Evolution of the Pfcrt and Pfmdr1 Markers and Revert of Chloroquine Sensitive Plasmodium falciparum in a Seasonal Malaria Chemoprevention Setting in Cameroon
Ngum Lesley Ngum1,3,5, Innocent M Ali 1,4, Palmer Masumbe Netongo1,2, Akindeh M Nji1,2, Jean Paul Kengne Chedjou1,6, Randolph Ngwafor1,8, Pacome Valery Tchuenkam kom1,4 Aristid M Ekollo1,7, Peter Thelma Ngwa Niba1,2, , Mbu’u Mbanwi Cyrille1,9, Calvino Tah Fomboh1,2, Nana William Dorian1,2 and Wilfred F Mbacham1,2,3*
1Biotechnology Center, University of Yaoundé 1, Cameroon
2Department of Biochemistry, Faculty of Science, University of Yaoundé 1, Cameroon
3Department of Biochemistry, Faculty of Medicine and Biomedical Science, University of Yaoundé 1, Cameroon
4Department of Biochemistry, Faculty of Science, University of Nchang, Cameroon
5Institute of Medicine and Medicinal Plants Studies, IMPM, Yaoundé, Cameroon
6Department of Biochemistry, Faculty of Science, University of Buea, Cameroon
7Department of Biochemistry, Faculty of Science, University of Ngoundere 1, Cameroon, National Malaria Control Program; Cameroon
8Department of microbiology, Faculty of science, university of Yaounde 1, Cameroon
*Corresponding author: Wilfred F Mbacham, Department of Biochemistry, Faculty of Medicine and Biomedical Science, University of Yaoundé 1, Cameroon
Received: 24 May 2022; Accepted: 01 June 2022; Published: 08 June 2022
Article Information
Citation: Ngum Lesley Ngum, Innocent M Ali, Palmer Masumbe Netongo, Akindeh M Nji, Jean Paul Kengne Chedjou, Randolph Ngwafor, Pacome Valery tchuenkam kom Aristid M Ekollo, Peter Thelma Ngwa Niba, Mbu’u Mbanwi Cyrille, Calvino Tah Fomboh, Nana William Dorian and Wilfred F Mbacham. Evolution of the Pfcrt and Pfmdr1 Markers and Revert of Chloroquine Sensitive Plasmodium Falciparum in A Seasonal Malaria Chemoprevention Setting in Cameroon. Fortune Journal of Health Sciences 5 (2022): 296-30
View / Download Pdf Share at FacebookAbstract
Plasmodium falciparum, the cause of the most lethal malaria in humans, which still remains a threat to global health, with children and pregnant women being the most affected. This burden is highly pronounced in areas were malaria transmission is seasonal. The emergence of drug-resistance poses a major obstacle to the control of malaria. However predicting decreased or increased sensitivity to anti-malarial and fixation of multidrug resistance genotypes is vital in the fight against malaria. In other to assess how drug policies can impact this burden in a seasonal malaria chemoprevention setting in Cameroon, we investigated molecular changes in Pfcrt and Pfmdr1 genes in pre and post seasonal malaria chemoprevention in Cameroon. To assess the evolution of these markers, finger-prick whole blood samples were collected on 3MM Whatman filter paper from over 405 children that were diagnosed positive for malaria. DNA was extracted from and dry blood spot and genotyped. Chi square test was used to test for significance in the distribution of the markers within the two time points or periods. Out of the 405 samples from different patients that were genotyped for Pfcrt and Pfmdr1. The proportion of pfcrt genotypes stood at 33.3% wild-types, 51.9% mutants and 14.2% mixed infection. That of Pfmdr1 stood at 25.3% wild-type, 63.5% mutants and 11.2% mixed infections. Pearson’s chi square/Fischer’s exact test revealed that the frequencies of the SNPs of the pfcrt and Pfmdr1 gene were significantly associated with their distribution within the two time points (pv ≤ 0.000001).
Keywords
<p>Plasmodium falciparum, molecular markers, status, seasonal malaria</p>
Article Details
1. Introduction
Human malaria is caused by an apicomplexan parasite of the genus plasmodium. Among the species responsible for human malaria, plasmodium falciparum is the most life threatening. This specie is responsible for the highest proportion of infectious disease in malaria endemic countries [1]. Malaria is the most important parasitic disease in terms of human suffering, resulting in the highest number of cases and deaths in malaria endemic countries. Globally there are estimated 229 million malaria cases, with the African region accounting for 94% of the total number of cases with majority of the case from sub-Saharan Africa [2].
Over the past years, Cameroon has implemented complimentary malaria control strategies base on WHO recommendations. Including the distributions of insecticide treated bed nets and implementation of intermittent preventive treatment in pregnant women [3, 4]. Replacing Chloroquine with Amodiaquine as first line treatment between 2002 and 2004 and the adoption of Artemisinine based combination therapy as first line treatment in 2004 [5, 6]. Despite all these, malaria is still one of the leading causes of morbidity and mortality in Cameroon with children under the age of 5 being the most affected especially in the north and far north regions [7, 8]. In Cameroon, malaria transmission varies with respect to the different ecological zones with a major seasonal peak from July to October in the Sahelian and sub Sahelian ecological regions [8]. know as seasonal malaria chemoprevention (SMC), which is based solely on preventing infection in children under the ages of five with Sulphodoxine Pyramitamine and Amodiaquine (SPAQ) [36]
The strategy is to administer monthly SPAQ doses to children under 5years during the malaria peak transmission season. Non-inclusion of children above the age of five as well as adult and the development of resistance against SPAQ by the parasite, are some of the challenges in the reduction of malaria burden in this zone, through this strategy. The unfortunate spread of resistance to SPAQ and to most, if not all anti-malaria drugs, some of which were once effective and safe, like Chloroquine, is the principal cause of long standing malaria burden especially in this regions [9]. Chloroquine resistance was first reported in the thai-cambodian borders in the late 1950s [10]. It then spread to other malaria endemic regions of the globe [11, 12].
In the 1990s almost all of Sub-Saharan African countries had reported Chloroquine resistant plasmodium falciparum. In Cameroon, the first Chloroquine treatment failure was documented in 1985 and was withdrawn as treatment for uncomplicated malaria in 2002 [5, 13]. Although significant progress has been made to understand the resistance mechanisms to administered anti-malarial drugs, the genetic basis of drug resistance is vital to implement strategies for efficient malaria control. Single nucleotide polymorphism in the genes “plasmodium falciparum multidrug resistance 1” (Pfmdr1), located in chromosome 5 [14, 15], “plasmodium falciparum chloroquine resistance transporter” (Pfcrt), on chromosome 7 Modulate the level of chloroquine resistance [16]. Several studies have confirmed a strong association between the mutant alleles Pfcrt-T76 and Pfmdr1-Y86 and high level of chloroquine resistant [17, 1 12, 13]. Before and after the replacement of Chloroquine as first line treatment, studies showed low Chloroquine failure rate in the North and Far North regions also known as seasonal malaria chemoprevention setting in Cameroon as compared to the Southern regions [5, 13].
These regions also account for the highest number of malaria cases and deaths in Cameroon with children under age of 5 being the most affected [7]. Chloroquine, was the most preferred antimalarial in many countries that were prone to malaria endemic plasmodium falciparum. The drug was safe, effective, widely available, remarkably inexpensive with a long half-life and protect from early relapse ( [12, 18]. Chloroquine could be administered to pregnant women as well as infants. The advantages of Chloroquine are simply too strong. With this high rate of prevalence of malaria in these regions before and even after the introduction seasonal malaria chemoprevention, we then set out to investigate the Evolution of the pfcrt T76 and pfmdr1 Y86 markers in a seasonal malaria chemoprevention setting in Cameroon. Reason being that the years of reliance on antimalarial other than Chloroquine could have led to the revert of Chloroquine sensitive plasmodium falciparum [12, 19, 20, 21], before or even after the introduction of seasonal malaria chemoprevention in Cameroon.
2. Materials and Methods
2.1 Study Area
The study was conducted in the North and Far North regions of Cameroon with Garoua and Maroua respectively as regional capitals. These regions fall within the major Sahelian geo-ecological region of Cameroon. Garoua is in the north region of Cameroon and lies at coordinates 6o24’N & 10O46’E. Garoua is bounded to the North by the Far North region, to the South by the Adamawa region, to the West by Nigeria, to the East by Tchad and Southeast by the Central African Republic. Garoua serves as a river port in years when the rainfall is abundant. The temperature here is on average at 31°C for most of the year and the vegetation is Guinea-Savanna. It receives an annual average rainfall of 380 mm with just about 4 months of rainy season [22]. The population is predominantly Muslim with agriculture as the prominent activity of the region.
Maroua is in the Far North region of Cameroon. And lies at the coordinates, 5o0’N & 12o27E. Mount Maroua dominates the skyline of the city. The climate of Maroua is Sahelian, being hot and dry for most of the year. Maroua lies along the same belt stretching from the West African country of Gambia through Burkina Faso and Niger to Northern Nigeria and across Tchad and towards Sudan. This region experiences low precipitation, and therefore prone to malaria epidemics. The predominant religion is Sufi Islam and most indigenes engage themselves in Agriculture (cotton farms) or raise cattle (Non-Fulani’s and Fulani’s respectively). They also engage in trade with neighboring Tchad and Nigeria [23].
2.2 Study population
Children between the ages of 6-120 months with primary diagnosis of malaria confirmed by a positive CareStart™ Malaria HRP2 pf (CAT NO: G0141, ACCESSBIO) Ag RDT and who fulfill all inclusion and none of the non-inclusion criteria, were enrolled in the study, after obtaining consent from their parents or guardians.
2.3 Study samples
Study participants were recruited from randomly selected health districts in the North and the Far North regions of Cameroon in 2016 prior to the implementation of SMC and in 2021 five years after the implementation of SMC. Febrile self-presenting patients who fulfilled all of the inclusion and none of the non-inclusion criteria’s were eligible for the study. An axillary temperature was measured to check a fever. Febrile patients were considered to have an axillary temperature of ≥ 37oC. A malaria rapid diagnostic test (RDT) was used to diagnose the patient for plasmodium falciparum infection. This was further confirmed by microscopy. Finger-prick blood samples were obtained from patients reported as being infected with plasmodium falciparum. No additional clinical assessment was carried out at the time of recruitment. Together, over 405 finger prink blood samples collected on Whatman#3mm filter paper were air-dried, sealed individually in zip luck bags and stored at room temperature for DNA extraction and further molecular analysis.
2.4 DNA extraction
Plasmodium falciparum genomic DNA was extracted from dried blood spots on 3mm Whatmann filter papers by chelex (Bio-Rad laboratories SIGMA) method as previously described by Plowe and coworkers [24]. The dry blood spot was cutout and placed in a micro-centrifuge labeled tube. 1 ml 0.5% Saponin was then added to it and incubated overnight at 4°C. The following day the saponin was replaced with 1 ml of phosphor buffer saline (1X PBS). After 20-30 minutes of incubation at 4°C, the PBS was discarded and the blood spot transferred to 20% hot chelex in a heating block. This mixture was then voltexed and centrifuge at 10,000 rpm for 2 minute and the supernatant transferred into a new tube. The tube was again centrifuge and the supernatant transferred to a final tube which was then stored at 20 oC.
2.5 DNA amplification
The extracted DNA was used for PCR amplification and the left over returned -20oC for storage. The Pfcrt and Pfmdr1 genes of the parasitic DNA were amplified using pairs of forward and reverse primers obtained from Inquaba Biotec (Pretoria, South Africa) flanking codons 76 and 86 of the respective genes in a two round PCR. Both codons were amplified in a T3 thermo- cycler (Biometra UK) [24]. The products of the first amplifications were used as substrates for the second amplification. For the amplification, reactions volumes of 25µl reaction mixtures were used, each of which contain; nuclease free water, 10 X thermopol buffers, 10mM dNTPs, Taq Polymerase and substrate. In the first round of the nested PCR for the gene Pfmdr1 the primers MDR1F (5’-GCGCGCGTTGAACAAAAAGAGTACCGCTG-3’) and MDR2R (5’GGGCCCTCGTACCAATTC CTGAACTCAC-3’) were used. Pre-denaturation was done at 950C for 5minutes followed by 30 cycles of denaturation at 950C for 30s annealing at 45oC for 30s and extension at 65OC for 45s with a final extension at 72oC for 5minutes.
The primers MDR3F (5’-TTTACCGTTTAAATGTTTACCTGC-3’) and MDR4R (5’-CCATCTTGATAAAAAACACTTCT-3’) were respectively used as forward and reverse primers for the second round of the nested PCR. Pre-denaturation was done at 950C for 3minutes, followed by 25 cycles of denaturation at 950C for 30s annealing at 45oC for 30s and extension at 65OC for 45s with a final extension at 65oC for 3minutes. For the Pfcrt gene, CRTP1 (5’-CCGTTAATAATAAATACACGCAG-3’) and CRTP2 (5’-CGGATGTTACAAAACTATAGTTACC-3’) were respectively used as forward and reverse primers for the first nested reaction. The PCR reaction mixture was subjected to the following conditions. Pre-denaturation was done at 940C for 3minutes, followed by 45 cycles of denaturation at 940C for 30s annealing at 56oC for 30s and extension at 60OC for 1 minute with a final extension at 60oC for 3minutes. For the second nested reaction, CRTD1 (5’-TGTGCTCATGTGTTTAAACTT-3’) and CRT (5’-CAAAACTATAGTTACCAATTTTG-3’) were respectively used as forward and reverse primers. The PCR reaction mixture was subjected to the following conditions. Pre-denaturation was done at 950C for 5minutes, followed by 30 cycles of denaturation at 920C for 30s annealing at 48oC for 30s and extension at 65OC for 45s with a final extension at 65oC for 5 minutes.
2.6 Restriction fragment length polymorphism
The PCR amplicon were then subjected to restriction fragment length polymorphism using site-specific restriction enzymes (New England Biolabs). The pfcrt alleles were analyzed by incubating the amplicon at 37oC overnight with the restriction enzyme Apo I. ApoI cleaves pfcrt-76K but not pfcrt-76T [24]. Analysis of the pfmdr1 alleles were performed by incubation the amplicon at 37oC overnight with the restriction enzyme Afl III. AfI III cleaves the coding sequence of allele pfmdr1-86N but not that of pfmdr1-86Y [25].
2.7 Agarose gel electrophoresis
The amplified DNA and the restricted products were subjected to electrophoresis in a 2 % agarose gel, stained with 0.5μl Ethidium Bromide and visualized under ultraviolet light. Chloroquine sensitive and resistant haplotypes were analyzed. Haplotypes were considered resistant if they all represented combinations of resistant related mutations and sensitive if they represented combinations of wild-types allele or mixed if both forms were present.
3. Results
Among the participants who were recruited in the study, 405 dried blood spots filter papers had sufficient information to be returned for further analysis. Dried blood spots from unidentifiable participants, or with duplicated patient code as well as those with incomplete patient’s code or information were not considered for further analysis. Among the 405 samples, 201/405(49.6%) were collected in 2016 and the remaining 204/405 (50.4%) in 2021. DNA was successfully extracted from all the 405 samples as confirmed by a Nano drop analysis. The Pfcrt and the Pfmdr1 genes were successfully amplified in the 201 samples, giving a genotyping success rate of 100%. For the samples collected in 2021, 144 pfcrt and 183 pfmdr1 genes were successfully amplified, giving a genotyping rate of 70.6% (144/204) and 89.7% (183/204) respectively. The amplified DNA was then assessed for single nucleotide polymorphisms (SNPs) at codons 76 and 86 of the Pfcrt and Pfmdr1 gene respectively.
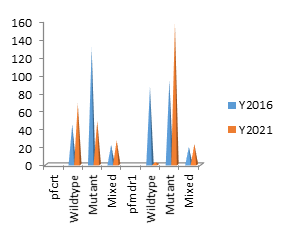
The observed frequencies of the investigated SNPs in Pfcrt and Pfmdr1 loci for both years are summarized in table 1. The Pfcrt gene harboring the wild type allele (K76) susceptible AAA nucleotide, coding for Lysine (Lys) was observed in 97/384 (25.3%) of the analyzed samples. Among which 94/201(46.8%) were sampled in 2016 and 3/183(1.6%) in 2021. The Pfcrt gene harboring the mutant allele (67T) susceptible ACA nucleotide coding for threonine (Thr) was observed in 244/384(63.5%). There were 87/201 (43.3%) sampled in 2016 and 157/183(85.8%) in 2021. Mixed infection containing both the Wild-type and mutant alleles were observed in 43/384(10.9%) among which 20/201(10%) were sampled in 2016 and 23/183 (10.9%) in 2021. The Pfmdr1 gene harboring the wild type allele (N86) susceptible TGA nucleotide, coding for Asparagine (Asn) was observed in 115/345 (33.3%) of the analyzed samples. among which 47/201(24.4%) were sampled in 2016 and 68/144(47.2%) in 2021.
Table 1: Proportion of single nucleotide polymorphism on Pfcrt and Pfmdr1 genes
A= Adenine, C= Cytosine, G= Guanine, T=Thymine, Asn=N =arginine, Tyr=Y= tyrosine, Lys=K= Lysine, Thr=T=Threonine, Pfcrt= Plasmodium falciparum Chloroquine resistant transporter gene, Pfmdr1 = Plasmodium falciparum multidrug resistant gene 1, CI=confidence interval, Y2016= year 2016, Y2021= year 2021
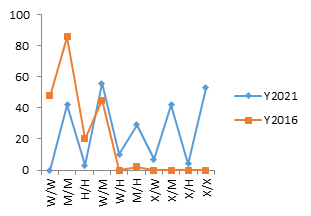
Table 2: Various Pfcrt and Pfmdr1 combination obtained from restriction analysis to determine single nucleotides polymorphisms in the respective genes
|
Category |
Y2016 |
Y2021 |
Total |
C I |
|
No mutation |
48(23.9) |
0(0.0) |
48(14.1) |
335.8 -286.8 |
|
Single mutant |
86(42.8) |
66(47.1) |
111(32.6) |
188.9 -77.9 |
|
double mutant |
108(53.7) |
74(52.9) |
182(53.4) |
300.2 -119.2 |
|
Total |
201 |
140 |
341 |
CI= Confidence interval
Pearson’s chi square/Fischer’s exact test of 31.7 and 105.5 for pfcrt and pfmdr1 genes respectively revealed that the frequencies of their SNPs were significantly associated with their distribution between the two years (pv ≤ 0.000001), with an increase in the mutant alleles between 2016 and 2021 figure 1. All mutants and SNPs were evaluated for both 2016 and 2021. A breakdown of the unique combination of mutants is shown in table 2 and the variation for each unique combination of SNPs for 2016 and 2021 is shown on figure 2.
4. Discussion
The aim of the current study was to investigate the evolution of Pfcrt and Pfmdr1 single nucleotide polymorphisms between pre SMC and post SMC in Cameroon. Pfmdr1 and Pfcrt are molecular markers of resistant to one of the drug use for seasonal malaria chemoprevention. These markers have also been proven to be strongly associated with a once effective and safer anti malaria drug call Chloroquine. Owing to the role Chloroquine has played in the fight against malaria and it comparatively low resistant in the seasonal malaria chemoprevention region before it replacement in 2002 as well as the introduction of seasonal malaria chemoprevention in Cameroon. Single nucleotides polymorphism was used to investigate the prevalence of mutations alleles in Pfcrt and Pfmdr1 genes, all of which are associated with Chloroquine resistance Plasmodium falciparum. A large proportion of Plasmodium falciparum isolates in this study 179/345(51.9%) carried the Chloroquine resistant Pfcrt-76T allele. Previously it has been reported that the Chloroquine resistant (CQR) mutant Pfcrt-K76T is a strong predictor of overall Chloroquine resistant [26] and that Pfmdr1 encodes a trans-membrane transporter and that point mutations in this gene modulate the level of Chloroquine resistance [16, 27].
We observed a high proportion of Pfcrt-K76T mutant alleles in 2016. This was not in line with previous study conducted in Cameroon by leonado et al which lead to the ban of Chloroquine importation and used in Cameroon in 2002. In this study they demonstrated a decreasing gradient in Chloroquine resistance plasmodium falciparum towards the Sahelian northern Cameroon [5]. This difference may be due to the continual used of quinolone based antimalarial or even the continual usage of Chloroquine after its withdrawal. The continual usage of this molecule may have enhances the prevalence of these resistant markers in the parasitic population. This results also differs from other studies conducted in Ethiopia, Zambia, Malawi , French guinea, new pupa guinea [12, 19, 21, 28] and 2 East African countries [20, 29] where the frequency of the Pfcrt-76T resistance allele was repopulated with the sensitivity ones after many years of Chloroquine withdrawal. Here the complete withdrawal of Chloroquine and quinoline related anti-malaria drugs lead to a reduction in drug pressure and an increase in Chloroquine susceptible plasmodium falciparum. It appears that CQ sensitive Plasmodium falciparum isolates are yet to be back in a seasonal malaria chemoprevention setting in Cameroon even though we experience a decrease in pfcrt mutant alleles between 2016 and 2021.
This decrease was disproportionately followed by a paralleled increased in pfmdr1 mutant alleles within the same period. Pfmdr1 is thought to transport and accumulate CQ in the parasites, food vacuole, mutations N86Y interfere with the transportation of the anti -malaria drugs leading to reduced CQ sensitivity [30-32]. Although genetic transformation experiments have found that pfmdr1 mutations can modulate the level of chloroquine resistance once it has been conferred by mutations in pfcrt, mutations in pfmdr1 are by them insufficient to confer chloroquine resistance [19]. Recent studies have reported that pfmdr1 mutations, in addition to pfcrt mutations, are no more strongly associated with chloroquine treatment failure than pfcrt mutations alone [6] and that pfmdr1 mutations do not add to the predictive value of pfcrt mutations for chloroquine treatment failure [20]. Even if pfmdr1 mutations do contribute to treatment failure, they require pfcrt mutations to exert an effect on the response to chloroquine.
Therefore, persistence of pfmdr1 mutations in a population with a very low prevalence of pfcrt mutations would not be expected to reduce chloroquine efficacy in that population [32-34]. The decrease in pfcrt mutant allele, might be explained by an advantage of the pfcrt K76 allele over its mutant form after removal of CQ pressure owing may be to the complete withdrawal of CQ from the non-official market. It can also be due to increase in awareness by the population on the efficacy of artemisinine based antimalarial drugs [30, 31]. A similar trend was observed in south Cameroon where it was concluded that chloroquine could not be reintroduce for the treatment of Plasmodium falciparum malaria since the pfcrt mutant allele was still comparatively high even though the study demonstrated a decrease in this pfcrt mutant allele towards the seasonal malaria chemoprevention region [13]. A similar decrease in pfcrt mutant allele in these regions has also been described earlier by Mbacham et al, Moye et al and Leonado et al [5, 6, 35] where they demonstrated a comparative reduction in Chloroquine resistance Plasmodium falciparum in these regions.
4. Conclusion
Chloroquine sensitive plasmodium falciparum is yet to return to the seasonal malaria chemoprevention setting in Cameroon.
Acknowledgement
This paper and the research behind it would not have been possible without financial support from;
The Global Fund to Fight AIDS, Tuberculosis and Malaria and the Malaria Research Capacity Development (MARCAD)
Conflicts of interest
We declare that there is no conflict of interest.
References
- Le Roch KG et al. “Discovery of gene function by expression profiling of the malaria parasite life cycle,” Science (80-. ) 301 (2003): 1503–1508.
- WHO, Global trends in the burden of malaria 1 (2020).
- Walker-Abbey A et al. “Malaria in pregnant Cameroonian women: The effect of age and gravidity on submicroscopic and mixed-species infections and multiple parasite genotypes,” J. Trop. Med. Hyg 72 (2005): 229–235.
- Nguela RL et al. “The effect of improved housing on indoor mosquito density and exposure to malaria in the rural community of Minkoameyos, Centre Region of Cameroon,” J 19 (2020): 1–16, 2020.
- Basco LK et al. “Molecular epidemiology of malaria in Cameroon. XXI. Baseline therapeutic efficacy of chloroquine, amodiaquine, and sulfadoxine-pyrimethamine monotherapies in children before national drug policy change,” J. Trop. Med. Hyg. 75 (2006): 388–395, 2006.
- Moyeh MN et al. “Effects of Drug Policy Changes on Evolution of Molecular Markers of Plasmodium falciparum Resistance to Chloroquine, Amodiaquine, and Sulphadoxine-Pyrimethamine in the South West Region of Cameroon,” Res. Treat (2018).
- National Malaria Control Programme, “Rapport D ’ Activites 2011 Du Programme National De Lutte,” (2011).
- US President and M Initiative, “PRESIDENT ’ S MALARIA INITIATIVE CAMEROON Malaria Operational Plan FY 2018 and FY 2019” (2019).
- Greenwood BM, Bojang K, Whitty CJM, and Targett GAT. “doi:10.1016/S0140-6736(05)66420-3,” Lance, 365 (2005): 1–12.
- Payne D. “Spread of chloroquine resistance in Plasmodium falciparum,” Today, 3 (1987): 241–246.
- Olatunde A, “Brief communications chloroquine-resistant plasmodium falciparum and malaria in africa,” R. Soc. Trop. Med. Hyg. 71 (1977): 80–81.
- Mekonnen SK et al. “Return of chloroquine-sensitive Plasmodium falciparum parasites and emergence of chloroquine-resistant Plasmodium vivax in Ethiopia,” J. 13 (2014): 1–9.
- Ndam NT et al. “Reemergence of chloroquine ? sensitive pfcrt K76 Plasmodium falciparum genotype in southeastern Cameroon,” J. (2017): 1–6.
- Wurtz N et al. “Prevalence of molecular markers of Plasmodium falciparum drug resistance in Dakar, Senegal,” J, 11 (2012): 1–10.
- Andriantsoanirina V et al., “Chloroquine clinical failures in P. falciparum malaria are associated with mutant Pfmdr-1, not Pfcrt in madagascar,” PLoS One 5 (2010).
- Valderramos SG and Fidock DA. “Transporters involved in resistance to antimalarial drugs,” Trends Pharmacol. Sci. 27 (2006): 594–601.
- Kublin JG et al, “Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi,” Infect. Dis 187 (2003): 1870–1875.
- Ganguly S et al. “In vivo therapeutic efficacy of chloroquine alone or in combination with primaquine against vivax malaria in kolkata, west bengal, india, and polymorphism in pvmdr1 and pvcrt-o genes,” Agents Chemother. 57 (2013): 1246–1251.
- Pelleau S et al., “Adaptive evolution of malaria parasites in French Guiana: Reversal of chloroquine resistance by acquisition of a mutation in pfcrt,” Natl. Acad. Sci. USA. 112 (2015): 11672–11677.
- Kiarie WC, Wangai L, Agola E, Kimani FT, and Hungu C. “Chloroquine sensitivity: Diminished prevalence of chloroquine-resistant gene marker pfcrt-76 13 years after cessation of chloroquine use in Msambweni, Kenya,” J. 14 (2015): 1–7.
- Mwanza S et al., “The return of chloroquine-susceptible Plasmodium falciparum malaria in Zambia,” J. 15 (2016): 1–6, 2016.
- Tabue RN et al. “Case Definitions of Clinical Malaria in Children from Three Health Districts in the North Region of Cameroon,” Biomed Res. Int. (2019).
- Arabi M, Xiao N, Taiwe KD and Liang S, “Cholera Incidence in the Far North Region of Cameroon: A Geographic Perspective,” African J. Soc. Sci 5 (2014): 141–156.
- Plowe CV, Djimde A, Bouare M, Doumbo O and Wellems TE. “Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: Polymerase chain reaction methods for surveillance in Africa,” J. Trop. Med. Hyg 52 (1995): 565–568.
- ABDOULAYE DJIMDÉ BS, PHARM D, OGOBARA K. DOUMBO, M.D., PH.D., JOSEPH F. CORTESE, M. D. KASSOUM KAYENTAO, M.D., SAFI DOUMBO, M.D., YACOUBA DIOURTÉ, PHARM.D.,* ALASSANE DICKO, P. D. XIN-ZHUAN SU, PH.D., TAKASHI NOMURA, M.D., PH.D., DAVID A. FIDOCK, and M. P. . THOMAS E. WELLEMS, M.D., PH.D., AND CHRISTOPHER V. PLOWE, M.D., “No Title,” Engl. J. Med., vol 344 (2001): 257–263.
- Griffing S et al., “pfmdr1 amplification and fixation of pfcrt chloroquine resistance alleles in Plasmodium falciparum in Venezuela,” Agents Chemother 54 (2010): 1572–1579.
- Duraisingh MT and Cowman AF. “Contribution of the pfmdr1 gene to antimalarial drug-resistance,” Acta Trop 94 (2005): 181–190.
- Sekihara M et al., “Lack of significant recovery of chloroquine sensitivity in Plasmodium falciparum parasites following discontinuance of chloroquine use in Papua New Guinea,” J 17 (2018): 1–10.
- Mohammed A, Ndaro A, Kalinga A, Manjurano A, Mosha JF and Mosha DF, “Trends in chloroquine resistance marker , Pfcrt-K76T mutation ten years after chloroquine withdrawal in Tanzania,” 12 (2013).
- Ly O et al., “Evolution of the pfcrt T76 and pfmdr1 Y86 markers and chloroquine susceptibility 8 years after cessation of chloroquine use in Pikine, Senegal,” Res 111 (2012): 1541–1546.
- Manirakiza A, Njuimo SP, Le Faou A, Malvy D, and Millet P. “Availability of antimalarial drugs and evaluation of the attitude and practices for the treatment of uncomplicated malaria in bangui, Central African Republic,” Trop. Med (2010).
- Reed MB, Saliba KJ, Caruana SR, Kirk K, and Cowman AF. “Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum,” Nature 403 (2000): 906–909.
- Plasmodium BC. “a Mol Ecul Ar Marke R F or Ch Loroquine-R Es Ista Nt Fa Lcipa Rum Ma L a Ria a Molecular Marker for Chloroquine-Resistant,” English J 344 (2001): 257–263.
- Jelinek T et al., “Diagnostic value of molecular markers in chloroquine-resistant falciparum malaria in southern Mauritania,” J. Trop. Med. Hyg 67 (2002): 449–453.
- Mbacham WF et al., “Efficacy of amodiaquine, sulphadoxine-pyrimethamine and their combination for the treatment of uncomplicated Plasmodium falciparum malaria in children in Cameroon at the time of policy change to artemisinin-based combination therapy,” J 9 (2010): 1–8.
- WHO, Seasonal Malaria Chemoprevention with Sulfadoxine–Pyrimethamine plus Amodiaquine in children; a field guide (2013).


 Impact Factor: * 6.2
Impact Factor: * 6.2 Acceptance Rate: 76.33%
Acceptance Rate: 76.33%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 