 58°C for 45 seconds, extension at 72°C for 2 minutes with final extension at 72°C for 10 minutes.
2.6.5. Agarose gel electrophoresis
PCR products were detected by electrophoresis on 1.5% agarose gel was prepared with 1X TBE buffer (TrisBorate EDTA). For 1.5% agarose gel preparation, 0.18 gram of agarose powder (LE, Analytical grade, Promega, Madison, USA) was mixed with 12.5ml TBE buffer. Mixture was boiled for few minutes to dissolve and cooled to 60-70°C. A comb was placed in gel tray and poured the agarose gel. After solidification, comb was removed and 6µl of amplicon was mixed with 1µl loading dye on para film and then loaded into the well of agarose gel. 2 µl of DNA ladder was mixed with 1µl loading dye and was loaded into one well. Gel containing amplicon and DNA ladder were then placed on the electrophoresis tank having 1X TBE buffer for 35 minutes at 100 volts. Positive control and negative control was also loaded in separate well.
2.6.6. Staining and de-staining of the gel
After electrophoresis, the gel was stained with ethidium bromide (20 µl ethidium bromide in 200ml distilled water) for 30 minutes. It was then de-stained with distilled water for 15 minutes.
2.6.7. Visualization and interpretation of results
The gel was observed under UV Trans-illuminator (Gel Doc, Major science, Taiwan) for DNA bands. The DNA bands were identified according to their molecular size by comparing with the molecular weight marker (100 bp DNA ladder) loaded in a separate lane. Samples showing the presence of corresponding bp band were considered positive for the presence of that organism.
2.6.8. Procedure of DNA sequencing
For sequencing of bacterial DNA, purification of amplified PCR product was done by using DNA purification kit (FAVOGEN, Biotech Corp). Purified PCR products of Staph. aureus were sent to 1st Base Laboratories, Malaysia for sequencing by capillary method (ABI PRISM 3500). BLAST analysis was performed to search for homologous sequences into the Gen Bank at www.ncbi.nlm.nih.gov.
2.6.9. Primers used in this study [26]
|
Gene |
Primer sequence (5’-3’) |
Size (bp) |
|
agr- I |
Forward: 5-ATG CAC ATG GTG CAC ATG C-3 |
441 |
|
Reverse: 5-GTC ACA AGT ACT ATA AGC TGC GAT-3 |
||
|
agr- II |
Forward: 5-ATG CAC ATG GTG CAC ATG C-3 |
575 |
|
Reverse: 5-TAT TAC TAA TTG AAA AGT GGC CAT AGC-3 |
||
|
agr- III |
Forward: 5-ATG CAC ATG GTG CAC ATG C-3 |
323 |
|
Reverse: 5-GTA ATGTAA TAG CTT GTA TAA TAA TAC CCA G-3 |
||
|
agr- IV |
Forward: 5-ATG CAC ATG GTG CAC ATG C-3 |
659 |
|
Reverse: 5-CGA TAA TGC CGT AAT ACC CG-3 |
3. Results
Out of 275 samples, 208 bacteria were isolated among which 45 (21.63%) Staph. aureus were identified.
Table I demonstrates detection of biofilm production by different methods. Among 45 Staph. aureus, TCP method were detected 32 (71.11%) and TM were detected 28 (62.22%) biofilm producers.
Table I: Detection of biofilm producers by TM and TCP method among isolated Staph.aureus. (N= 45)
|
TM n (%) |
TCP |
P value |
||
|
BP n (%) |
BN n (%) |
|||
|
BP |
28 (62.22) |
26 (57.78) |
2 (4.44) |
>0.05 |
|
BN |
17 (37.78) |
6 (13.33) |
11 (24.45) |
|
|
Total |
45 (100) |
32 (71.11) |
13 (28.89) |
|
BP = Biofilm positive, BN = Biofilm negative, N = Total number of bacteria, n= Number of positive.
Table II demonstrates among 45 isolates of Staph. aureus11.11%, 28.89 % and 31.11% of them were strong, medium and weak biofilm producer by the TCP method and 6.67%, 22.22% and 33.33% of them were strong, medium and weak biofilm producer by the TM respectively.
Table II: Screening of the isolates of Staph. aureus for biofilm formation by TCP method and TM.(N= 45)
|
Biofilm formation |
TCP n (%) |
TM n (%) |
P value |
|
Strong |
5 (11.11) |
3 (6.67) |
>0.05 |
|
Moderate |
13 (28.89) |
10 (22.22) |
>0.05 |
|
Weak |
14 (31.11) |
15 (33.33) |
>0.05 |
|
Total |
32 (71.11) |
28 (62.22) |
N = Total number of Staph. aureus, n= Number of biofilm producers
Table III shows comparison of antibiotic resistance pattern between biofilm-producing and non-biofilm-producing Staph. aureus. Among 32 isolated biofilm-producing Staph. aureus 53.13% were resistant to oxacillin, 46.88% were resistant to cefoxitin and ampicillin whereas out of 13 non biofilm-producing Staph. aureus 23.08% were resistant to oxacillin and ampicillin, 30.77% were resistant to cefoxitin. 21.88% biofilm-producers were resistant to vancomycin and 18.75% were resistant to linezolid but 61.53% non-biofilm-producers were resistant to vancomycin and linezolid. The resistance rate of biofilm producing Staph. Aureus were more in Azithromycin (62.50%), Clindamycin (59.38%), Erythromycin (56.25%), Oxacillin (53.13%), Cefoxitin and Ampicillin (46.88%) followed by Amikacin and Vancomycin (21.88%) and Linezolid (18.75%).
Table III: Comparison of antibiotic resistance pattern between biofilm-producing and non-biofilm-producing Staph. aureus.
|
Antimicrobials |
Biofilm producers (N=32) n (%) |
Non-biofilm producers (N=13) n (%) |
P value |
|
Azithromycin |
20 (62.50) |
11 (84.61) |
>0.05 |
|
Clindamycin |
19 (59.38) |
11 (84.61) |
>0.05 |
|
Erythromycin |
18 (56.25) |
13 (100.00) |
>0.05 |
|
Oxacillin |
17 (53.13) |
3 (23.08) |
>0.05 |
|
Cefoxitin |
15 (46.88) |
4 (30.77) |
>0.05 |
|
Ampicillin |
15 (46.88) |
3 (23.08) |
>0.05 |
|
Amikacin |
7 (21.88) |
5 (38.46) |
>0.05 |
|
Vancomycin |
7 (21.88) |
8 (61.53) |
<0.05 |
|
Linezolid |
6 (18.75) |
8 (61.53) |
<0.05 |
|
Teicoplanin |
3 (9.38) |
8 (61.53) |
<0.05 |
N = Total number of biofilm-producing Staph. aureus, n = Number of resistant bacteria
Table IV demonstrates the proportion of QS genes among biofilm producing Staph. aureus. Out of 32 biofilm-producing Staph. aureus, agr-I was the predominant one, followed by agr-III, agr-II and agr-IV.
Table IV: Proportion of agr genes among the isolated biofilm producing Staph. aureus. (N= 32)
|
QS genes |
Biofilm producing Staph. aureus n (%) |
|
agr-I |
15 (46.87) |
|
agr-II |
2 (6.25) |
|
agr-III |
7 (21.88 |
|
agr-IV |
2 (6.25) |
|
No agr gene was found |
6 (18.75) |
|
Total |
32 (100.00) |
N = Total number of biofilm-producing Staph. aureus, n= Number of positive
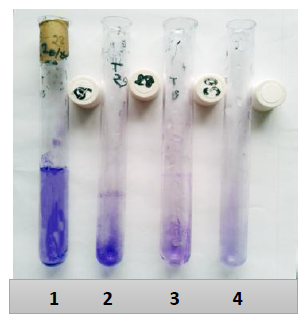
Figure 1: Photograph of detection of biofilm producing Staphylococcus aureus by tube method. 1, 2, 3 are strong, moderate and weak biofilm positive strain respectively and 4 is biofilm negative strain.

Figure 2: Photograph of screening of biofilm forming Staph. aureus by TCP method.
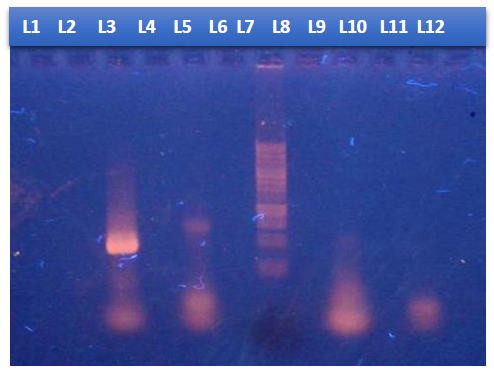
Figure 3: Photograph of gel electrophoresis: negative control without DNA (Lan 1), negative control Staphylococcus aureus ATCC 25923 (Lan 2), amplified DNA of 441 bp for agr-I gene (Lan 3), amplified DNA of 575 bp for agr-II gene (Lan 5), hundred bp DNA ladder (Lan 7), negative sample (Lan 12)
4. Discussion
Staph. aureus is the leading cause of biofilm-mediated life threatening infections, as biofilm production influences the efficacy of antibiotic therapy [2].
In the present study, 11.11% of the Staph. aureus were highly virulent showing strong adherence, 28.89% , 31.11% and 28.89% showing moderate, weak and non-adherence, respectively. This finding was almost similar to the related previous studies [27, 28]. Environmental factors like sugars (glucose or lactose) or proteases present in the growth medium, surface area, type of surface (rough/smooth), and porosity, charge of the surface and the genetic makeup of the Staph. aureus isolate affect biofilm formation [29].
Another study showed that 14.51%, 50.38% and 35.11% were strong, moderate and negative for biofilm formation respectively [30]. This difference in biofilm formation patterns among bacterial isolates may be due to difference in strain types, number of bacterial isolates, sample size, geographic locations and methodological variations to assess biofilm formation [31].
In this current study, antimicrobial resistance pattern was higher among non-biofilm-producing Staph. aureus than biofilm-producers. In this study, among biofilm-producing Staph. aureus 56.25%, 46.88%, 28.13% and 9.38% were resistant to erythromycin, cefoxitin, clindamycin and teicoplanin, respectively . This finding was nearly close to the previous studies [27, 32].
In our study, 46.88% biofilm-producing Staph. aureus were resistant to ampicillin. A previous study reported that 86.7% biofilm-producing Staph. aureus were resistant to ampicillin which was much higher than the present study [27]. The higher resistance of isolates to ampicillin may be attributed to the fact that it is the commonly used antibiotic for treatment of skin and nasal infections [33]. Differences in the antibiotic resistance patterns varies widely between different geographic regions. Many factors contribute to those patterns including local infection control programs implemented, antibiotic prescribing policies, epidemiology of the studied strains themselves and uncontrolled use of antibiotic in agriculture and livestock [34, 35].
In the present study, the total number of QS genes among 32 biofilm producing Staph. aureus isolates were 26(81.25%). Similar previous study reported that 50.82% QS genes were detected [36]. In this current study, among 32 biofilm producing Staph. aureus a majority (46.88%) of isolates belonged to agr-I, followed by agr-III (21.88%), agr-II (6.25%), and agr-IV (6.25%). This finding was nearly close to the previous studies [37, 38]. Another study reported that agr-II was the predominant one which was isolated from milk [39].This difference of obtained results may be due to differences of geographical location and source of isolation [36].
DNA sequence of amplified PCR product and translated nucleotide base sequence of agr-I showed point mutations and deletion including base substitution at multiple positions. Large and small deletions are the main source of gene-inactivating mutations followed by insertions/duplications and a considerable number of point mutation had been detected, including truncation by nonsense and missense mutations [40, 41].
In terms of the annual burden of morbidity and mortality it imposes on society,Staph. aureusis potentially the most significant bacterial pathogen. This is particularly so in the case of chronicStaph. aureusbiofilm infections. Staph. aureusis a clinically relevant pathogen due to its antimicrobial resistance and evasion of the host immune system [42]. So, biofilm detection may be included in routine laboratory tests for the better management of chronic or device associated infections.
Some studies have shown that some specific diseases were associated with specific agr types. One previous study showed that agr-I group was associated with invasive infections, especially bacteremia, agr-II group with invasive disease and agr-III group with noninvasive infection and TSST [43, 44, 45].
The resulting information would be very helpful for finding association of chronic Staph. aureusbiofilm infections with agr specific types as well as in targeting agr for the development of anti-staphylococcal drugs.
5. Conclusion
The clinical isolates of Staph. aureus of hospitalized patients exhibit a high degree of biofilm formation. We can conclude from our study that TCP is a reliable method to detect biofilm forming microorganisms. TCP method can be recommended as a routine test for detection of biofilm producing bacteria in laboratories.
Higher rate of antimicrobial resistance is demonstrated by non-biofilm producers than biofilm-producers. Although biofilm-positive strains have a higher tendency to show resistance to oxacillin, cefoxitin, ampicillin compared to biofilm-negative strains but not statistically significant (>0.05).
Among biofilm-producing Staph. aureus agr-I gene was predominant than other agr genes.
6. Acknowledgement
The author is thankful to all the teachers and staff of Microbiology department, Dhaka Medical College, Dhaka. This study was partially funded by Bangladesh Medical Research Council (BMRC).
7. Conflict of interest
There is no conflict of interest.
8. Ethics approval
This study was approved by Research Review Committee (RRC) of Department of Microbiology and Ethical Review Committee (ERC) of Dhaka Medical College, Dhaka, Bangladesh (Reference number: MEU-DMC/ECC/2019/171).
9. References
- Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clinical microbiology reviews 15 (2002): 167-93.
- Götz F. Staphylococcus and biofilms. Molecular microbiology 43 (2002): 1367-78.
- Del Papa MF, Hancock LE, Thomas VC, Perego M. Full activation of Enterococcus faecalis gelatinase by a C-terminal proteolytic cleavage. Journal of bacteriology 189 (2007): 8835-43.
- Kanwar I, K Sah A, K Suresh P. Biofilm-mediated antibiotic-resistant oral bacterial infections: mechanism and combat strategies. Current pharmaceutical design 23 (2017): 2084-95.
- Murugan K, Usha M, Malathi P, Al-Sohaibani AS, Chandrasekaran M. Biofilm forming multi drug resistant Staphylococcus spp. among patients with conjunctivitis. Polish journal of microbiology 59 (2010): 233.
- Gowrishankar S, Duncun Mosioma N, Karutha Pandian S. Coral-associated bacteria as a promising antibiofilm agent against methicillin-resistant and-susceptible Staphylococcus aureus biofilms. Evidence-Based Complementary and Alternative Medicine 2012 (2012).
- Atshan SS, Shamsudin MN, Thian Lung LT, Sekawi Z, Ghaznavi-Rad E, Pei Pei C. Comparative characterization of genotypically different clones of MRSA in the production of biofilms. Journal of Biomedicine and Biotechnology 2012 (2012).
- Mataraci E, Dosler S. In vitro activities of antibiotics and antimicrobial cationic peptides alone and in combination against methicillin-resistant Staphylococcus aureus biofilms. Antimicrobial agents and chemotherapy 56 (2012): 6366-71.
- Gamage AM, Shui G, Wenk MR, Chua KL. N-Octanoylhomoserine lactone signalling mediated by the BpsI–BpsR quorum sensing system plays a major role in biofilm formation of Burkholderia pseudomallei. Microbiology 157 (2011):1176-86.
- Hong SH, Hegde M, Kim J, Wang X, Jayaraman A, Wood TK. Synthetic quorum-sensing circuit to control consortial biofilm formation and dispersal in a microfluidic device. Nature communications 3 (2012):1-8.
- Fowler Jr VG, Sakoulas G, McIntyre LM, et al. Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. The Journal of infectious diseases 190 (2004): 1140-9.
- Khan BA, Yeh AJ, Cheung GY, Otto M. Investigational therapies targeting quorum-sensing for the treatment of Staphylococcus aureus infections. Expert opinion on investigational drugs 24 (2015): 689-704.
- Han M, Gu J, Gao GF, Liu WJ. China in action: national strategies to combat against emerging infectious diseases. Science China Life Sciences 60 (2017): 1383-5.
- Liu J, Yu H, Huang Y, et al. Complete genome sequence of a novel bacteriophage infecting Bradyrhizobium diazoefficiens USDA110. Science China Life Sciences 61 (2018): 118-21.
- Cheesbrough M. Microscopical techniques used in Microbiology, culturing bacterial pathogens, biochemical tests to identify bacteria. In: Cheesbrough M. (editor). District Laboratory Practice in Tropical Countries. Part 2, 2nd ed, Cambridge University Press, India (2009): 35-70.
- Christensen GD, Simpson WA, Younger JJ, et al. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. Journal of clinical microbiology 22(1985): 996-1006.
- Christensen GD, Simpson WA, Bisno AL, Beachey EH. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infection and immunity 37 (1982): 318-26.
- Stepanovic S, Vukovic D, Hola V, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by Staphylococci. Apmis 115 (2007): 891-9.
- Bauer AW. Antibiotic susceptibility testing by a standardized single disc method. American Journal of Clinical Pathology45 (1966): 149-58.
- Clinical and Laboratory Standard Institute (CLSI) performance standards for susceptibility testing; Twenty-ninth informational supplement. CLSI document M100- S28 (2019).
- Clinical and Laboratory Standard Institute (CLSI). Performance standards for susceptibility testing. Twenty-eight Informational Supplement. CLSI document M 100- S27 (2015).
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. Version 8.0 (2018).
- Khan F, Shukla I, Rizvi M, Mansoor T, Sharma SC. Detection of biofilm formation in Staphylococcus aureus. Does it have a role in treatment of MRSA infections? Trends Med Res 6 (2011): 116-23.
- Farzana R, Shamsuzzaman SM, Mamun KZ, Shears P. Antimicrobial susceptibility pattern of extended spectrum (beta-lactamase producing gram-negative bacteria isolated from wound and urine in a tertiary care hospital, Dhaka city, Bangladesh.Southeast Asian Journal of Tropical Medicine & Public Health 44(2013): 96-103.
- Khosravi AD, Barazandeh B. Investigation of genetic heterogeneity in Mycobacterium tuberculosis isolates from tuberculosis patients using DNA finger printing. Indian Journal of Medical Sciences 59 (2005): 253-258.
- Shopsin B, Mathema B, Alcabes P, et al. Prevalence of agr specificity groups among Staphylococcus aureus strains colonizing children and their guardians.Journal of clinical microbiology41 (2003): 456-459.
- Neopane P, Nepal HP, Shrestha R, Uehara O, Abiko Y. In vitro biofilm formation by Staphylococcus aureus isolated from wounds of hospital-admitted patients and their association with antimicrobial resistance. International journal of general medicine 11 (2018): 25.
- Ghellai L, Hassaine H, Klouche N, et al. Detection of biofilm formation of a collection of fifty strains of Staphylococcus aureus isolated in Algeria at the University Hospital of Tlemcen. Journal of Bacteriology Research 6 (2014): 1-6.
- Christensen BB, Sternberg C, Andersen JB, et al. Establishment of new genetic traits in a microbial biofilm community. Applied and Environmental Microbiology 64 (1998): 2247-55.
- Khan F, Shukla I, Rizvi M, Mansoor T, Sharma SC. Detection of biofilm formation in Staphylococcus aureus. Does it have a role in treatment of MRSA infections? Trendsin Medical Research 6 (2011): 116-23.
- Awoke N, Kassa T, Teshager L. Magnitude of biofilm formation and antimicrobial resistance pattern of bacteria isolated from urinary catheterized inpatients of jimma university medical center, Southwest Ethiopia. International journal of microbiology 2019 (2019).
- Manandhar S, Singh A, Varma A, Pandey S, Shrivastava N. Biofilm producing clinical Staphylococcus aureus isolates augmented prevalence of antibiotic resistant cases in tertiary care hospitals of Nepal. Frontiers in microbiology 9 (2018): 2749.
- Savariraj WR, Ravindran NB, Kannan P, et al. Prevalence, antimicrobial susceptibility and virulence genes of Staphylococcus aureus isolated from pork meat in retail outlets in India.Journal of food safety 39(2019): 12589.
- Dhanalakshmi TA, Umapathy BL, Mohan DR. Prevalence of Methicillin, Vancomycin and Multidrug Resistance among Staphylococcus aureus. Journal of Clinical and Diagnostic Research 6(2012): 974-7.
- Okeke IN, Lamikanra A, Edelman R. Socioeconomic and behavioral factors leading to acquired bacterial resistance to antibiotics in developing countries. Emerging infectious diseases 5(1999): 18.
- Turkey AM, Barzani KK, Suleiman AA, Abed JJ. Molecular assessment of accessory gene regulator (agr) quorum sensing system in biofilm forming Staphylococcus aureus and study of the effect of silver nanoparticles on agr system. Iranian journal of microbiology 10 (2018): 14.
- Bibalan MH, Shakeri F, Javid N, Ghaemi A, Ghaemi EA. Accessory gene regulator types of Staphylococcus aureus isolated in Gorgan, North of Iran. Journal of Clinical and Diagnostic Research: JCDR 8 (2014): 7.
- Javdan S, Narimani T, Abadi MS, Gholipour A. Agr typing of Staphylococcus aureus species isolated from clinical samples in training hospitals of Isfahan and Shahrekord. BMC research notes 12 (2019): 1-6.
- Momtaz H, Tajbakhsh E, Abbasian B, Momeni M. Investigation of accessory gene regulator (agr) in Staphylococcus aureus isolated from clinical and subclinical bovine mastitis in Iran. African Journal of Microbiology Research 4 (2010): 471-4.
- Gracia-Aznarez FJ, Fernandez V, Pita G, et al. Whole exome sequencing suggests much of non-BRCA1/BRCA2 familial breast cancer is due to moderate and low penetrance susceptibility alleles. PloS one 8 (2013).
- Rodríguez-Hernández MJ, Pachón J, Pichardo C, et al. Imipenem, doxycycline and amikacin in monotherapy and in combination in Acinetobacter baumannii experimental pneumonia. Journal of Antimicrobial Chemotherapy 45 (2000): 493-501.
- Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence 2 (2011): 445-59.
- Ayed SB, Boubaker IB, Samir E, Redjeb SB. Prevalence of agr specificity groups among methicilin resistant Staphylococcus aureus circulating at Charles Nicolle hospital of Tunis. Pathologie Biologie 54 (2006): 435-8.
- Rasmussen G, Monecke S, Ehricht R, Söderquist B. Prevalence of clonal complexes and virulence genes among commensal and invasive Staphylococcus aureus isolates in Sweden. PloS one 8 (2013): e77477.
- Jarraud S, Lyon GJ, Figueiredo AM, et al. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. Journal of Bacteriology 182 (2000): 6517-22.


 Impact Factor: * 6.2
Impact Factor: * 6.2 Acceptance Rate: 76.33%
Acceptance Rate: 76.33%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 