Genetic Polymorphism of Alcohol Dehydrogenase 2 (ADH1B) in Association with Alcohol Consumption in Nepalese Population
Rabina Ramtel1, Vijay Kumar Sharma2, Rahul Pathak3, Binod Kumar Yadav2, Eans Tara Tuladhar2, Mithileshwor Raut2, Aseem Bhattarai2, Raju Kumar Dubey2, Apeksha Niraula2, Alisha Sapkota2, Hari Sharan Makaju2, Anant Neupane2, Pujan KC4, Kiran Poudel4
1Department of Biochemistry, Kist Medical College and Teaching Hospital, Lalitpur, Nepal
2Department of Biochemistry, Institute of Medicine, Kathmandu, Nepal
3Department of Gastroenterology medicine, Institute of Medicine, Kathmandu, Nepal
4National Public Health Laboratory, Nepal
*Corresponding author: Vijay Kumar Sharma, Maharajgunj Medical Campus, Maharajgunj, 44600, Kathmandu, Nepal.
Received: 30 November 2023; Accepted: 11 December 2023; Published: 20 December 2023
Article Information
Citation: Rabina Ramtel, Vijay Kumar Sharma, Rahul Pathak, Binod Kumar Yadav, Eans Tara Tuladhar, Mithileshwor Raut, Aseem Bhattarai, Raju Kumar Dubey, Apeksha Niraula, Alisha Sapkota, Hari Sharan Makaju, Anant Neupane, Pujan KC, Kiran Poudel. Genetic Polymorphism of Alcohol Dehydrogenase 2 (ADH1B) in Association with Alcohol Consumption in Nepalese Population. Fortune Journal of Health Sciences 6 (2023): 527-534.
View / Download Pdf Share at FacebookAbstract
Background and Objective: Alcohol is metabolized by the alcohol dehydrogenase (ADH) enzyme system. ADH1B is mostly ethnic and race dependent. Functional polymorphisms within ADH1B gene, alters the enzymatic metabolism of ethanol consequently making a difference in the alcohol elimination rates. Thus, it’s necessary to assess SNP of ADH1B gene in subjects consuming alcohol to explore their genotypic influence on alcohol metabolism. This study was aimed to determine the genetic polymorphisms of ADH1B (Arg47His) in patients with chronic liver disease due to alcohol consumption and healthy non-alcoholic subjects without liver disease, it’s impact on duration of alcohol consumption, and to observe the distribution of its polymorphic variants within different ethnic groups encountered during the study.
Methods and Methodology: A total of 82 EDTA blood samples were taken from alcoholic and non-alcoholic subjects. Molecular analysis was done for detection of ADH1B polymorphism by PCR-CTPP method and products were visualized by 1.5% agarose gel electrophoresis.
Results: Overall, the homozygous form i.e. ADH1B*1/*1 genotype have the highest prevalence rate. The frequency of ADH1B*1 allele was 97.55% and ADH1B*2 allele was 2.45%. Similarly, ADH1B*1/*1 genotype was found to be highest in aadibasi/janajati ethnicity. Furthermore, individuals consuming alcohol for a longer duration have ADH1B*1/*1 genotype.
Conclusion: It can be concluded that the presence of ADH1B*1/*1, ADH1B*1/*2 and ADH1B*2/*2 genotype alters alcohol metabolism, its consumption and tolerance. The presence of ADH1B*1/*1 genotype increases the tolerance towards alcohol and makes individual more liable to alcoholism preceding the onset of alcohol use disorders. Thus, result might indicate the presence of ADH1B*2 allele to be protective against alcoholism and its subsequent consequence.
Keywords
<p>ADH1B polymorphism, SNP, PCR-CTTP, genotype frequency, allele frequency</p>
Article Details
1. Introduction
Alcohol, world’s third largest risk factor for disease burden, is answerable for about 4.5% Disability- Adjusted Life Years (DALY) [1]. Over 90 % of ingested ethanol is metabolized by the enzyme system of liver; alcohol dehydrogenase (ADH), aldehyde dehydrogenase (ALDH) and cytochrome P450 2E1 (CYP2E1). Ethanol biodegradation involves its oxidation to acetaldehyde, catalyzed by alcohol dehydrogenases (ADHs). Acetaldehyde is then oxidized to acetic acid by aldehyde dehydrogenase (ALDH) [2, 3]. Alcohol dehydrogenase, a cytosolic enzyme have seven genes: ADH1A, ADH1B, ADH1C, ADH4, ADH5, ADH6, and ADH7 that encode medium-chain alcohol dehydrogenases (ADHs). Genetic polymorphism in ADH genes leads to production of enzymes with different kinetic properties that modulates individual differences in alcohol and acetaldehyde oxidizing capacity in the body [4-6].
ADH1B have three different alleles in which there is alterations in the amino acid sequence of the encoded β subunit. The ADH1B*1 allele encodes the β1 subunit that has arginine (Arg) at positions 47 and ADH1B*2 encodes the β2 subunit that has an amino acid histidine (His) at position 47, both being common in Asian population. ADH1B*3 encodes the β3 subunit and have cysteine (Cys) at position 370; this allele is primarily found in people of African descent. The well-known genetic polymorphism in ADH1B is the alteration of arginine (ADH1B*1) to histidine (ADH1B*2) at codon 47th position due to a G/A base transition in exon 3 [6-10].
The ADH1B Arg47His polymorphism affects the enzyme activity substantially. The differences within the amino acid sequences encoded by the assorted ADH1B alleles cause differences in the predicted rates of ethanol metabolism within the liver [11]. The ADH1B is the low Km class enzyme in the seven ADH isoforms and exhibits higher activity than the other six ADH isoforms in catalyzing ethanol to acetaldehyde. Uncommon genotype ADH1B*2/2 encoding super active subunit of ADH1B has a significantly 40-fold higher enzyme activity than common genotype ADH1B*1/1 [10, 12]. Individuals carrying the ADH1B*2 allele and ALDH2*2 alleles show high blood acetaldehyde concentrations after the intake of only moderate alcohol amounts Individuals carrying these ADH1B*2 and ALDH2*2 alleles also drink less alcohol because of unpleasant physiological and psychological effects, such as cardiovascular effects, dysphoria, palpitation, dry mouth, headache, nausea, and facial warning, which are attributed to acetaldehyde accumulation [13]. Therefore, these alleles as previously mentioned seem to be associated with the protective effect against alcohol toxicity and alcohol dependence [14].
2. Materials and Methods
2.1 Study design
This was a prospective clinical study conducted in Department of Biochemistry and Department of Gastroenterology Medicine, Tribhuvan University Teaching Hospital, Institute of Medicine (IOM), Maharajgunj, Kathmandu, Nepal from July 2021 to December 2021. Study population included patients with chronic liver disease due to alcoholism as cases and non- alcoholic patients with no history of liver disease as a control.
2.2 Selection of cases
41 Patients within the age 20 - 65 consuming more than 40 g (men) and 20 g (women) of alcohol per day and with alcoholic liver disease were included in the study. Likewise, for the control 41 subjects with no history of alcohol consumption, liver disease or any chronic illness during the time of sample collection were enrolled. Participants were interviewed by principal investigator to fill up a self-designed semi structured pro-forma sheet. Demographic characteristics like age, sex, ethnicity, alcoholism, and duration of alcohol intake were all noted. Following the interview, 5ml of the blood sample was drawn from each patients after a written consent.
2.3 Molecular analysis
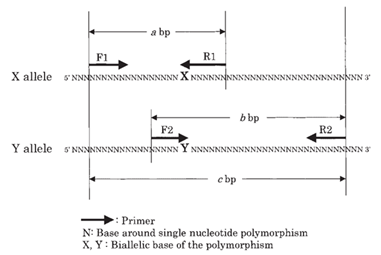
Genomic DNA was extracted from the whole blood sample using Spin Star TM total DNA kit 2.0 (ADT Biotech Sdn Bhd). Concentration and purity of extracted DNA was assessed by nanodrop. Isolated DNA was amplified by polymerase chain reaction-confronting two pair primer (PCR-CTPP) technique, which amplifies allele-specific DNA for two independent, single-nucleotide polymorphisms, at the same time in one tube, using four allele-specific primers designed as: F1 for the sense primer of allele X; R1 for the anti-sense primer of allele X with the anti-sense base of polymorphism site at the 3′ end; F2 for the sense primer of allele Y with the sense base of polymorphism site at the 3′ end; and R2 for the anti-sense primer of allele Y as shown in figure C. The difference in the size of amplified DNA allows to genotype the single nucleotide polymorphism [15].
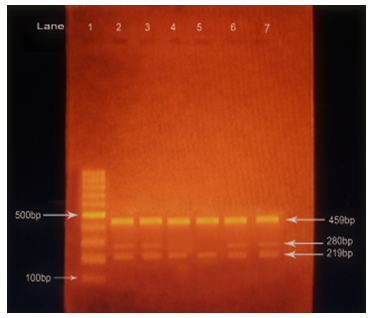
Two- paired primers, outer primer (forward 1 (F1) and Reverse2 (R2) and inner primer (Forward 2 (F2) and reverse1 (R1), provided by Macrogen, South Korea was used (listed in table 4) for genotyping of ADH1B Arg47His polymorphism. PCR assay was performed in a 25µl reaction mixture which comprehends 2x PCR master mix 12.5 µl, Forward primer 1(F1) (10pmol/ µl) 1 µl, Reverse primer 1(R1) (10pmol/ µl) 1 µl, Forward primer 2(F2) (10pmol/ µl) 1 µl, Reverse primer 2(R2) (10pmol/ µl) 1 µl, DNA template 1 µl, and nuclease free water 8.5 µl. For amplification, the reaction mixture was subjected to thermal cycling in T100 thermal cycler (Bio-Rab Laboratories) as shown in table 5 as determined after multiple steps of optimization. The PCR products were then analyzed by 1.5% agarose gel electrophoresis, mixing 3µl of PCR product with 1µl of loading dye. (bromophenol blue) 100bp of DNA ladder (solis bioDyne) was used as a marker to identify PCR products. The amplified DNA are 219 base pair (bp) for 47Arg (ADH1B*1), 280 bp for 47His (ADH1B*2) as well as a common band with 459 bp was visualized for ADH1B. For the heterozygous genotype, ADH1B*1/*2 three bands were visualized at 459bp, 280bp and 219bp and for the homozygous genotype, ADH1B*1/*1 two bands were visualized at 459bp and 219bp as is shown in figure D.
Table 4: Primers for Alcohol Dehydrogenase 2 (ADH2)/(ADH1B) Arg47His Genotyping
|
Polymorphism |
Primer sequence |
Sequence |
|
ADH2 (ADH1B) Arg47His |
Outer F1 primer |
5’-GGG CTT TAG ACT GAA TAA CCT TGG-3’ |
|
Inner R1 primer |
5’-AAC CAC GTG GTC ATC TGT GC-3’ |
|
|
Inner F2 primer |
5’- GGT GGC TGT AGG AAT CTG TCA-3’, |
|
|
Outer R2 primer |
5’-AGG GAA AGA GGA AAC TCC TGA A-3’ |
Table 5: PCR thermal cycling condition
|
Step |
Temperature (°C) |
Time |
Number of cycles |
|
Initial denaturation |
94 |
10 minutes |
1 |
|
Denaturation |
94 |
30 seconds |
35 |
|
Annealing |
58 |
1 minute |
35 |
|
Extension |
72 |
1 minute |
35 |
|
final extension |
72 |
5 minutes |
1 |
Figure D: Gel picture showing amplified PCR products in 1.5% agarose gel electrophoresis. Here lane indicates the well of gel where 1 = ladder, 2, 3, 4, 5, 6, 7 indicates the products of PCR from the samples of study population. In the figure bands in lane 2, 3, 6, and 7 at 459bp, 280bp, and 219bp signifies the heterozygous form of ADH1B*1/*2 genotype and bands in lane 4 and 5 at 459bp and 219bp signifies the homozygous form of ADH1B*1/*1 genotype.
2.4 Statistical Analysis
The Statistical Package for Social Sciences (SPSS) version 22.0 software for Windows was used for the statistical analysis. Data were expressed in the form of tables and bar diagrams. Chi-square test of significance was used to obtain the frequencies of ADH1B alleles and genotypes.
3. Results
A total of 82 participants were investigated, 41 were cases 31(75.60%) male and 10(24.30%) female and 41 were control in which 29(70.70 %) were male and 12 (29.20%) were female. The mean ± SD age of alcoholic subjects was 51.61±10.14 years. The maximum number of patients and controls belong to age group 51-60.
3.1 Distribution of genotype and allele frequency in cases and controls.
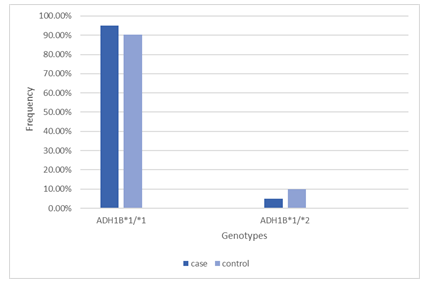
The genotype frequency of the ADH1B polymorphism in alcoholic (cases) subjects was 95.1% for homozygous ADH1B*1/*1 genotype, 4.9% for heterozygous ADH1B*1/*2 genotype. In control (non-alcoholic) subjects, 90.2% was for homozygous ADH1B*1/*1 genotype and 9.8% was for heterozygous ADH1B*1/*2. Even so, individuals with ADH1B*2/*2 genotype was not detected in our study populations. The genotype frequency and allele frequency is shown in table 1 and figure A.
Table 1: Genotype and allele frequency in cases and controls
|
Genotypes Frequency |
Allele Frequency |
P value |
|||||
|
Group |
N |
ADH1B*1/*1 (%) |
ADH1B*1/*2 (%) |
ADH1B*2/*2 (%) |
ADH1B*1 allele (%) |
ADH1B*2 allele (%) |
|
|
Case |
41 |
39 (95.1%) |
2 (4.9%) |
0 |
97.55% |
2.45% |
|
|
Control |
41 |
37 (90.2%) |
4 (9.8%) |
0 |
95.10% |
4.90% |
0.396 |
|
Total |
82 |
76 (92.7%) |
6 (7.3%) |
0 |
96.33% |
3.67% |
(NS) |
*in the table represents the variants of ADH1B gene
3.2 Distribution of ADH1B*1/*1 and ADH1B*1/*2 Genotypes of Alcoholics and Non-alcoholics in Different Ethnic Groups
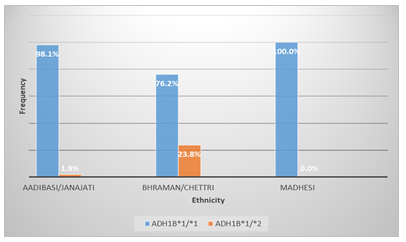
The ADH1B*1/*1 genotype frequency in aadibasi/janajati, bhraman/ chhetri and madhesi was found to be 98.1%, 76.2% and 100% respectively. ADH1B*1/*2 genotype was found in a greater percentage i.e 23.8% in bhraman/ chhetri as compared to 1.9% of aadibasi/janajati as shown in Figure B
3.3 Duration of Alcohol Consumption and Genotypic Frequency
The ADH1B*1/*1 genotype was more commonly prevalent in alcoholic individuals with longer history of alcohol consumption. (Table 2)
Table 2: Duration of alcohol consumption and genotypic frequency
|
Genotype frequency |
Duration of alcohol consumption in years |
||||
|
0 – 10 years |
11 – 20 years |
21 – 30 years |
31 – 40 years |
Total |
|
|
ADH1B*1/*1 |
8 (20.5%) |
14 (35.9%) |
12 (30.8%) |
5 (12.8%) |
39 (100%) |
|
ADH1B*1/*2 |
0 |
2 (100%) |
0 |
0 |
2 (100%) |
|
Total |
8 (19.5%) |
16 (39%) |
12 (29.3%) |
5 (12.2%) |
41 (100%) |
4. Discussion
Genetic polymorphisms have been reported in ADH1B genes and genetic variations in this alcohol metabolizing enzymes is allied to alterations in alcohol metabolism, response, consumption and related problems [16, 17]. In the human evolutionary history, it is known that single nucleotide polymorphisms which are localized in autosomal chromosome are changed by effects of many factors. Determination of those polymorphisms may be a crucial data in terms of relations between societies. Nepal, a Southeast Asian country have diverse ethnic group resulting a clear heterogeneity and genetic admixture. On that ground, special importance was given in our study to investigate the allele frequencies and genotype distributions of ADH1B gene in Nepalese population. To our knowledge, our present study is the first study to determine the genotype and allele frequencies of ADH1B polymorphisms in Nepalese population. In our present study, the ADH1B*1/*1 genotype was found to be most common of all genotypes in both alcoholic and non-alcoholic subjects whereas the frequency of ADH1B*1/*2 genotype was found to be low in both alcoholics and control groups. Thus, there was no significant differences in the genotype and allele frequencies in between cases (alcoholics) and the control (non-alcoholic) groups.
A frequency of 95.1% of ADH1B*1/*1 genotype in our result is consistent with the study conducted [11] in which ADH1B*1/*1 was 93.0%. Likewise, 4.9% of ADH1B*1/*2 genotype was found to be prevalent in our study which is also similar to the findings of [11, 18] in which 6.40% and 6.7% was ADH1B*1/*2 genotype among alcoholics respectively. However, no any individuals with ADH1B*2/*2 genotype was detected in our study alike to the study conducted by Kayaalti Z et.al., 2010 [12] in which no prevalence of ADH1B*2/*2 was found among 211 turkish alcoholic subjects.
In our study, frequency of ADH1B*1(ADH2.1) i.e Arg allele and ADH1B*2 (ADH2.2) i.e His allele was found to be 97.5% and 2.45% respectively in alcoholic cases and 95.1% and 4.9% respectively in control groups. These results were similar to the results reported by Salman E et.al., 2007 [11] in which Arg allele and His allele were 96% and 4% respectively in alcoholic patients and 99% and 1% respectively in control groups. On the contrary, Kayaalti Z et.al., 2010 [12] reported 91.9% and 8.1% frequency of Arg allele and His allele respectively. Thus, in keeping with our study Arg allele (ADH1B*1) was more commonly seen in both alcoholic and non-alcoholic Nepalese populations. The study participants were stratified into three ethnic group i.e Aadibasi/Janajati, Bhraman/Chhetri and Madeshi. No significant differences were found within the presence of ADH1B*1/*1 genotype in all three ethnicities whereas ADH1B*1/*2 was found to be highest in Bhraman/Chhetri with a frequency of 13.8% and a lowest frequency of 1.9% was noted in aadibasi/janajati, and wasn’t noted in madhesi ethnicity. The prevalence reported by some authors varies among different ethnic groups, ADH1B*1 allele was found to be predominant in Caucasian populations (~ 90%). Whereas ADH1B*2 occurs in Japanese, Chinese, and other Oriental population at about 85% percentage [12]. This is in contrast to our study where ADH1B*1 allele was found to be 97.55% and ADH1B*2 was found to be 2.45%. ADH1B*1/*1 and ADH1B*1/*2 genotype frequencies in Nepalese were not statistically different from Turkish populations Salman E et.al., 2007 [11], Finnish (Goedde et al., 1992) [19], Mexican (Konishi et al., 2003) [20]. However, ADH1B*2/2 genotype frequencies were statistically lower than Uzbek (Ahn et al., 2009)[21], Mongolian (Shen et al., 1997) [22], Jewish (Neumark et al., 2004) [23], Vietnamese (Iron et al., 1992) [24], Chinese (Guo et al., 2008) [25], and Japanese (Matsuo et al., 2006) [8] populations which exhibited the highest frequency values. The frequency of ADH1B*1/*1 and ADH1B*1/*2 in the abovementioned ethnic groups is shown in table 3. Our present study, also reported that, alcoholic patients having longer history of alcoholism (since more than 10 years up to 40 years) have higher frequency of ADH1B*1/*1 genotype and ADH1B*1 allele. This resembles a study done by Yin G et.al 2016 which revealed that alcohol intake were greater in subjects with the ADH1B 47Arg/Arg (ADH1B*1/*1) compared with each ADH1B 47His allele in men and women [26]. Similarly Yin G et.al., 2011 showed that both current alcohol drinkers (hatched bar) and heavy alcohol drinkers (black bar) were slightly more frequent with increasing numbers of the ADH1B*47Arg allele and disclosed the effect modification of the ADH1B Arg47His (ADH1B*1/*2) polymorphism on association with alcohol consumption [27].
Thus, it can be inferred from the study that the presence of ADH1B*1 allele and its genotype is responsible for inducing alcohol tolerance and promote alcoholism, as ADH1B*1 allele are associated with slower ADH activity. This is consistent with the result reported by Salman E et.al., 2007 [11] in which patients with history of maximum years of alcohol consumption and having alcohol induced liver cirrhosis have the higher frequency of ADH1B*1/*1 genotype. Similarly, population with Chinese, Mongolian and Japanese ethnicities have higher rate of prevalence of the heterozygous genotype (ADH1B*1/*2) and ADH1B*2 allele of ADH1B gene. Thereafter, it indicates the sensitivity of these ethnicities towards alcohol as ADH1B*2 are associated with higher ADH activity and increases the offensive effects of alcohol.
Table 3: Comparison of the genotype frequency of ADH1B in Nepalese alcoholic population and previously published data in different populations
|
Population |
N |
Genotypes of ADH1B |
P |
||
|
ADH1B*1/*1 |
ADH1B*1/2 |
ADH1B*2/2 |
|||
|
Nepalese (present study) |
82 |
92.70% |
7.30% |
0 |
0.396 (NS) |
|
European |
|||||
|
Five European population: French, Spanish, German, Swedish, polish. (Borras etal., 2000) [28] |
451 |
92.50% |
7% |
0 |
0.001* |
|
Asian |
|||||
|
Indian (Goedde et.al., 1992) [19] |
167 |
85.00% |
10% |
5% |
0.002* |
|
Turkish (Kayaalti etal., 2010) [12] |
211 |
83.90% |
16.10% |
0 |
0.372 (NS) |
|
Chinese (Guo et al., 2008) [25] |
480 |
5% |
35% |
60% |
0.001* |
|
Mangolian (Shen et al.,1997) [22] |
35 |
17% |
40% |
43% |
0.001* |
|
American |
|||||
|
Mexican (Konishi et al 2003) [20] |
104 |
92.3 |
6.7 |
1 |
0.026* |
|
Africans: Black Africans (Niger) (Iron et al., 1992) [24] |
45 |
75.60% |
24.4 |
0 |
0.183* |
5. ?onclusion
To my knowledge this is the first study conducted in Nepal, to demonstrate the frequency of SNP in ADH1B gene (i.e frequency of ADH1B*1 allele and ADH1B*2 allele and genotypic frequency of ADH1B*1/*1, ADH1B*1/*2 and ADH1B*2/*2) and its effect in the metabolism of alcohol and its tolerance. The study demonstrate the allele frequency of ADH1B*1 allele (Arg allele) is 97.5% and ADH1B*2 allele (His allele) is 2.45%. Also the genotype frequency of ADH1B*1/*1, ADH1B*1/*2 and ADH1B*2/*2 was found to be 95.1%, 4.9% and 0% respectively in alcoholic patients. The study found that the ADH1B*1/*1 genotype was most prevalent in all three ethinic groups i.e aadibasi/janajati, bhraman/chhetri, and madhesi. Similarly, the study also reported that patients having longer history of alcohol consumption have higher frequency of ADH1B*1/*1 genotype. Based on the results obtained from the study, our study concluded that the presence of ADH1B*1/*1, ADH1B*1/*2 and ADH1B*2/*2 genotype affects the metabolism of alcohol, consumption of alcohol and its tolerance. It was reported that presence of ADH1B*1/*1 genotype (have slow ADH activity), increases the tolerance towards alcohol making individual more susceptible towards alcoholism and precede the onset of alcohol use disorders. The presence of ADH1B*2/*2 genotype (with high ADH activity), exceeds ethanol metabolism and results in the accumulation of acetaldehyde, a highly toxic metabolite which exhibits unpleasant effects like facial flushing, headache, tachycardia and hypotension even after consumption of minute quantity of ethanol. Thus, these limiting factors might be helpful to refrain from alcoholism and its subsequent consequences for individuals with ADH1B*2/*2 genotype.
Glossary
Alcohol dehydrogenase
Alcohol Dehydrogenase 2
Alcohol dehydrogenase 1 B
Aldehyde dehydrogenase
Arginine allele
Alcohol use disorders identification test
Ethylene diamine tetra-acetic acid
Ethidium Bromide
Histidine allele.
Polymerase chain reaction with confronting two- pair primers
Pico mole per liter
Single Nucleotide Polymorphism
Tris acetate EDTA
Funding Statement
This study was partially funded by Nepal Health Research Council.
Approval of the Study
This study has been approved for Master’s level thesis by Department of Biochemistry and Department of Gastroenterology Medicine., Institute of Medicine.
Ethical Approval
The study has given ethical clearance by the Institutional Review Committee, Institute of Medicine and the letter has been submitted along with the manuscript submission.
Deceleration of Conflict of Interest: None
References
- Shakya D, Shyangwa P, Sen B. Alcohol dependence syndrome: a study of sociodemographic profile, psychiatric morbidity and help seeking behaviour in BPKIHS [dissertation], Department of psychiatry, BPKIHS, Dharan, Nepal July (2005).
- Katsarou MS, Karakonstantis K, Demertzis N, Vourakis E, Skarpathioti A, Nosyrev AE, et al. Effect of single?nucleotide polymorphisms in ADH 1B, ADH 4, ADH 1C, OPRM 1, DRD 2, BDNF, and ALDH 2 genes on alcohol dependence in a Caucasian population, Pharmacology Research Perspectives 5 (2017): e00326.
- Zakhari S. Overview: how is alcohol metabolized by the body? Alcohol research & health 29 (2006): 245.
- Hoang YTT, Nguyen YT, Nguyen HD, Le ATP, H.T.T. Bui, N.P. Vu, H.H. Nguyen, Single Nucleotide Polymorphisms of ADH1B, ADH1C and ALDH2 Genes in 235 People Living in Thai Nguyen Province of Vietnam, Asian Pacific Journal of Cancer Prevention, 23 (2022): 4243-4251.
- Lin CH, Nfor ON, Ho CC, Hsu SY, Tantoh DM, Liaw YC, et al. Association of ADH1B polymorphism and alcohol consumption with increased risk of intracerebral hemorrhagic stroke, Journal of Translational Medicine, 19 (2021): 1-8.
- Edenberg HJ. The genetics of alcohol metabolism, Alcohol Research, 30 (2007): 5.
- Hurley TD, Edenberg HJ, Li TK, Pharmacogenomics of alcoholism, Pharmacogenomics: The search for individualized therapies (2002): 417-441.
- Matsuo K, Wakai K, Hirose K, Ito H, Saito T, Suzuki T, et al. A gene–gene interaction between ALDH2 Glu487Lys and ADH2 His47Arg polymorphisms regarding the risk of colorectal cancer in Japan, Carcinogenesis 27 (2006): 1018-1023.
- Wu CF, Wu DC, Hsu HK, Kao E-L, Lee J-M, Lin C-C, et al. Relationship between genetic polymorphisms of alcohol and aldehyde dehydrogenases and esophageal squamous cell carcinoma risk in males, World journal of gastroenterology: WJG 11 (2005): 5103.
- Tóth R, Fiatal S, Petrovski B, McKee M, Ádány R. Combined effect of ADH1B RS1229984, RS2066702 and ADH1C RS1693482/RS698 alleles on alcoholism and chronic liver diseases, Disease markers 31 (2011): 267-277.
- Salman E. Alcohol Dehydrogenase and Aldehyde Dehydrogenase Gene Polymorphism in Turkish Alcohololic People and Control Group, in, Izmir Institute of Technology (Turkey) (2007).
- Kayaalt? Z, Söylemezo?lu T. Distribution of ADH1B, ALDH2, CYP2E1∗ 6, and CYP2E1∗ 7B genotypes in Turkish population, Alcohol 44 (2010): 415-423.
- Kaya A, Grivel M, Clinton L. Under-Researched Demographics: Heavy Episodic Drinking and Alcohol-Related Problems Among Asian Americans, Alcohol Research 38 (2016): E1.
- Edenberg HJ, McClintick JN. Alcohol dehydrogenases, aldehyde dehydrogenases, and alcohol use disorders: a critical review, Alcoholism: Clinical and Experimental Research 42 (2018): 2281-2297.
- Tamakoshi A, Hamajima N, Kawase H, Wakai K, Katsuda N, Saito T, et al. Duplex Polymerase Chain Reaction with Confronting Two-Pair Primers (PCR–CTPP) for Genotyping Alcohol Dehydrogenase Β Subunit (Adh2) and Aldehyde Dehydrogenase 2 (Aldh2), Alcohol and Alcoholism 38 (2003): 407-410.
- Sun F, Tsuritani I, Honda R, Ma Z-Y, Yamada Y. Association of genetic polymorphisms of alcohol-metabolizing enzymes with excessive alcohol consumption in Japanese men, Human genetics 105 (1999): 295-300.
- Brennan P, Lewis S, Hashibe M, Bell DA, Boffetta P, Bouchardy C, et al. Pooled analysis of alcohol dehydrogenase genotypes and head and neck cancer: a HuGE review, American journal of epidemiology 159 (2004): 1-16.
- Chinnaswamy P, Vijayalakshmi V. Subtypes of ADH2 gene in alcoholics, Indian Journal of Clinical Biochemistry 20 (2005): 104-109.
- Goedde H, Agarwal D, Fritze G, Meier-Tackmann D, Singh S, Beckmann G, et al. Distribution of ADH 2 and ALDH2 genotypes in different populations, Human genetics 88 (1992): 344-346.
- Konishi T, Calvillo M, Leng A-S, Feng J, Lee T, Lee H, et al. The ADH3* 2 and CYP2E1 c2 alleles increase the risk of alcoholism in Mexican American men, Experimental and Molecular Pathology 74 (2003): 183-189.
- Ahn KS, Abdiev S, Rahimov B, Malikov Y, Bahramov S, Okada R, et al. Alcohol dehydrogenase 1B and Aldehyde dehydrogenase 2 Polymorphisms in Uzbekistan, Asian Pac J Cancer Prev 10 (2009): 17-20.
- Shen YC, Fan JH, Edenberg HJ, Li TK, Cui YH, Wang YF, et al Polymorphism of ADH and ALDH genes among four ethnic groups in China and effects upon the risk for alcoholism, Alcoholism: Clinical and Experimental Research 21 (1997): 1272-1277.
- Neumark YD, Friedlander Y, Durst R, Leitersdorf E, Jaffe D, Ramchandani VA, et al. Alcohol dehydrogenase polymorphisms influence alcohol?elimination rates in a male Jewish population, Alcoholism: Clinical and Experimental Research 28 (2004): 10-14.
- Iron A, Groppi A, Fleury B, Begueret J, Cassaigne A, Couzigou P. Polymorphism of class I alcohol dehydrogenase in French, Vietnamese and Niger populations: genotyping by PCR amplification and RFLP analysis on dried blood spots, in: Annales de Genetique 35 (1992): 152-156.
- Guo Y-M, Wang Q, Liu Y-Z, Chen H-M, Qi Z, Guo Q-H, Genetic polymorphisms in cytochrome P4502E1, alcohol and aldehyde dehydrogenases and the risk of esophageal squamous cell carcinoma in Gansu Chinese males, World journal of gastroenterology: WJG 14 (2008): 1444.
- Yin G, Naito M, Wakai K, Morita E, Kawai S, Hamajima N, et al. ALDH2 polymorphism is associated with fasting blood glucose through alcohol consumption in Japanese men, Nagoya journal of medical science 78 (2016): 183.
- Yin G, Ohnaka K, Morita M, Tabata S, Tajima O, Kono S. Genetic polymorphisms of alcohol dehydrogenase and aldehyde dehydrogenase: alcohol use and type 2 diabetes in japanese men, Epidemiology Research International (2011).
- Borràs E, Coutelle C, Rosell A, Fernández?Muixi F, Broch M, Crosas B, et al. Genetic polymorphism of alcohol dehydrogenase in europeans: TheADH2* 2 allele decreases the risk for alcoholism and is associated with ADH3, Hepatology 31 (2000): 984-989.


 Impact Factor: * 6.2
Impact Factor: * 6.2 Acceptance Rate: 76.33%
Acceptance Rate: 76.33%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 